Advances in Applying Computer-Aided Drug Design for Neurodegenerative Diseases
Abstract
1. Introduction
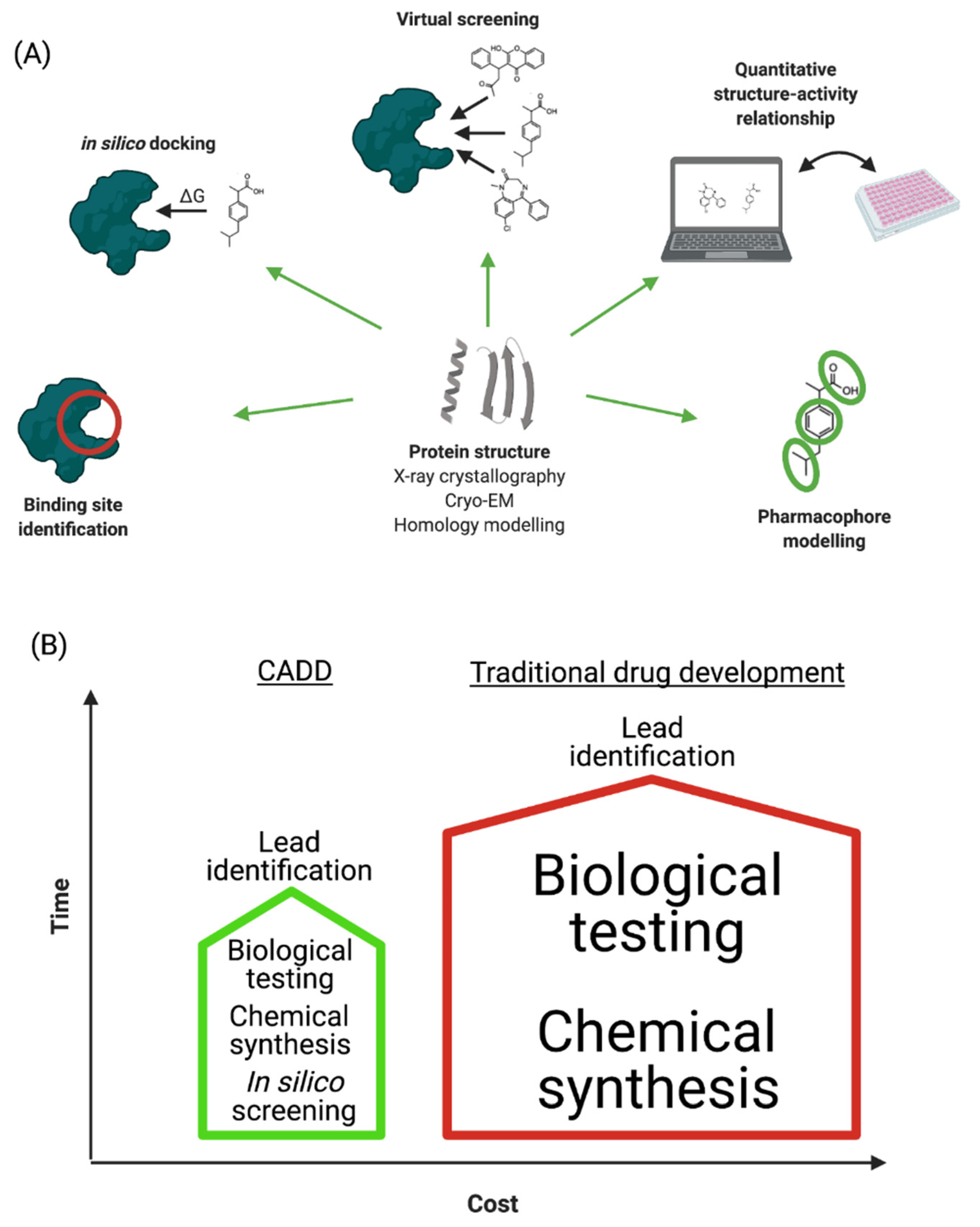
2. Computer-Aided Drug Design
2.1. Drug Target Selection
2.2. Determination of the Protein Structure
2.3. Homology Modelling
2.4. Identification of Binding Sites
2.5. Molecular Dynamics Simulation
2.6. Molecular Docking Studies
2.7. Virtual Screening
2.8. Quantitative Structure—Activity Relationship Study
2.9. Pharmacophore Modelling
3. Neurodegenerative Diseases
3.1. Alzheimer’s Disease (AD)
3.1.1. Macromolecular Targets in AD
Acetylcholinesterase
Beta-Secretase and Gamma-Secretase Enzymes
Caspases
Acetylcholine (ACh) Receptors
N-Methyl-D-Aspartate Receptor
ROCK-I and NOX2 Enzymes
3.2. Parkinson’s Disease (PD)
3.2.1. Macromolecular Targets in PD
COMT (Catechol-O-Methyltransferase) Inhibitors
Dopamine Agonists
Gene Variants
Glutamate Antagonists
MAO-B
3.3. Amyotrophic Lateral Sclerosis (ALS)
3.3.1. Macromolecular Targets in ALS
SOD1
MAPK
Casein Kinase 1 (CK-1) Inhibitors
3.4. Huntington’s Disease
3.4.1. Macromolecular Targets in HD
4-Aminobutyrate Aminotransferase
4. A Roadmap for Implementing CADD in ND Drug Design
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maciejczyk, M.; Zalewska, A. Salivary redox biomarkers in selected neurodegenerative diseases. J. Clin. Med. 2020, 9, 497. [Google Scholar] [CrossRef] [PubMed]
- Rabanel, J.-M.; Perrotte, M.; Ramassamy, C. Nanotechnology at the Rescue of Neurodegenerative Diseases: Tools for Early Diagnostic. In Nanobiotechnology in Neurodegenerative Diseases; Springer: Berlin/Heidelberg, Germany, 2019; pp. 19–48. [Google Scholar]
- Sehgal, S.A.; Hammad, M.A.; Tahir, R.A.; Akram, H.N.; Ahmad, F. Current Therapeutic Molecules and Targets in Neurodegenerative Diseases Based on in silico Drug Design. Curr. Neuropharmacol. 2018, 16, 649–663. [Google Scholar] [CrossRef]
- Livingston, G.; Huntley, J.; Sommerlad, A.; Ames, D.; Ballard, C.; Banerjee, S.; Brayne, C.; Burns, A.; Cohen-Mansfield, J.; Cooper, C. Dementia prevention, intervention, and care: 2020 report of the Lancet Commission. Lancet 2020, 396, 413–446. [Google Scholar] [CrossRef]
- Prince, M.; Knapp, M.; Guerchet, M.; McCrone, P.; Prina, M.; Comas-Herrera, A.; Wittenberg, R.; Adelaja, B.; Hu, B.; King, D. Dementia UK: Update; Alzheimers Society: Belfast, UK, 2014; pp. 1–136. [Google Scholar]
- Prime Minister’s Challenge on Dementia. Available online: https://www.gov.uk/government/publications/prime-ministers-challenge-on-dementia-2020 (accessed on 14 April 2020).
- Shukla, R.; Singh, T.R. Virtual screening, pharmacokinetics, molecular dynamics and binding free energy analysis for small natural molecules against cyclin-dependent kinase 5 for Alzheimer’s disease. J. Biomol. Struct. Dyn. 2020, 38, 248–262. [Google Scholar] [CrossRef]
- Mouchlis, V.D.; Melagraki, G.; Zacharia, L.C.; Afantitis, A. Computer-Aided Drug Design of β-Secretase, γ-Secretase and Anti-Tau Inhibitors for the Discovery of Novel Alzheimer’s Therapeutics. Int. J. Mol. Sci. 2020, 21, 703. [Google Scholar] [CrossRef]
- Am Ende, D.J.; Am Ende, M.T. Chemical engineering in the pharmaceutical industry: An introduction. Chem. Eng. Pharm. Ind. Drug Prod. Des. Dev. Modeling 2019, 1–17. [Google Scholar] [CrossRef]
- Talele, T.T.; Khedkar, S.A.; Rigby, A.C. Successful applications of computer aided drug discovery: Moving drugs from concept to the clinic. Curr. Top. Med. Chem. 2010, 10, 127–141. [Google Scholar] [CrossRef]
- Kaushik, A.C.; Kumar, A.; Bharadwaj, S.; Chaudhary, R.; Sahi, S. Structure-Based Approach for In-silico Drug Designing. In Bioinformatics Techniques for Drug Discovery; Springer: Berlin/Heidelberg, Germany, 2018; pp. 21–25. [Google Scholar]
- Hamad, O.; Amran, S.; Sabbah, A. Drug Discovery-Yesterday and Tomorrow: The Common Approaches in Drug Design and Cancer. Cell Cell. Life Sci. J. 2018, 3, 000119. [Google Scholar]
- Lu, W.; Zhang, R.; Jiang, H.; Zhang, H.; Luo, C. Computer-Aided Drug Design in Epigenetics. Front. Chem. 2018, 6. [Google Scholar] [CrossRef]
- Kapetanovic, I. Computer-aided drug discovery and development (CADDD): In silico-chemico-biological approach. Chem. Biol. Interact. 2008, 171, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Macalino, S.J.Y.; Gosu, V.; Hong, S.; Choi, S. Role of computer-aided drug design in modern drug discovery. Arch. Pharmacal. Res. 2015, 38, 1686–1701. [Google Scholar] [CrossRef] [PubMed]
- Xiang, M.; Cao, Y.; Fan, W.; Chen, L.; Mo, Y. Computer-aided drug design: Lead discovery and optimization. Comb. Chem. High Throughput Screen. 2012, 15, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.; Blaney, J.; Blundell, T.; Clark, D.; Davis, A.M.; Ealick, S.; Kim, S.-H.; McCammon, J.A.; Verdonk, M.; Wijnand, M. Computational and Structural Approaches to Drug Discovery: Ligand-Protein Interactions; Royal Society of Chemistry: London, UK, 2007. [Google Scholar]
- Hauri, S.; Khakzad, H.; Happonen, L.; Teleman, J.; Malmström, J.; Malmström, L. Rapid determination of quaternary protein structures in complex biological samples. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Baig, M.H.; Ahmad, K.; Rabbani, G.; Danishuddin, M.; Choi, I. Computer aided drug design and its application to the development of potential drugs for neurodegenerative disorders. Curr. Neuropharmacol. 2018, 16, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Makhouri, F.R.; Ghasemi, J.B. In Silico studies in drug research against neurodegenerative diseases. Curr. Neuropharmacol. 2018, 16, 664–725. [Google Scholar] [CrossRef] [PubMed]
- Fawzi, M.M.; Abdallah, H.H.; Suroowan, S.; Jugreet, S.; Zhang, Y.; Hu, X. In Silico Exploration of Bioactive Phytochemicals Against Neurodegenerative Diseases via Inhibition of Cholinesterases. Curr. Pharm. Des. 2020, 26, 4151–4162. [Google Scholar] [CrossRef]
- Schmiedel, J.M.; Lehner, B. Determining protein structures using deep mutagenesis. Nat. Genet. 2019, 51, 1177. [Google Scholar] [CrossRef]
- Kumar, J.; Ranjan, T.; Kumar, R.R.; Ansar, M.; Rajani, K.; Kumar, M.; Kumar, V.; Kumar, A. In silico Characterization and Homology Modelling of Potato Leaf Roll Virus (PLRV) Coat Protein. Curr. J. Appl. Sci. Technol. 2019, 1–8. [Google Scholar] [CrossRef]
- Morales-Navarro, S.; Prent-Peñaloza, L.; Rodríguez Núñez, Y.A.; Sánchez-Aros, L.; Forero-Doria, O.; González, W.; Campilllo, N.E.; Reyes-Parada, M.; Martínez, A.; Ramírez, D. Theoretical and Experimental Approaches AiMed. at Drug Design Targeting Neurodegenerative Diseases. Processes 2019, 7, 940. [Google Scholar] [CrossRef]
- Wang, B.; Dai, P.; Ding, D.; Del Rosario, A.; Grant, R.A.; Pentelute, B.L.; Laub, M.T. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 2019, 15, 141–150. [Google Scholar] [CrossRef]
- Chan, H.S.; Li, Y.; Dahoun, T.; Vogel, H.; Yuan, S. New binding sites, new opportunities for GPCR drug discovery. Trends Biochem. Sci. 2019, 44, 312–330. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiao, S.; Ji, S.; Han, N.; Liu, D.; Zhou, J. Identification of DNA–protein binding sites by bootstrap multiple convolutional neural networks on sequence information. Eng. Appl. Artif. Intell. 2019, 79, 58–66. [Google Scholar] [CrossRef]
- Ye, W.; Wang, W.; Jiang, C.; Yu, Q.; Chen, H. Molecular dynamics simulations of amyloid fibrils: An in silico approach. Acta Biochim. Biophys. Sin. 2013, 45, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Makarasen, A.; Kuno, M.; Patnin, S.; Reukngam, N.; Khlaychan, P.; Deeyohe, S.; Intachote, P.; Saimanee, B.; Sengsai, S.; Boonsri, P. Molecular Docking Studies and Synthesis of Amino-oxy-diarylquinoline Derivatives as Potent Non-nucleoside HIV-1 Reverse Transcriptase Inhibitors. Drug Res. 2019, 69, 671–682. [Google Scholar] [CrossRef]
- Vilar, S.; Sobarzo-Sánchez, E.; Uriarte, E. In Silico Prediction of P-glycoprotein Binding: Insights from Molecular Docking Studies. Curr. Med. Chem. 2019, 26, 1746–1760. [Google Scholar] [CrossRef] [PubMed]
- Nunes, R.R.; Fonseca, A.L.D.; Pinto, A.C.D.S.; Maia, E.H.B.; Silva, A.M.D.; Varotti, F.D.P.; Taranto, A.G. Brazilian malaria molecular targets (BraMMT): Selected receptors for virtual high-throughput screening experiments. Memórias Do Inst. Oswaldo Cruz 2019, 114, e180465. [Google Scholar] [CrossRef] [PubMed]
- Zerroug, A.; Belaidi, S.; BenBrahim, I.; Sinha, L.; Chtita, S. Virtual screening in drug-likeness and structure/activity relationship of pyridazine derivatives as Anti-Alzheimer drugs. J. King Saud Univ. Sci. 2019, 31, 595–601. [Google Scholar] [CrossRef]
- Vieira, T.; Magalhaes, R.; Sousa, S. Tailoring specialized scoring functions for more efficient virtual screening. Frontiers 2019, 2, 1–4. [Google Scholar]
- Ray, R. Understanding the Structural Importance of the Non-Binding and Binding Parts of Bedaquiline and Its Analogues with ATP Synthase Subunit C Using Molecular Docking, Molecular Dynamics Simulation and 3D-QSAR Techniques. In Proceedings of the International Conference on Drug Discovery (ICDD) 2020, Hyderabad, India, 19 February 2020. [Google Scholar]
- Kotzabasaki, M.I.; Sotiropoulos, I.; Sarimveis, H. QSAR modeling of the toxicity classification of superparamagnetic iron oxide nanoparticles (SPIONs) in stem-cell monitoring applications: An integrated study from data curation to model development. RSC Adv. 2020, 10, 5385–5391. [Google Scholar] [CrossRef]
- Gbeddy, G.; Egodawatta, P.; Goonetilleke, A.; Ayoko, G.; Chen, L. Application of quantitative structure-activity relationship (QSAR) model in comprehensive human health risk assessment of PAHs, and alkyl-, nitro-, carbonyl-, and hydroxyl-PAHs laden in urban road dust. J. Hazard. Mater. 2020, 383, 121154. [Google Scholar] [CrossRef]
- Du, M.; Qiu, Y.; Li, Q.; Li, Y. Efficacy coefficient method assisted quadruple-activities 3D-QSAR pharmacophore model for application in environmentally friendly PAE molecular modification. Environ. Sci. Pollut. Res. Int. 2020, 27, 24103–24114. [Google Scholar] [CrossRef]
- Fan, F.; Warshaviak, D.T.; Hamadeh, H.K.; Dunn, R.T. The integration of pharmacophore-based 3D QSAR modeling and virtual screening in safety profiling: A case study to identify antagonistic activities against adenosine receptor, A2A, using 1,897 known drugs. PLoS ONE 2019, 14, e0204378. [Google Scholar] [CrossRef]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Espinoza-Moraga, M.; Caballero, J.; Gaube, F.; Winckler, T.; Santos, L.S. 1-Benzyl-1,2,3,4-tetrahydro-β-carboline as channel blocker of N-methyl-D-aspartate receptors. Chem. Biol. Drug Des. 2012, 79, 594–599. [Google Scholar] [CrossRef]
- Mishra, C.B.; Kumari, S.; Manral, A.; Prakash, A.; Saini, V.; Lynn, A.M.; Tiwari, M. Design, synthesis, in-silico and biological evaluation of novel donepezil derivatives as multi-target-directed ligands for the treatment of Alzheimer’s disease. Eur. J. Med. Chem. 2017, 125, 736–750. [Google Scholar] [CrossRef]
- Samadi, A.; de la Fuente Revenga, M.; Pérez, C.; Iriepa, I.; Moraleda, I.; Rodríguez-Franco, M.I.; Marco-Contelles, J. Synthesis, pharmacological assessment, and molecular modeling of 6-chloro-pyridonepezils: New dual AChE inhibitors as potential drugs for the treatment of Alzheimer’s disease. Eur. J. Med. Chem. 2013, 67, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.H.H.; Fatima, A.; Gaurav, A. In Silico investigation of flavonoids as potential trypanosomal nucleoside hydrolase inhibitors. Adv. Bioinform. 2015, 2015, 1–10. [Google Scholar] [CrossRef] [PubMed]
- De Andrade Teles, R.B.; Diniz, T.C.; Costa Pinto, T.C.; de Oliveira Júnior, R.G.; Gama E Silva, M.; de Lavor, É.M.; Fernandes, A.W.C.; de Oliveira, A.P.; de Almeida Ribeiro, F.P.R.; da Silva, A.A.M.; et al. Flavonoids as Therapeutic Agents in Alzheimer’s and Parkinson’s Diseases: A Systematic Review of Preclinical Evidences. Oxidative Med. Cell. Longev. 2018, 2018, 1–21. [Google Scholar] [CrossRef]
- Wei, H.; Wu, G.; Chen, J.; Zhang, X.; Xiong, C.; Lei, Y.; Chen, W.; Ruan, J. (2S)-5, 2′, 5′-trihydroxy-7-methoxyflavanone, a natural product from abacopteris penangiana, presents neuroprotective effects in vitro and in vivo. Neurochem. Res. 2013, 38, 1686–1694. [Google Scholar] [CrossRef]
- Fjelldal, M.F.; Freyd, T.; Evenseth, L.M.; Sylte, I.; Ring, A.; Paulsen, R.E. Exploring the overlapping binding sites of ifenprodil and EVT -101 in GluN2B-containing NMDA receptors using novel chicken embryo forebrain cultures and molecular modeling. Pharmacol. Res. Perspect. 2019, 7, e00480. [Google Scholar] [CrossRef]
- Kumar, S.; Chowdhury, S.; Kumar, S. In silico repurposing of antipsychotic drugs for Alzheimer’s disease. Bmc Neurosci. 2017, 18, 76. [Google Scholar] [CrossRef]
- Thomas, S.J.; Grossberg, G.T. Memantine: A review of studies into its safety and efficacy in treating Alzheimer’s disease and other dementias. Clin. Interv. Aging 2009, 4, 367. [Google Scholar]
- Memantine for Treatment of Cognitive Impairment in Patients with Parkinson’s Disease and Dementia. Available online: https://clinicaltrials.gov/ct2/show/NCT00294554 (accessed on 14 April 2021).
- Remya, C.; Dileep, K.V.; Tintu, I.; Variyar, E.J.; Sadasivan, C. Design of potent inhibitors of acetylcholinesterase using morin as the starting compound. Front. Life Sci. 2012, 6, 107–117. [Google Scholar] [CrossRef]
- Du, Y.; Qu, J.; Zhang, W.; Bai, M.; Zhou, Q.; Zhang, Z.; Li, Z.; Miao, J. Morin reverses neuropathological and cognitive impairments in APPswe/PS1dE9 mice by targeting multiple pathogenic mechanisms. Neuropharmacology 2016, 108, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tadayon, M.; Garkani-Nejad, Z. In silico study combining QSAR, docking and molecular dynamics simulation on 2,4-disubstituted pyridopyrimidine derivatives. J. Recept. Signal Transduct. 2019, 39, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.K.; Tota, S.; Tripathi, T.; Chander, S.; Nath, C.; Saxena, A.K. Lead optimization studies towards the discovery of novel carbamates as potent AChE inhibitors for the potential treatment of Alzheimer’s disease. Bioorganic. Med. Chem. 2012, 20, 6313–6320. [Google Scholar] [CrossRef]
- Samadi, A.; Estrada, M.; Pérez, C.; Rodríguez-Franco, M.I.; Iriepa, I.; Moraleda, I.; Chioua, M.; Marco-Contelles, J. Pyridonepezils, new dual AChE inhibitors as potential drugs for the treatment of Alzheimer’s disease: Synthesis, biological assessment, and molecular modeling. Eur. J. Med. Chem. 2012, 57, 296–301. [Google Scholar] [CrossRef]
- Popugaeva, E.; Chernyuk, D.; Zhang, H.; Postnikova, T.Y.; Pats, K.; Fedorova, E.; Poroikov, V.; Zaitsev, A.V.; Bezprozvanny, I. Derivatives of Piperazines as potential therapeutic agents for Alzheimer’s disease. Mol. Pharmacol. 2019, 95, 337–348. [Google Scholar] [CrossRef]
- Varadaraju, K.R.; Kumar, J.R.; Mallesha, L.; Muruli, A.; Mohana, K.N.S.; Mukunda, C.K.; Sharanaiah, U. Virtual Screening and Biological Evaluation of Piperazine Derivatives as Human Acetylcholinesterase Inhibitors. Int. J. Alzheimers Dis. 2013, 2013, 653962. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, Y.; Jin, L.; Liang, G. Revealing the mechanism of EGCG, Genistein, Rutin, Quercetin, and Silibinin against hIAPP aggregation via computational simulations. Interdiscip. Sci. Comput. Life Sci. 2020, 12, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Ramalingayya, G.V.; Cheruku, S.P.; Nayak, P.G.; Kishore, A.; Shenoy, R.; Rao, C.M.; Krishnadas, N. Rutin protects against neuronal damage in vitro and ameliorates doxorubicin-induced memory deficits in vivo in Wistar rats. Drug Des. Dev. Ther. 2017, 11, 1011. [Google Scholar] [CrossRef]
- Vancraenenbroeck, R.; De Raeymaecker, J.; Lobbestael, E.; Gao, F.; De Maeyer, M.; Voet, A.; Baekelandt, V.; Taymans, J.M. In silico, in vitro and cellular analysis with a kinome-wide inhibitor panel correlates cellular LRRK2 dephosphorylation to inhibitor activity on LRRK2. Front. Mol. Neurosci. 2014, 7, 51. [Google Scholar] [CrossRef]
- West, A.B. Achieving neuroprotection with LRRK2 kinase inhibitors in Parkinson disease. Exp. Neurol. 2017, 298, 236–245. [Google Scholar] [CrossRef]
- Padhi, A.K.; Banerjee, K.; Gomes, J.; Banerjee, M. Computational and Functional Characterization of Angiogenin Mutations, and Correlation with Amyotrophic Lateral Sclerosis. PLoS ONE 2014, 9, e111963. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-J.; Chang, T.-T.; Chen, H.-Y.; Chen, C.Y.-C. Finding inhibitors of mutant superoxide dismutase-1 for amyotrophic lateral sclerosis therapy from traditional chinese medicine. Evid. Based Complement. Altern. Med. 2014, 2014, 156276. [Google Scholar] [CrossRef]
- Bello, O.S.; Gonzalez, J.; Capani, F.; Barreto, G.E. In silico docking reveals possible Riluzole binding sites on Nav1. 6 sodium channel: Implications for amyotrophic lateral sclerosis therapy. J. Theor. Biol. 2012, 315, 53–63. [Google Scholar] [CrossRef]
- Benavides-Serrato, A.; Saunders, J.T.; Holmes, B.; Nishimura, R.N.; Lichtenstein, A.; Gera, J. Repurposing Potential of Riluzole as an ITAF Inhibitor in mTOR Therapy Resistant Glioblastoma. Int. J. Mol. Sci. 2020, 21, 344. [Google Scholar] [CrossRef]
- Huang, N.K.; Lin, J.H.; Lin, J.T.; Lin, C.I.; Liu, E.M.; Lin, C.J.; Chen, W.P.; Shen, Y.C.; Chen, H.M.; Chen, J.B.; et al. A new drug design targeting the adenosinergic system for Huntington’s disease. PLoS ONE 2011, 6, e20934. [Google Scholar] [CrossRef]
- Frydryskova, K.; Masek, T.; Pospisek, M. Changing faces of stress: Impact of heat and arsenite treatment on the composition of stress granules. Wiley Interdiscip. Rev. RNA 2020, e1596. [Google Scholar] [CrossRef]
- Hebert, L.E.; Weuve, J.; Scherr, P.A.; Evans, D.A. Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology 2013, 80, 1778–1783. [Google Scholar] [CrossRef]
- Prince, M.; Knapp, M.; Guerchet, M.; McCrone, P.; Prina, M.; Comas-Herrera, A.; Wittenberg, R.; Adelaja, B.; Hu, B.; King, D. Dementia UK: Overview; Alzheimers Society: Belfast, UK, 2014. [Google Scholar]
- Mayrhofer, A.; Shora, S. Psychosocial Interventions for Younger People Diagnosed with Dementia: A Focus on Communities. In Proceedings of the Young Dementia Annual Conference 2019, London, UK, 20 November 2019; Available online: https://careinfo.org/event/young-dementia-2019/ (accessed on 28 April 2021).
- Nelson, P.T.; Alafuzoff, I.; Bigio, E.H.; Bouras, C.; Braak, H.; Cairns, N.J.; Castellani, R.J.; Crain, B.J.; Davies, P.; Tredici, K.D. Correlation of Alzheimer disease neuropathologic changes with cognitive status: A review of the literature. J. Neuropathol. Exp. Neurol. 2012, 71, 362–381. [Google Scholar] [CrossRef]
- Bellenguez, C.; Grenier-Boley, B.; Lambert, J.-C. Genetics of Alzheimer’s disease: Where we are, and where we are going. Curr. Opin. Neurobiol. 2020, 61, 40–48. [Google Scholar] [CrossRef]
- Bordone, M.P.; Salman, M.M.; Titus, H.E.; Amini, E.; Andersen, J.V.; Chakraborti, B.; Diuba, A.V.; Dubouskaya, T.G.; Ehrke, E.; Espindola de Freitas, A. The energetic brain–A review from students to students. J. Neurochem. 2019, 151, 139–165. [Google Scholar] [CrossRef]
- Nedergaard, M.; Goldman, S.A. Glymphatic failure as a final common pathway to dementia. Science 2020, 370, 50–56. [Google Scholar] [CrossRef]
- Kitchen, P.; Salman, M.M.; Halsey, A.M.; Clarke-Bland, C.; MacDonald, J.A.; Ishida, H.; Vogel, H.J.; Almutiri, S.; Logan, A.; Kreida, S. Targeting Aquaporin-4 Subcellular Localization to Treat Central Nervous System Edema. Cell 2020, 181, 784–799.e719. [Google Scholar] [CrossRef]
- Salman, M.M.; Sheilabi, M.A.; Bhattacharyya, D.; Kitchen, P.; Conner, A.C.; Bill, R.M.; Woodroofe, M.N.; Conner, M.T.; Princivalle, A.P. Transcriptome analysis suggests a role for the differential expression of cerebral aquaporins and the MAPK signalling pathway in human temporal lobe epilepsy. Eur. J. Neurosci. 2017, 46, 2121–2132. [Google Scholar] [CrossRef]
- Sylvain, N.J.; Salman, M.M.; Pushie, M.J.; Hou, H.; Meher, V.; Herlo, R.; Peeling, L.; Kelly, M.E. The effects of trifluoperazine on brain edema, aquaporin-4 expression, and metabolic markers during the acute phase of stroke using photothrombotic mouse model. Biochim. Et Biophys. Acta (BBA) Biomembr. 2021, 1863, 183573. [Google Scholar] [CrossRef] [PubMed]
- Abir-Awan, M.; Kitchen, P.; Salman, M.M.; Conner, M.T.; Conner, A.C.; Bill, R.M. Inhibitors of mammalian aquaporin water channels. Int. J. Mol. Sci. 2019, 20, 1589. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, P.; Day, R.E.; Taylor, L.H.; Salman, M.M.; Bill, R.M.; Conner, M.T.; Conner, A.C. Identification and molecular mechanisms of the rapid tonicity-induced relocalization of the aquaporin 4 channel. J. Biol. Chem. 2015, 290, 16873–16881. [Google Scholar] [CrossRef] [PubMed]
- Salman, M.M.; Kitchen, P.; Woodroofe, M.N.; Brown, J.E.; Bill, R.M.; Conner, A.C.; Conner, M.T. Hypothermia increases aquaporin 4 (AQP4) plasma membrane abundance in human primary cortical astrocytes via a calcium/transient receptor potential vanilloid 4 (TRPV4)- and calmodulin-mediated mechanism. Eur. J. Neurosci. 2017, 46, 2542–2547. [Google Scholar] [CrossRef] [PubMed]
- Akincioglu, H.; Gulcin, I. Potent Acetylcholinesterase Inhibitors: Potential Drugs for Alzheimer’s Disease. Mini Rev. Med. Chem. 2020. [Google Scholar] [CrossRef]
- Tang, H.; Song, P.; Li, J.; Zhao, D. Effect of Salvia miltiorrhiza on acetylcholinesterase: Enzyme kinetics and interaction mechanism merging with molecular docking analysis. Int. J. Biol. Macromol. 2019, 135, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Ram, H.; Jaipal, N.; Kumar, P.; Deka, P.; Kumar, S.; Kashyap, P.; Kumar, S.; Singh, B.P.; Alqarawi, A.A.; Hashem, A.; et al. Dual Inhibition of DPP-4 and Cholinesterase Enzymes by the Phytoconstituents of the Ethanolic Extract of Prosopis cineraria Pods: Therapeutic Implications for the Treatment of Diabetes-associated Neurological Impairments. Curr. Alzheimer Res. 2019, 16, 1230–1244. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, M.K.; Choudhury, S. Tea polyphenols as multi-target therapeutics for Alzheimer’s disease: An in silico study. Med. Hypotheses 2019, 125, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Vuree, S.; Goud, H.; Hussain, T.; Nayarisseri, A.; Singh, S.K. Identification of High-affinity Small Molecules Targeting Gamma Secretase for the Treatment of Alzheimer’s Disease. Curr. Top. Med. Chem. 2019, 19, 1173–1187. [Google Scholar] [CrossRef] [PubMed]
- D’Amelio, M.; Sheng, M.; Cecconi, F. Caspase-3 in the central nervous system: Beyond apoptosis. Trends Neurosci. 2012, 35, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Imbriani, P.; Tassone, A.; Meringolo, M.; Ponterio, G.; Madeo, G.; Pisani, A.; Bonsi, P.; Martella, G. Loss of Non-Apoptotic Role of Caspase-3 in the PINK1 Mouse Model of Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 3407. [Google Scholar] [CrossRef]
- Kavanagh, E.; Rodhe, J.; Burguillos, M.A.; Venero, J.L.; Joseph, B. Regulation of caspase-3 processing by cIAP2 controls the switch between pro-inflammatory activation and cell death in microglia. Cell Death Dis. 2014, 5, e1565. [Google Scholar] [CrossRef]
- Snigdha, S.; Smith, E.D.; Prieto, G.A.; Cotman, C.W. Caspase-3 activation as a bifurcation poInt. between plasticity and cell death. Neurosci. Bull. 2012, 28, 14–24. [Google Scholar] [CrossRef]
- Cancela, S.; Canclini, L.; Mourglia-Ettlin, G.; Hernández, P.; Merlino, A. Neuroprotective effects of novel nitrones: In vitro and in silico studies. Eur. J. Pharmacol. 2020, 871, 172926. [Google Scholar] [CrossRef]
- Goubau, C.; Jaeken, J.; Levtchenko, E.N.; Thys, C.; Di Michele, M.; Martens, G.A.; Gerlo, E.; De Vos, R.; Buyse, G.M.; Goemans, N.; et al. Homozygosity for aquaporin 7 G264V in three unrelated children with hyperglyceroluria and a mild platelet secretion defect. Genet Med. 2013, 15, 55–63. [Google Scholar] [CrossRef]
- Greig, N.H.; Reale, M.; Tata, A.M. New pharmacological approaches to the cholinergic system: An overview on muscarinic receptor ligands and cholinesterase inhibitors. Recent Pat. CNS Drug Discov. 2013, 8, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Volpato, D.; Holzgrabe, U. Designing hybrids targeting the cholinergic system by modulating the muscarinic and nicotinic receptors: A concept to treat Alzheimer’s disease. Molecules 2018, 23, 3230. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Bonus, M.; Gohlke, H.; Klöcker, N. Isoform-specific Inhibition of N-methyl-D-aspartate Receptors by Bile Salts. Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Jensen, M.O.; Jogini, V.; Stein, R.A.; Lee, C.H.; McHaourab, H.S.; Shaw, D.E.; Gouaux, E. Mechanism of NMDA receptor channel block by MK-801 and memantine. Nature 2018, 556, 515–519. [Google Scholar] [CrossRef]
- Geerts, H.; Grossberg, G.T. Pharmacology of acetylcholinesterase inhibitors and N-methyl-D-aspartate receptors for combination therapy in the treatment of Alzheimer’s disease. J. Clin. Pharmacol. 2006, 46, 8s–16s. [Google Scholar] [CrossRef]
- Singh, R.; Ganeshpurkar, A.; Kumar, D.; Kumar, D.; Kumar, A.; Singh, S.K. Identifying potential GluN2B subunit containing N-Methyl-D-aspartate receptor inhibitors: An integrative in silico and molecular modeling approach. J. Biomol. Struct. Dyn. 2019, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Waqar, M.; Batool, S. In silico analysis of binding interaction of conantokins with NMDA receptors for potential therapeutic use in Alzheimer’s disease. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 42. [Google Scholar] [CrossRef]
- Alokam, R.; Singhal, S.; Srivathsav, G.S.; Garigipati, S.; Puppala, S.; Sriram, D.; Perumal, Y. Design of dual inhibitors of ROCK-I and NOX2 as potential leads for the treatment of neuroinflammation associated with various neurological diseases including autism spectrum disorder. Mol. Biosyst. 2015, 11, 607–617. [Google Scholar] [CrossRef]
- Simpson, D.S.; Oliver, P.L. ROS generation in microglia: Understanding oxidative stress and inflammation in neurodegenerative disease. Antioxidants 2020, 9, 743. [Google Scholar] [CrossRef]
- Sturchio, A.; Marsili, L.; Mahajan, A.; Grimberg, B.; Kauffman, M.A.; Espay, A.J. How Have Advances in Genetic Technology Modified Movement Disorders Nosology? Eur. J. Neurol. 2020. [Google Scholar] [CrossRef]
- Paul, D.A.; Qureshi, A.R.M.; Rana, A.Q. Peripheral neuropathy in Parkinson’s disease. Neurol. Sci. 2020. [Google Scholar] [CrossRef]
- Coundouris, S.P.; Terrett, G.; Laakso, L.; Schweitzer, D.; Kneebone, A.; Rendell, P.G.; Henry, J.D. A meta-analytic review of prospection deficits in Parkinson’s disease. Neurosci. Biobehav. Rev. 2020, 108, 34–47. [Google Scholar] [CrossRef]
- Kelly, J.; Moyeed, R.; Carroll, C.; Albani, D.; Li, X. Gene expression meta-analysis of Parkinson’s disease and its relationship with Alzheimer’s disease. Mol. Brain 2019, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Henderson, E.J.; Lyell, V.; Bhimjiyani, A.; Amin, J.; Kobylecki, C.; Gregson, C.L. Management of fracture risk in Parkinson’s: A revised algorithm and focused review of treatments. Parkinsonism Relat. Disord. 2019, 64, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Haunton, V.J. Movement disorders: A themed collection. Age Ageing 2020, 49, 12–15. [Google Scholar] [CrossRef]
- Félix, J.P.; Vieira, F.H.T.; Cardoso, Á.A.; Ferreira, M.V.G.; Franco, R.A.P.; Ribeiro, M.A.; Araújo, S.G.; Corrêa, H.P.; Carneiro, M.L. A Parkinson’s Disease Classification Method: An Approach Using Gait Dynamics and Detrended Fluctuation Analysis. In Proceedings of the 2019 IEEE Canadian Conference of Electrical and Computer Engineering (CCECE), Edmonton, AB, Canada, 5–8 May 2019; pp. 1–4. [Google Scholar]
- Chakraborty, A.; Brauer, S.; Diwan, A. A review of possible therapies for Parkinson’s disease. J. Clin. Neurosci. 2020. [Google Scholar] [CrossRef]
- Ma, Z.; Liu, H.; Wu, B. Structure-based drug design of catechol-O-methyltransferase inhibitors for CNS disorders. Br. J. Clin. Pharmacol. 2014, 77, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Borovac, J.A. Focus: The Aging Brain: Side effects of a dopamine agonist therapy for Parkinson’s disease: A mini-review of clinical pharmacology. Yale J. Biol. Med. 2016, 89, 37. [Google Scholar]
- Thobois, S. Proposed dose equivalence for rapid switch between dopamine receptor agonists in Parkinson’s disease: A review of the literature. Clin. Ther. 2006, 28, 1–12. [Google Scholar] [CrossRef]
- Missale, C.; Nash, S.R.; Robinson, S.W.; Jaber, M.; Caron, M.G. Dopamine receptors: From structure to function. Physiol. Rev. 1998, 78, 189–225. [Google Scholar] [CrossRef] [PubMed]
- Le Foll, B.; Collo, G.; Rabiner, E.A.; Boileau, I.; Pich, E.M.; Sokoloff, P. Dopamine D3 receptor ligands for drug addiction treatment: Update on recent findings. In Progress in Brain Research; Elsevier: Amsterdam, The Netherlands, 2014; Volume 211, pp. 255–275. [Google Scholar]
- Maggio, R.; Aloisi, G.; Silvano, E.; Rossi, M.; Millan, M.J. Heterodimerization of dopamine receptors: New insights into functional and therapeutic significance. Parkinsonism Relat. Disord. 2009, 15, S2–S7. [Google Scholar] [CrossRef]
- Connor-Robson, N.; Booth, H.; Martin, J.G.; Gao, B.; Li, K.; Doig, N.; Vowles, J.; Browne, C.; Klinger, L.; Juhasz, P. An integrated transcriptomics and proteomics analysis reveals functional endocytic dysregulation caused by mutations in LRRK2. Neurobiol. Dis. 2019, 127, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Jin, W. Novel Insights into PARK7 (DJ-1), a Potential Anti-Cancer Therapeutic Target, and Implications for Cancer Progression. J. Clin. Med. 2020, 9, 1256. [Google Scholar] [CrossRef] [PubMed]
- Bartonikova, T.; Mensikova, K.; Kolarikova, K.; Vodicka, R.; Vrtel, R.; Otruba, P.; Kaiserova, M.; Vastik, M.; Mikulicova, L.; Ovecka, J.; et al. New endemic familial parkinsonism in south Moravia, Czech Republic and its genetical background. Medicine 2018, 97, e12313. [Google Scholar] [CrossRef]
- Schmidt, S.H.; Knape, M.J.; Boassa, D.; Mumdey, N.; Kornev, A.P.; Ellisman, M.H.; Taylor, S.S.; Herberg, F.W. The dynamic switch mechanism that leads to activation of LRRK2 is embedded in the DFGpsi motif in the kinase domain. Proc. Natl. Acad. Sci. USA 2019, 116, 14979–14988. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, N.; Chellam, J.; Kannan, R.R. Exploring the functional impact of mutational drift in LRRK2 gene and identification of specific inhibitors for the treatment of Parkinson disease. J. Cell. Biochem. 2018, 119, 4878–4889. [Google Scholar] [CrossRef] [PubMed]
- Flor, P.J.; Battaglia, G.; Nicoletti, F.; Gasparini, F.; Bruno, V. Neuroprotective activity of metabotropic glutamate receptor ligands. In Molecular and Cellular Biology of Neuroprotection in the CNS; Springer: Berlin/Heidelberg, Germany, 2003; pp. 197–223. [Google Scholar]
- Montastruc, J.; Rascol, O.; Senard, J. Glutamate antagonists and Parkinson’s disease: A review of clinical data. Neurosci. Biobehav. Rev. 1997, 21, 477–480. [Google Scholar] [CrossRef]
- Marzo, A.; Dal Bo, L.; Monti, N.C.; Crivelli, F.; Ismaili, S.; Caccia, C.; Cattaneo, C.; Fariello, R.G. Pharmacokinetics and pharmacodynamics of safinamide, a neuroprotectant with antiparkinsonian and anticonvulsant activity. Pharmacol. Res. 2004, 50, 77–85. [Google Scholar] [CrossRef]
- Stocchi, F.; Arnold, G.; Onofrj, M.; Kwiecinski, H.; Szczudlik, A.; Thomas, A.; Bonuccelli, U.; Van Dijk, A.; Cattaneo, C.; Sala, P. Improvement of motor function in early Parkinson disease by safinamide. Neurology 2004, 63, 746–748. [Google Scholar] [CrossRef]
- Wijesekera, L.C.; Leigh, P.N. Amyotrophic lateral sclerosis. Orphanet J. Rare Dis. 2009, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, A.; Palovcak, M. The role of augmentative communication devices in the medical management of ALS. NeuroRehabilitation 2007, 22, 445–450. [Google Scholar] [CrossRef]
- Martin, L.J.; Liu, Z. Opportunities for neuroprotection in ALS using cell death mechanism rationales. Drug Discov. Today Dis. Models 2004, 1, 135–143. [Google Scholar] [CrossRef]
- Aebischer, P.; Kato, A.C. Playing defense against Lou Gehrig’s disease. Sci. Am. 2007, 297, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, Y.; Torres, A.S.; O’Halloran, T.V. Oxygen-induced maturation of SOD1: A key role for disulfide formation by the copper chaperone CCS. EMBO J. 2004, 23, 2872–2881. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, K.; Fujiwara, Y.; Terada, T.; Shimizu, K.; Wada, K.; Kabuta, T. Virtual screening identification of novel chemical inhibitors for aberrant interactions between pathogenic mutant SOD1 and tubulin. Neurochem. Int. 2019, 126, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, E.; Rajasekaran, R. Rational design of linear tripeptides against the aggregation of human mutant SOD1 protein causing amyotrophic lateral sclerosis. J. Neurol. Sci. 2019, 405, 116425. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, E.; Rajasekaran, R. Quantum chemical and molecular mechanics studies on the assessment of interactions between resveratrol and mutant SOD1 (G93A) protein. J. Comput. Aided Mol. Des. 2018, 32, 1347–1361. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, E.; Rajasekaran, R. Computational investigation of curcumin, a natural polyphenol that inhibits the destabilization and the aggregation of human SOD1 mutant (Ala4Val). RSC Adv. 2016, 6, 102744–102753. [Google Scholar] [CrossRef]
- Srinivasan, E.; Rajasekaran, R. Comparative binding of kaempferol and kaempferide on inhibiting the aggregate formation of mutant (G85R) SOD1 protein in familial amyotrophic lateral sclerosis: A quantum chemical and molecular mechanics study. Biofactors 2018, 44, 431–442. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Mei, Z.Q.; Wu, J.W.; Wang, Z.X. Enzymatic activity and substrate specificity of mitogen-activated protein kinase p38alpha in different phosphorylation states. J. Biol. Chem. 2008, 283, 26591–26601. [Google Scholar] [CrossRef] [PubMed]
- Thalhamer, T.; McGrath, M.; Harnett, M. MAPKs and their relevance to arthritis and inflammation. Rheumatology 2008, 47, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Chico, L.K.; Van Eldik, L.J.; Watterson, D.M. Targeting protein kinases in central nervous system disorders. Nat. Rev. Drug Discov. 2009, 8, 892–909. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, V.; Sharma, D.; Kalia, K.; Tiwari, V. Neuroprotective effects of silibinin: An in silico and in vitro study. Int. J. Neurosci. 2018, 128, 935–945. [Google Scholar] [CrossRef]
- Laev, S.S.; Salakhutdinov, N.F.; Lavrik, O.I. Tyrosyl-DNA phosphodiesterase inhibitors: Progress and potential. Bioorganic. Med. Chem. 2016, 24, 5017–5027. [Google Scholar] [CrossRef]
- Salado, I.G.; Redondo, M.; Bello, M.L.; Perez, C.; Liachko, N.F.; Kraemer, B.C.; Miguel, L.; Lecourtois, M.; Gil, C.; Martinez, A.; et al. Protein Kinase CK-1 Inhibitors As New Potential Drugs for Amyotrophic Lateral Sclerosis. J. Med. Chem. 2014, 57, 2755–2772. [Google Scholar] [CrossRef]
- Bissaro, M.; Moro, S. Rethinking to riluzole mechanism of action: The molecular link among protein kinase CK1δ activity, TDP-43 phosphorylation, and amyotrophic lateral sclerosis pharmacological treatment. Neural. Regen. Res. 2019, 14, 2083–2085. [Google Scholar] [CrossRef] [PubMed]
- Cozza, G.; Gianoncelli, A.; Montopoli, M.; Caparrotta, L.; Venerando, A.; Meggio, F.; Pinna, L.A.; Zagotto, G.; Moro, S. Identification of novel protein kinase CK1 delta (CK1δ) inhibitors through structure-based virtual screening. Bioorganic. Med. Chem. Lett. 2008, 18, 5672–5675. [Google Scholar] [CrossRef]
- Cozza, G.; Pinna, L.A.; Moro, S. Kinase CK2 inhibition: An update. Curr. Med. Chem. 2013, 20, 671–693. [Google Scholar] [CrossRef]
- Kearney, J.A.; Buchner, D.A.; De Haan, G.; Adamska, M.; Levin, S.I.; Furay, A.R.; Albin, R.L.; Jones, J.M.; Montal, M.; Stevens, M.J. Molecular and pathological effects of a modifier gene on deficiency of the sodium channel Scn8a (Nav1. 6). Hum. Mol. Genet. 2002, 11, 2765–2775. [Google Scholar] [CrossRef]
- Caldwell, J.H.; Schaller, K.L.; Lasher, R.S.; Peles, E.; Levinson, S.R. Sodium channel Nav1. 6 is localized at nodes of Ranvier, dendrites, and synapses. Proc. Natl. Acad. Sci. USA 2000, 97, 5616–5620. [Google Scholar] [CrossRef]
- Gunasekaran, R.; Narayani, R.S.; Vijayalakshmi, K.; Alladi, P.A.; Shobha, K.; Nalini, A.; Sathyaprabha, T.; Raju, T. Exposure to cerebrospinal fluid of sporadic amyotrophic lateral sclerosis patients alters Nav1. 6 and Kv1. 6 channel expression in rat spinal motor neurons. Brain Res. 2009, 1255, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Saba, L.; Viscomi, M.T.; Martini, A.; Caioli, S.; Mercuri, N.B.; Guatteo, E.; Zona, C. Modified age-dependent expression of NaV1. 6 in an ALS model correlates with motor cortex excitability alterations. Neurobiol. Dis. 2019, 130, 104532. [Google Scholar] [CrossRef]
- Seki, S.; Yamamoto, T.; Quinn, K.; Spigelman, I.; Pantazis, A.; Olcese, R.; Wiedau-Pazos, M.; Chandler, S.H.; Venugopal, S. Circuit-Specific Early Impairment of Proprioceptive Sensory Neurons in the SOD1G93A Mouse Model for ALS. J. Neurosci. 2019, 39, 8798–8815. [Google Scholar] [CrossRef] [PubMed]
- Grant, P.; Song, J.Y.; Swedo, S.E. Review of the use of the glutamate antagonist riluzole in psychiatric disorders and a description of recent use in childhood obsessive-compulsive disorder. J. Child Adolesc. Psychopharmacol. 2010, 20, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Anzai, I.; Toichi, K.; Tokuda, E.; Mukaiyama, A.; Akiyama, S.; Furukawa, Y. Screening of drugs inhibiting in vitro oligomerization of Cu/Zn-superoxide dismutase with a mutation causing amyotrophic lateral sclerosis. Front. Mol. Biosci. 2016, 3, 40. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.B.; Gusella, J.F. Huntingtons disease. N. Engl. J. Med. 1986, 315, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Novak, M.J.; Tabrizi, S.J. Huntington’s disease. BMJ 2010, 340, c3109. [Google Scholar] [CrossRef] [PubMed]
- Wexler, N.S.; Collett, L.; Wexler, A.R.; Rawlins, M.D.; Tabrizi, S.J.; Douglas, I.; Smeeth, L.; Evans, S.J. Incidence of adult Huntington’s disease in the UK: A UK-based primary care study and a systematic review. BMJ Open 2016, 6, e009070. [Google Scholar] [CrossRef]
- Kavanaugh, M.S.; Cho, C.; Maeda, H.; Swope, C. “I am no longer alone”: Evaluation of the first North American camp for youth living in families with Huntington’s disease. Child. Youth Serv. Rev. 2017, 79, 325–332. [Google Scholar] [CrossRef]
- Ayala, K.M. Role of the SLP in management of Huntington’s disease: A literature review. 2017. Available online: https://repositories.lib.utexas.edu/handle/2152/62346 (accessed on 14 April 2021). [CrossRef]
- Travessa, A.M.; Rodrigues, F.B.; Mestre, T.A.; Ferreira, J.J. Fifteen years of clinical trials in Huntington’s disease: A very low clinical drug development success rate. J. Huntingt. Dis. 2017, 6, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Tabrizi, S. Targeting Huntingtin Expression in Patients with Huntington’s Disease. N. Engl. J. Med. 2019, 380, 2307–2316. [Google Scholar] [CrossRef]
- Iftikhar, H.; Batool, S.; Deep, A.; Narasimhan, B.; Sharma, P.C.; Malhotra, M. In silico analysis of the inhibitory activities of GABA derivatives on 4-aminobutyrate transaminase. Arab. J. Chem. 2017, 10, S1267–S1275. [Google Scholar] [CrossRef]
- Boonstra, E.; de Kleijn, R.; Colzato, L.S.; Alkemade, A.; Forstmann, B.U.; Nieuwenhuis, S. Neurotransmitters as food supplements: The effects of GABA on brain and behavior. Front. Psychol. 2015, 6, 1520. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, S.; Kasthuri, G.; Prabhu, S.; Manogar, P.; Parameswari, N. Screening and identification of novel inhibitors against human 4-aminobutyrate-aminotransferase: A computational approach. Egypt J. Basic Appl. Sci. 2018, 5, 210–219. [Google Scholar] [CrossRef]
- Schneider, G. Virtual screening: An endless staircase? Nat. Rev. Drug Discov. 2010, 9, 273–276. [Google Scholar] [CrossRef]
- Verkhivker, G.M.; Bouzida, D.; Gehlhaar, D.K.; Rejto, P.A.; Arthurs, S.; Colson, A.B.; Freer, S.T.; Larson, V.; Luty, B.A.; Marrone, T. Deciphering common failures in molecular docking of ligand-protein complexes. J. Comput. Aided Mol. Des. 2000, 14, 731–751. [Google Scholar] [CrossRef]
- Josephs, D.; Spicer, J.; O’Doherty, M. Molecular imaging in clinical trials. Target. Oncol. 2009, 4, 151–168. [Google Scholar] [CrossRef]
- Cheatham, T.E., III; Young, M.A. Molecular dynamics simulation of nucleic acids: Successes, limitations, and promise. Biopolym. Orig. Res. Biomol. 2000, 56, 232–256. [Google Scholar] [CrossRef]
- Klebe, G. Virtual ligand screening: Strategies, perspectives and limitations. Drug Discov. Today 2006, 11, 580–594. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Korb, O.; Olsson, T.S.; Bowden, S.J.; Hall, R.J.; Verdonk, M.L.; Liebeschuetz, J.W.; Cole, J.C. Potential and limitations of ensemble docking. J. Chem. Inf. Modeling 2012, 52, 1262–1274. [Google Scholar] [CrossRef]
- MacDonald, D.; Breton, R.; Sutcliffe, R.; Walker, J. Uses and limitations of quantitative structure-activity relationships (QSARs) to categorize substances on the Canadian Domestic Substance List as persistent and/or bioaccumulative, and inherently toxic to non-human organisms. Sar Qsar Environ. Res. 2002, 13, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Papakonstantinou, E.; Megalooikonomou, V.; Vlachakis, D. Dark Suite: A comprehensive toolbox for computer-aided drug design. Embnet. J. 2020, 25, e928. [Google Scholar] [CrossRef]
- Berdigaliyev, N.; Aljofan, M. An overview of drug discovery and development. Future Med. Chem. 2020, 12, 939–947. [Google Scholar] [CrossRef]
- Shi, J.; Zha, W. Predicting Human Pharmacokinetics: Physiologically Based Pharmacokinetic Modeling and In Silico ADME Prediction in Early Drug Discovery. Eur. J. Drug Metab. Pharmacokinet. 2019, 44, 135–137. [Google Scholar] [CrossRef]
- Van De Waterbeemd, H.; Gifford, E. ADMET in silico modelling: Towards prediction paradise? Nat. Rev. Drug Discov. 2003, 2, 192–204. [Google Scholar] [CrossRef]
- Faller, B.; Winiwarter, S.; Chang, G.; Desai, P.; Menzel, C.; Rieko, A.; Keefer, C.; Broccatelli, F. Prediction of fraction unbound in microsomal and hepatocyte incubations–a comparison of methods across industry data sets (by the IQ in silico ADME working group). Mol. Pharm. 2019, 16, 4077–4085. [Google Scholar]
- Blomme, E.A.; Will, Y. Toxicology strategies for drug discovery: Present and future. Chem. Res. Toxicol. 2016, 29, 473–504. [Google Scholar] [CrossRef] [PubMed]
- Pruss, R.M. Phenotypic screening strategies for neurodegenerative diseases: A pathway to discover novel drug candidates and potential disease targets or mechanisms. CNS Neurol. Disord. Drug Targets Former. Curr. Drug Targets CNS Neurol. Disord. 2010, 9, 693–700. [Google Scholar] [CrossRef]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. CSS-Palm 2.0: An updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 2008, 21, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Nikzad, H.; Karimian, M.; Sareban, K.; Khoshsokhan, M.; Colagar, A.H. MTHFR-Ala222Val and male infertility: A study in Iranian men, an updated meta-analysis and an in silico-analysis. Reprod. Biomed. Online 2015, 31, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Glykos, N.M. Software news and updates carma: A molecular dynamics analysis program. J. Comput. Chem. 2006, 27, 1765–1768. [Google Scholar] [CrossRef] [PubMed]
- CCDC. What’s New. Available online: https://www.ccdc.cam.ac.uk/solutions/whats-new/ (accessed on 14 April 2021).
- Zuo, Z.; MacMillan, D.W. Decarboxylative arylation of α-amino acids via photoredox catalysis: A one-step conversion of biomass to drug pharmacophore. J. Am. Chem. Soc. 2014, 136, 5257–5260. [Google Scholar] [CrossRef]
- Vázquez, K.; Espinosa-Bustos, C.; Soto-Delgado, J.; Tapia, R.A.; Varela, J.; Birriel, E.; Segura, R.; Pizarro, J.; Cerecetto, H.; González, M. New aryloxy-quinone derivatives as potential anti-Chagasic agents: Synthesis, trypanosomicidal activity, electrochemical properties, pharmacophore elucidation and 3D-QSAR analysis. RSC Adv. 2015, 5, 65153–65166. [Google Scholar] [CrossRef]
- Bennett, B.C.; Wan, Q.; Ahmad, M.F.; Langan, P.; Dealwis, C.G. X-ray structure of the ternary MTX NADPH complex of the anthrax dihydrofolate reductase: A pharmacophore for dual-site inhibitor design. J. Struct. Biol. 2009, 166, 162–171. [Google Scholar] [CrossRef]
- François, P.; Hakim, V. Design of genetic networks with specified functions by evolution in silico. Proc. Natl. Acad. Sci. USA 2004, 101, 580–585. [Google Scholar] [CrossRef]
- Fischer, B.; Fukuzawa, K.; Wenzel, W. Receptor-specific scoring functions derived from quantum chemical models improve affinity estimates for in-silico drug discovery. Proteins Struct. Funct. Bioinform. 2008, 70, 1264–1273. [Google Scholar] [CrossRef]
- Khatami, S.G.; Mubeen, S.; Hofmann-Apitius, M. Data science in neurodegenerative disease: Its capabilities, limitations, and perspectives. Curr. Opin. Neurol. 2020, 33, 249. [Google Scholar] [CrossRef]
- Ferro, M.P.; Heilshorn, S.C.; Owens, R.M. Materials for blood brain barrier modeling in vitro. Mater. Sci. Eng. R Rep. 2020, 140, 100522. [Google Scholar] [CrossRef]
- May, J.-N.; Golombek, S.K.; Baues, M.; Dasgupta, A.; Drude, N.; Rix, A.; Rommel, D.; von Stillfried, S.; Appold, L.; Pola, R. Multimodal and multiscale optical imaging of nanomedicine delivery across the blood-brain barrier upon sonopermeation. Theranostics 2020, 10, 1948. [Google Scholar] [CrossRef]
- Juthani, R.; Madajewski, B.; Yoo, B.; Zhang, L.; Chen, P.-M.; Chen, F.; Turker, M.Z.; Ma, K.; Overholtzer, M.; Longo, V.A. Ultrasmall Core-Shell Silica Nanoparticles for Precision Drug Delivery in a High-Grade Malignant Brain Tumor Model. Clin. Cancer Res. 2020, 26, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Blauwendraat, C.; Heilbron, K.; Vallerga, C.L.; Bandres-Ciga, S.; von Coelln, R.; Pihlstrøm, L.; Simón-Sánchez, J.; Schulte, C.; Sharma, M.; Krohn, L. Parkinson’s disease age at onset genome-wide association study: Defining heritability, genetic loci, and α-synuclein mechanisms. Mov. Disord. 2019, 34, 866–875. [Google Scholar] [CrossRef]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019, 18, 1091–1102. [Google Scholar] [CrossRef]
- Ciani, M.; Benussi, L.; Bonvicini, C.; Ghidoni, R. Genome Wide Association Study and Next Generation Sequencing: A glimmer of light towards new possible horizons in Frontotemporal Dementia research. Front. Neurosci. 2019, 13, 506. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, L.; Farias, F.H.; Dube, U.; Mihindukulasuriya, K.A.; Harari, O. Polygenic risk scores in neurodegenerative diseases: A review. Curr. Genet. Med. Rep. 2019, 7, 22–29. [Google Scholar] [CrossRef]
- Adams, H.H.; Evans, T.E.; Terzikhan, N. The Uncovering Neurodegenerative Insights Through Ethnic Diversity consortium. Lancet Neurol. 2019, 18, 915. [Google Scholar] [CrossRef]
- Mancuso, R.; Fryatt, G.; Cleal, M.E.; Obst, J.; Pipi, E.; Monzon-Sandoval, J.; Winchester, L.; Ribe, E.; Webber, C.; Nevado, A. CSF1R inhibition by JNJ-40346527 alters microglial proliferation and phenotype and results in attenuation of neurodegeneration in P301S mice. Brain 2019, 142, 3243–3264. [Google Scholar] [CrossRef]
- Rahman, S.; Datta, M.; Kim, J.; Jan, A.T. CRISPR/Cas: An intriguing genomic editing tool with prospects in treating neurodegenerative diseases. Semin. Cell Dev. Biol. 2019, 96, 22–31. [Google Scholar] [CrossRef]
- Offen, D.; Perets, N.; Betzer, O.; Popovtzer, R.; Shapira, R.; Ashery, U. Targeting damages in the brain: Exosomes derived from MSC present migration and homing abilities to different neurodegenerative and neuropsychiatric locations. Cytotherapy 2019, 21, e6. [Google Scholar] [CrossRef]
- Raikwar, S.P.; Kikkeri, N.S.; Sakuru, R.; Saeed, D.; Zahoor, H.; Premkumar, K.; Mentor, S.; Thangavel, R.; Dubova, I.; Ahmed, M.E. Next generation precision medicine: CRISPR-mediated genome editing for the treatment of neurodegenerative disorders. J. Neuroimmune Pharmacol. 2019, 14, 608–641. [Google Scholar] [CrossRef]
- Aldewachi, H.; Al-Zidan, R.N.; Conner, M.T.; Salman, M.M. High-Throughput Screening Platforms in the Discovery of Novel Drugs for Neurodegenerative Diseases. Bioengineering 2021, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Mittal, R.; Woo, F.W.; Castro, C.S.; Cohen, M.A.; Karanxha, J.; Mittal, J.; Chhibber, T.; Jhaveri, V.M. Organ-on-chip models: Implications in drug discovery and clinical applications. J. Cell. Physiol. 2019, 234, 8352–8380. [Google Scholar] [CrossRef] [PubMed]
- Salman, M.M.; Marsh, G.; Kusters, I.; Delincé, M.; Di Caprio, G.; Upadhyayula, S.; de Nola, G.; Hunt, R.; Ohashi, K.G.; Gray, T.; et al. Design and Validation of a Human Brain Endothelial Microvessel-on-a-Chip Open Microfluidic Model Enabling Advanced Optical Imaging. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J. Functional MRI of brain physiology in aging and neurodegenerative diseases. Neuroimage 2019, 187, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Muir, E.R.; Biju, K.; Cong, L.; Rogers, W.E.; Hernandez, E.T.; Duong, T.Q.; Clark, R.A. Functional MRI of the mouse olfactory system. Neurosci. Lett. 2019, 704, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Zheng, C.; Cui, B.; Qi, Z.; Zhao, Z.; An, Y.; Qiao, L.; Han, Y.; Zhou, Y.; Lu, J. Multiparametric imaging hippocampal neurodegeneration and functional connectivity with simultaneous PET/MRI in Alzheimer’s disease. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2440–2452. [Google Scholar] [CrossRef]

| No. | Software | No. of Citations to Published Studies | Score | Features | Accessibility | Website |
|---|---|---|---|---|---|---|
| 1 | HADDOCK | 26,490 | 4.7323 | Docks protein−protein based on biochemical or biophysical information | Free | https://wenmr.science.uu.nl/haddock2.4/ |
| 2 | AutoDock Autodock 1 Autodock 2.4 Autodock 3 Autodock 4 Autodock 4.2 Autodock Vina AutoDockFR AutoDockTools | 22,422 | 4.6599 | Automated docking tools | Free | http://autodock.scripps.edu/ |
| 3 | Glide Glide 1.8 Glide 2 Glide 2.5 | 22,091 | 4.6535 | Rapid, accurate docking and scoring approach | Subscription | https://www.schrodinger.com/glide |
| 4 | FlexX | 19,987 | 4.6100 | Predicts the geometry of the protein–ligand complex and estimates the binding affinity | Free | https://www.biosolveit.de/FlexX/ |
| 5 | LigandFit | 19,890 | 4.6079 | Presents a shape-based approach for docking ligands into the active site of the protein | Subscription | https://www.phenix-online.org/documentation/reference/ligandfit.html |
| 6 | AmberTools | 14,572 | 4.4728 | A suite of biomolecular simulation programs | Subscription | https://ambermd.org/ |
| 7 | ENCoM | 13,145 | 4.4280 | A coarse-grained normal mode analysis method utilized for different residues in proteins or nucleotides in RNA | Free | http://biophys.umontreal.ca/nrg/resources.html |
| 8 | PROCHECK-NMR | 10,783 | 4.3420 | Checks the stereochemical quality of a protein structure solved by NMR | Free | https://www.ebi.ac.uk/thornton-srv/software/PROCHECK/ |
| 9 | MCDOCK | 10,603 | 4.3347 | Allows for a full flexibility of ligands in the docking calculations | Free | DOI: 10.1021/jm990129n |
| 10 | ICM ICM 2.8 ICM-Dock | 10,271 | 4.3209 | A new method for protein modelling and design applications to docking and structure prediction | Subscription | http://www.molsoft.com/docking.html |
| 11 | Dock Dock2 Dock3 Dock4 Dock5 Dock6 Dock7 Dock8 Dock9 | 8181 | 4.2221 | Based on a geometric matching algorithm | Free | http://dock.compbio.ucsf.edu/ |
| 12 | SOFT Docking | 7474 | 4.1828 | Predicts the sites of interaction between two cognate molecules based on their 3D structures | Subscription | https://doi.org/10.1016/0022-2836(91)90859-5 |
| 13 | FDS | 7188 | 4.1659 | Cluster analysis based on distance similarities | Free | http://www.scfbio-iitd.res.in/dock/fds.jsp |
| 14 | DockVision | 6950 | 4.1512 | Increases capability to generate laudable results | Free | http://dockvision.sness.net/overview/overview.html |
| 15 | PRODOCK | 6442 | 4.1183 | Renders the programming easier and the definition of molecular flexibility more straightforward | Subscription | https://doi.org/10.1002/(SICI)1096-987X(199903)20:4<412::AID-JCC3>3.0.CO;2-N |
| 16 | YASARA YASARA Dynamics YASARA Model YASARA NMR Module YASARA Structure YASARA View YASARA Virtual Reality Workstation YASARA/WHAT IF Twinset | 5870 | 4.0779 | A molecular-graphics, -modelling, and -simulation program | Free | http://www.yasara.org/products.htm |
| 17 | KBDOCK | 5820 | 4.0742 | A program that proposes structural templates for protein docking | Free | http://kbdock.loria.fr/ |
| 18 | TreeDock | 5796 | 4.0724 | A docking tool that is able to explore all clash-free orientations at very fine resolution in a reasonable time | Subscription | https://doi.org/10.1021/ja011240x |
| 19 | LePro | 5639 | 4.0605 | Generates a docking input file for LeDock with refined protein atoms within 0.4 nm of any atom of the ligand | Free | http://www.lephar.com/download.htm |
| 20 | DockoMatic | 5594 | 4.0570 | A software that docks secondary ligands, used to assist inverse virtual screening | Free | https://doi.org/10.1186/1756-0500-3-289 |
| 21 | SYBYL_ChemScore SYBYL_D-Score SYBYL_F-Score SYBYL_G-Score | 5486 | 4.0485 | A conformational sampling and scoring function | Subscription | https://doi.org/10.1021/jm0203783 |
| 22 | ZDOCK ZDOCKpro | 5415 | 4.0429 | A new scoring function for the initial stage of unbound docking | Subscription | http://zdock.umassmed.edu/ |
| 23 | AADS | 5087 | 4.0157 | An automated active site identification, docking, and scoring protocol | Free | http://www.scfbio-iitd.res.in/dock/ActiveSite_new.jsp |
| 24 | Surflex Dock | 4896 | 3.9991 | An automatic and flexible molecular docking algorithm for rapid in silico drug-screening applications | Subscription | https://doi.org/10.1007/s10822-007-9114-2 |
| 25 | PyMOL PyMOL 1.4.1 PyMOL 2.1.1 PyMOL 2.4 | 4805 | 3.9910 | An open-source, user-sponsored, molecular visualization system | Subscription | http://www.pymol.org |
| 26 | FlipDock | 4614 | 3.9733 | Allows the automated docking of flexible ligand molecules into active sites of flexible receptor molecules | Free | http://flipdock.scripps.edu/ |
| 27 | SymmDock | 4545 | 3.9668 | A flexible induced-fit backbone refinement in molecular docking | Free | http://bioinfo3d.cs.tau.ac.il/FiberDock/php.php |
| 28 | ClusPro | 4360 | 3.9487 | A widely used tool for protein–protein docking | Free | http://nrc.bu.edu/cluster |
| 29 | Surflex | 4180 | 3.9304 | A robust screening tool | Subscription | https://pubmed.ncbi.nlm.nih.gov/12570372/ |
| 30 | ConsDock | 4001 | 3.9114 | A pose within 2 Ao RMSD of the X-ray structure can be performed with this software | Subscription | https://doi.org/10.1002/prot.10119 |
| NDs | Molecular Docking Targets | Molecule | Software | Assay Type |
|---|---|---|---|---|
| Alzheimer’s disease | Acetylcholinesterase, Beta-secretase enzymes, Muscarinic and nicotinic ACh receptors, N-methyl-D-aspartate receptor, Tau proteins | 1-benzy-l1,2,3,4-tetrahydro- b-carboline), 3-substituted-1H-indoles, 6-triazolyl amidine derivatives [40] | ICM | cell-based assay [40] |
| Chloropyridonepezil [41] | Autodock Vina | In vitro blood–brain barrier model [42] | ||
| Flavone, 5-hydroxyflavone, 7-hydroxyflavone, chrysin, apigenin, kaempferol, fisetin, and quercetin [43] | AutoDock | Mice and rats models [44,45] | ||
| Ifenprodil [46] | Schrödinger Suite | Primary cultures from chicken embryo forebrain (E10) [46] | ||
| Memantine [47,48] | Glide | Human clinical trial [49] | ||
| Morin [50] | Glide | In APPswe/PS1dE9 mice [51] | ||
| Pyridopyrimidine derivatives [52] | Auto grid and auto dock | In vitro enzyme inhibitory model [53] | ||
| Pyridonepezil [54] | Autodock Vina | In vitro blood–brain barrier model [42] | ||
| Piperazine derivatives [55] | PASS software | Tested on AChE in vitro by using Ellman’s method [56] | ||
| Rutin [57] | AutoDock and Autodock Vina | Doxorubicin (DOX)-treated neuroblastoma cells (IMR32) and doxorubic-induced cognitive dysfunction in Wistar rats [58] | ||
| Parkinson’s disease | Dopamine receptors, expression and mitochondrial localization, Mutant LRRK2, Mutated, PINK1, PARK2, DJ1 SNCA Motif | LRRK2 kinase inhibitors (9-methyl-N-phenylpurine-2,8-diamine, N-phenylquinazolin-4-amine, and 1,3-dihydroindol-2-one) [59] | MOE | Both in vitro and in vivo studies were established [60] |
| Amyotrophic lateral sclerosis | Mutant SODI, SODI oligomerization, CASP-3, CASP-8, TDP-43, p38 MAPK Nav1.6 sodium channel | Angiogenin [61] | AmberTools20 | HeLa cells (Nuclear translocation assay) [61]) |
| Hesperidin and THSG [62]) | (Molecular Dynamics (MD) Simulation | High affinity to mutant SOD1 [62] | ||
| Riluzole [63] | PROCHECK program | FDA-approved drug for ALS [64] | ||
| Huntington’s disease | FIP-2 Specificity protein, 1HTT Interacting proteins Mutant HTT, Infant Testing Nuclear receptor corepressor, Postsynaptic density-95 | T1–11 (synthesized in a high yield by the substitution reaction) [65] | AutoDockTools | PC12 cells [65] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salman, M.M.; Al-Obaidi, Z.; Kitchen, P.; Loreto, A.; Bill, R.M.; Wade-Martins, R. Advances in Applying Computer-Aided Drug Design for Neurodegenerative Diseases. Int. J. Mol. Sci. 2021, 22, 4688. https://doi.org/10.3390/ijms22094688
Salman MM, Al-Obaidi Z, Kitchen P, Loreto A, Bill RM, Wade-Martins R. Advances in Applying Computer-Aided Drug Design for Neurodegenerative Diseases. International Journal of Molecular Sciences. 2021; 22(9):4688. https://doi.org/10.3390/ijms22094688
Chicago/Turabian StyleSalman, Mootaz M., Zaid Al-Obaidi, Philip Kitchen, Andrea Loreto, Roslyn M. Bill, and Richard Wade-Martins. 2021. "Advances in Applying Computer-Aided Drug Design for Neurodegenerative Diseases" International Journal of Molecular Sciences 22, no. 9: 4688. https://doi.org/10.3390/ijms22094688
APA StyleSalman, M. M., Al-Obaidi, Z., Kitchen, P., Loreto, A., Bill, R. M., & Wade-Martins, R. (2021). Advances in Applying Computer-Aided Drug Design for Neurodegenerative Diseases. International Journal of Molecular Sciences, 22(9), 4688. https://doi.org/10.3390/ijms22094688










