Structure and Expression Analysis of Sucrose Phosphate Synthase, Sucrose Synthase and Invertase Gene Families in Solanum lycopersicum
Abstract
1. Introduction
2. Results
2.1. Identification of SPS, SUS and INV Genes in Tomato Genome
2.2. Phylogenetic Analysis of the SPS, SUS and INV Families
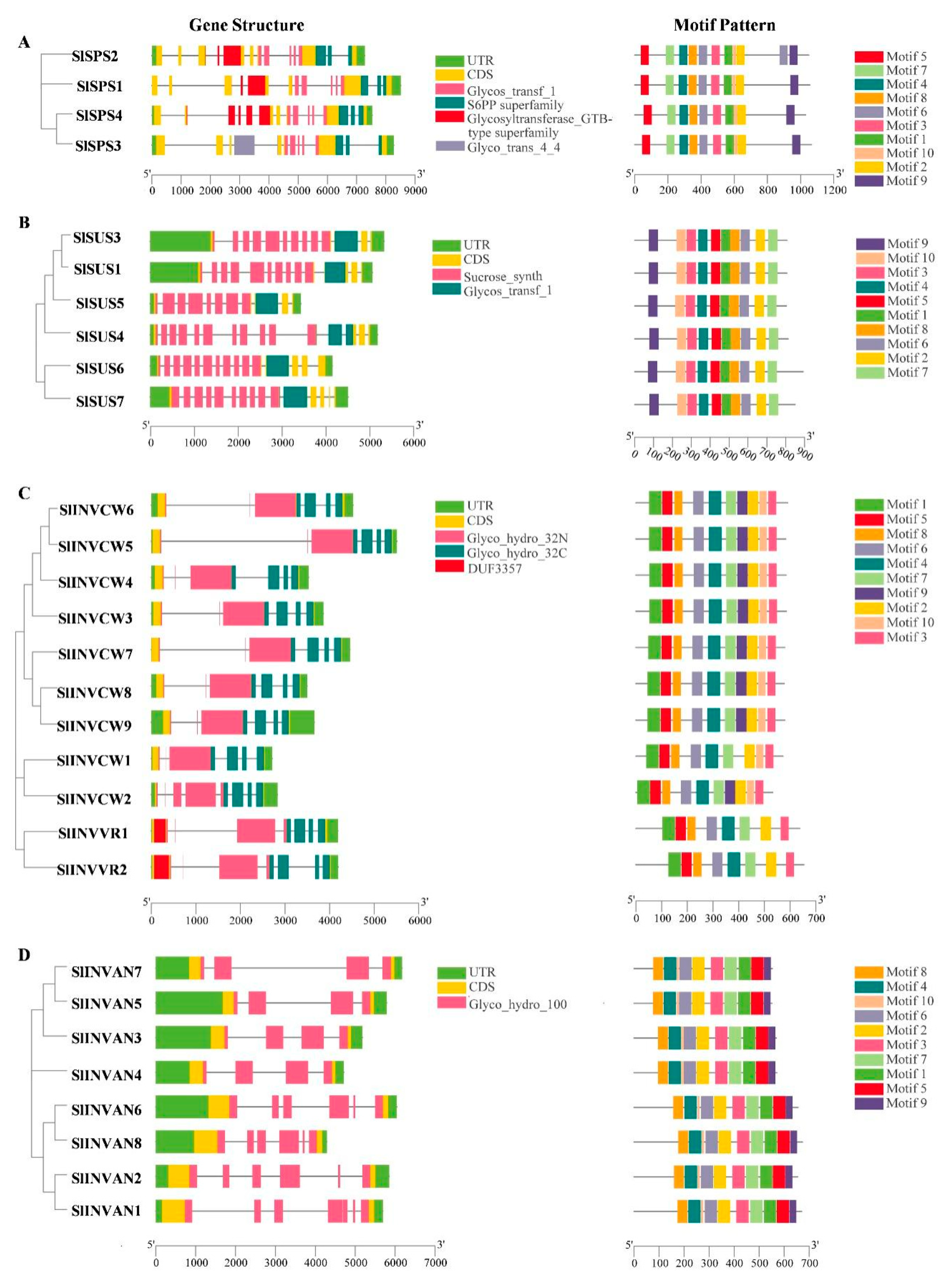
2.3. Gene Structure and Conserved Protein Motif Analysis of SPS, SUS and INV Gene Families
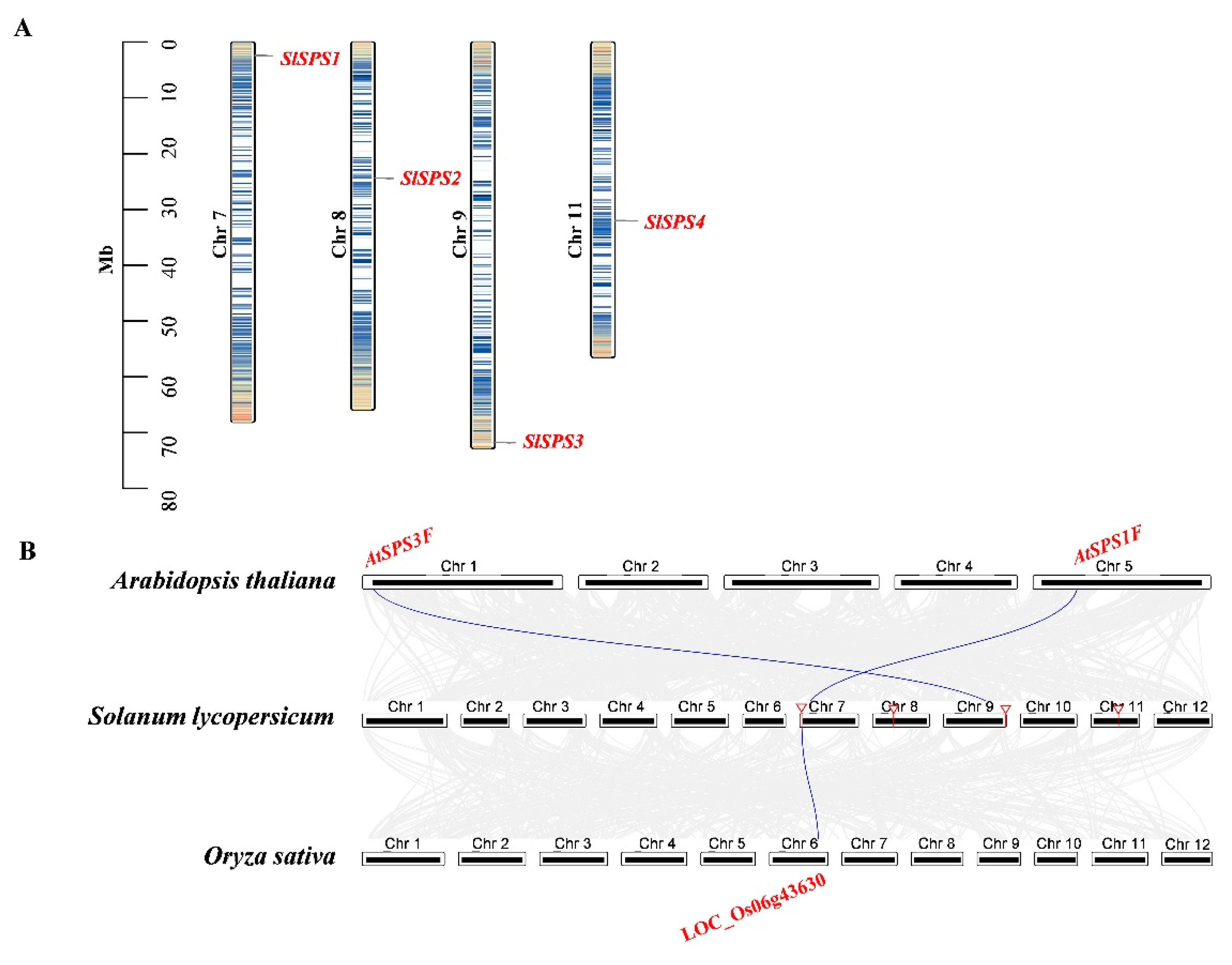
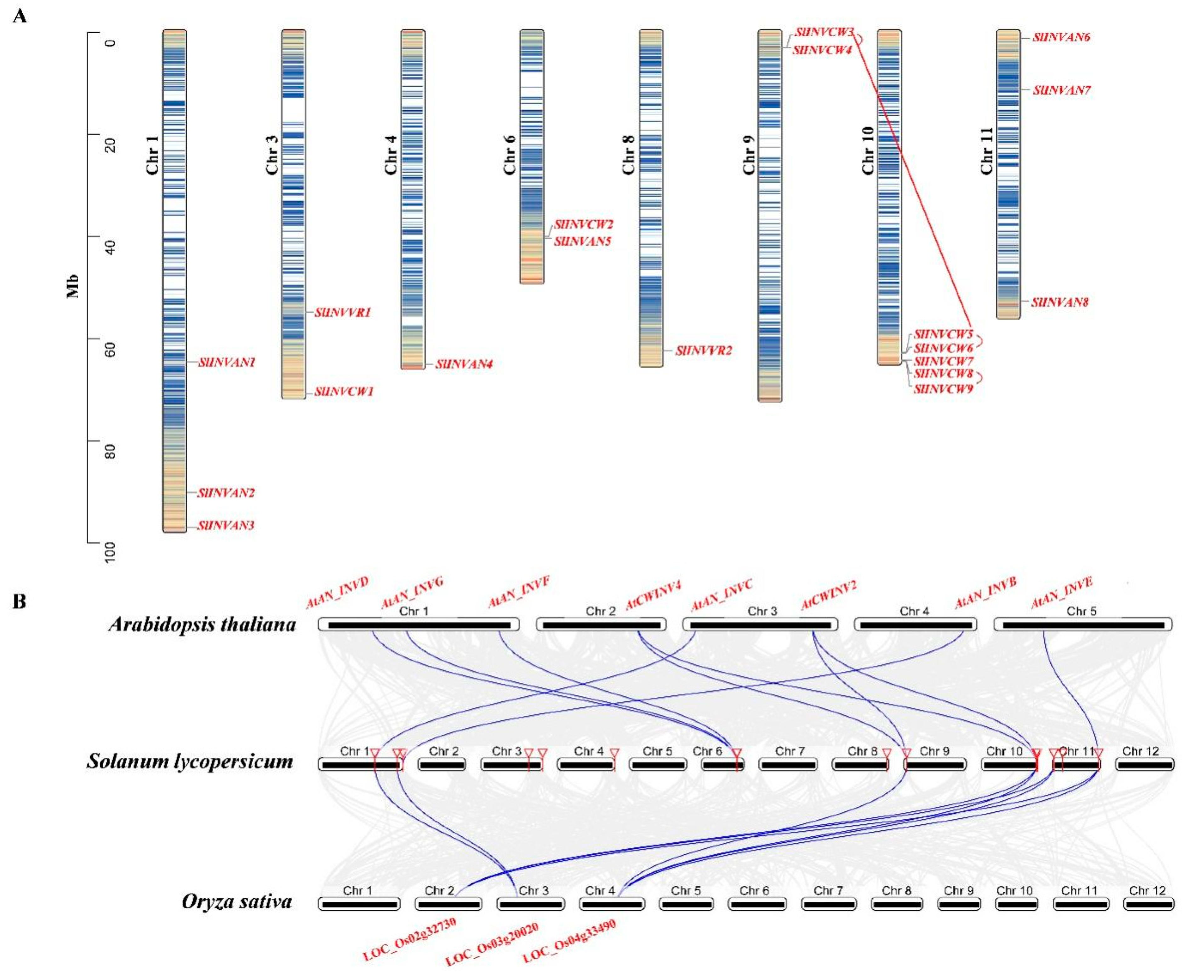
2.4. Chromosome Distribution, Synteny Analysis of SPS, SUS and INV Genes
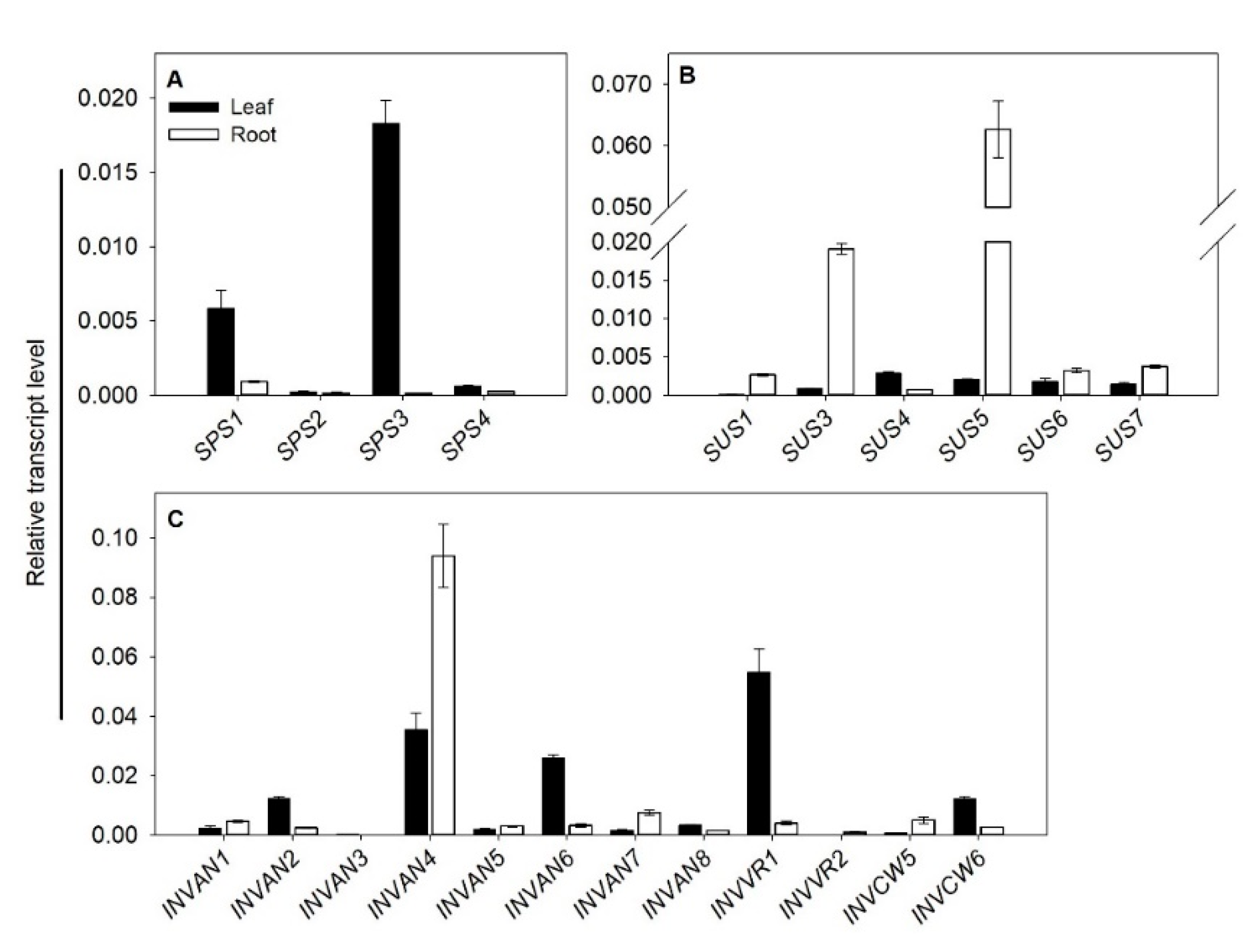
2.5. Tissue Expression Profile of SUS, INV and SPS in Tomato
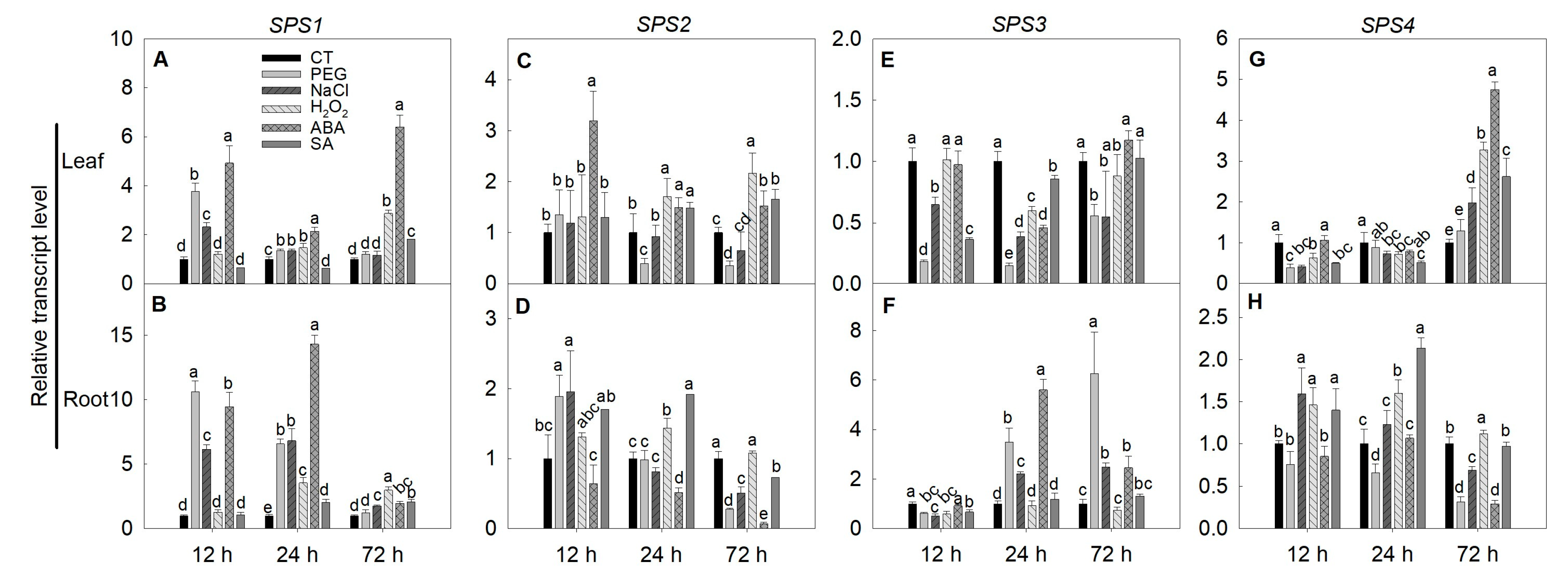
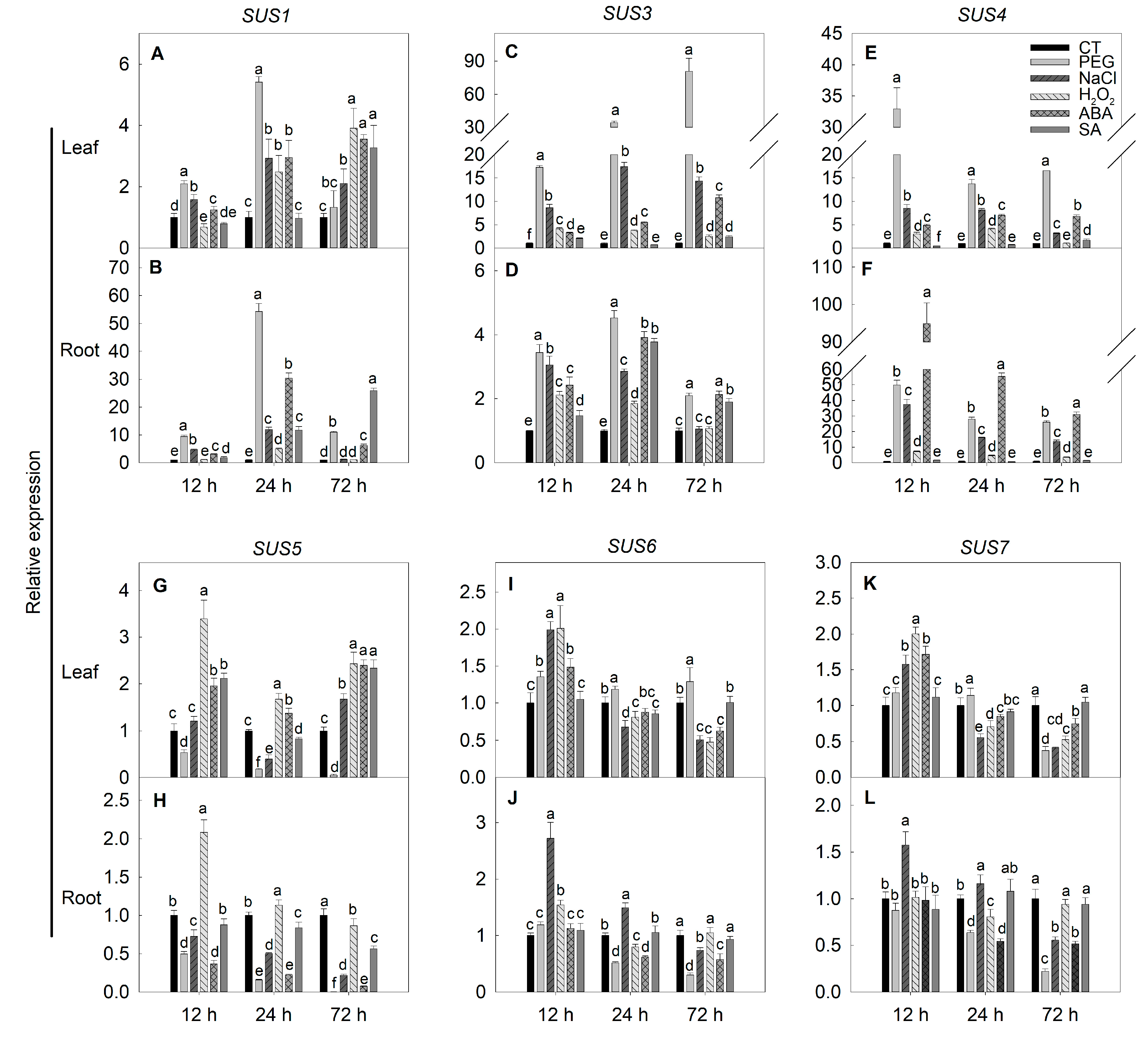
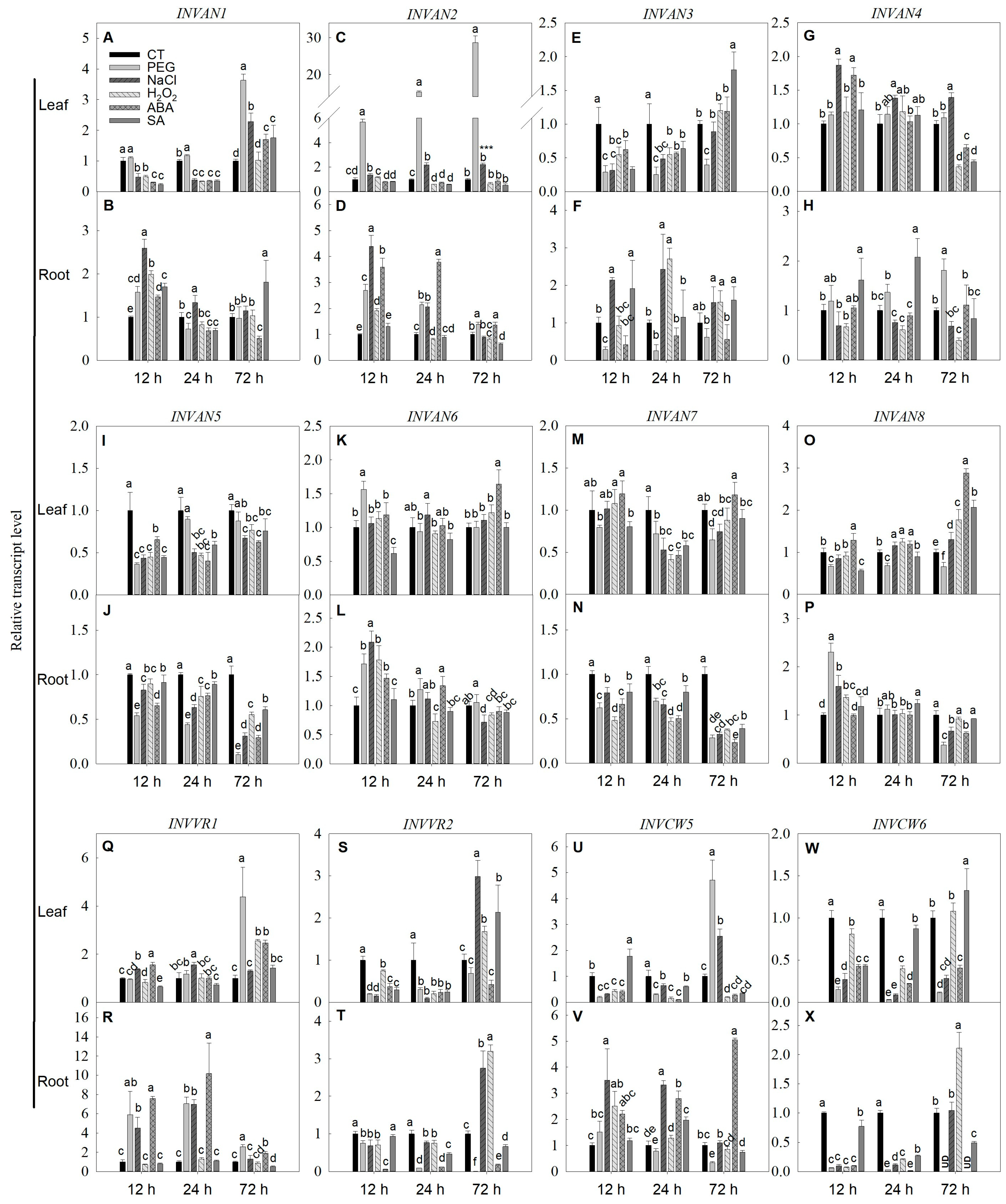
2.6. Expression of SPS, SUS and INV Genes under Simulated Stress Conditions in Tomato
3. Discussion
3.1. Evolution and Structure of SPS Genes
3.2. Evolution and Structure of SUS Genes
3.3. Evolution, Structure and Classification of INV Genes
3.4. Tissue Expression Pattern of SPS, SUS and INV Genes
3.5. Expressions of SPS, SUS and INV Genes under Stresses
4. Materials and Methods
4.1. Identification of SPS, SUS and INV Genes in Tomato
4.2. Phylogenetic Relationship, Gene Structure and Protein Motif Analysis
4.3. Chromosome Distribution, Gene Duplication and Syntenic Relationship Analysis
4.4. Plant Preparation and Chemical Treatment
4.5. RNA Extraction and Quantitative RT-PCR Analysis
4.6. Tissue Expression Characteristics of SPSs, SUSs and INVs in Tomato
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Stein, O.; Granot, D. An overview of sucrose synthases in plants. Front. Plant. Sci. 2019, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Guo, Q.; Chai, S.; Cheng, Y.; Ruan, M.; Ye, Q.; Wang, R.; Yao, Z.; Zhou, G.; Li, Z.; et al. Evolutionary conservation and expression patterns of neutral/alkaline invertases in solanum. Biomolecules 2019, 9, 763. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, J.; Li, C.; Zhang, Z.; Ma, F.; Li, M. Response of sugar metabolism in apple leaves subjected to short-term drought stress. Plant Physiol. Biochem. 2019, 141, 164–171. [Google Scholar] [CrossRef]
- Ruan, Y.-L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Ann. Rev. Plant. Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Wu, L.; Yang, Y.; Zhou, G.; Ruan, Y.-L. Evolution of sucrose metabolism: The dichotomy of invertases and beyond. Trends Plant. Sci. 2017, 23, 163–177. [Google Scholar] [CrossRef]
- Langenkämper, G.; Fung, R.W.M.; Newcomb, R.D.; Atkinson, R.G.; Gardner, R.C.; MacRae, E.A. Sucrose phosphate synthase genes in plants belong to three different families. J. Mol. Evol. 2002, 54, 322–332. [Google Scholar] [CrossRef]
- Okamura, M.; Aoki, N.; Hirose, T.; Yonekura, M.; Ohto, C.; Ohsugi, R. Tissue specificity and diurnal change in gene expression of the sucrose phosphate synthase gene family in rice. Plant Sci. 2011, 181, 159–166. [Google Scholar] [CrossRef]
- Castleden, C.K.; Aoki, N.; Gillespie, V.J.; Macrae, E.A.; Quick, W.P.; Buchner, P.H.; Foyer, C.H.; Furbank, R.T.; Lunn, J.E. Evolution and function of the sucrose-phosphate synthase gene families in wheat and other grasses. Plant Physiol. 2004, 135, 1753–1764. [Google Scholar] [CrossRef]
- Li, M.; Feng, F.; Cheng, L. Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS ONE 2012, 7, 3. [Google Scholar]
- Ma, P.; Zhang, X.; Chen, L.; Zhao, Q.; Zhang, Q.; Hua, X.; Wang, Z.; Tang, H.; Yu, Q.; Zhang, M.; et al. Comparative analysis of sucrose phosphate synthase (SPS) gene family between Saccharum officinarum and Saccharum spontaneum. BMC Plant. Biol. 2020, 20, 1–15. [Google Scholar] [CrossRef]
- Xu, X.; Yang, Y.; Liu, C.; Sun, Y.; Zhang, T.; Hou, M.; Huang, S.; Yuan, H. The evolutionary history of the sucrose synthase gene family in higher plants. BMC Plant. Biol. 2019, 19, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wei, P.; Wu, M.; Xu, Y.; Li, F.; Luo, Z.; Zhang, J.; Chen, A.; Xie, X.; Cao, P.; et al. Analysis of the sucrose synthase gene family in tobacco: Structure, phylogeny, and expression patterns. Planta 2015, 242, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.; Cao, Y.; Cheng, X.; Meng, D.; Chen, Y.; Shakoor, A.; Gao, J.; Cai, Y. The sucrose synthase gene family in chinese pear (Pyrus bretschneideri rehd.): Structure, expression, and evolution. Molecules 2018, 23, 1144. [Google Scholar] [CrossRef]
- Ji, X.; Ende, W.V.; Laere, A.V.; Cheng, S.; Bennett, J. Structure, evolution, and expression of the two invertase gene families of rice. J. Mol. Evol. 2005, 60, 615–634. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zheng, Y.; Ding, S.; Zhang, Q.; Chen, Y.; Zhang, J. Molecular cloning, structure, phylogeny and expression analysis of the invertase gene family in sugarcane. BMC Plant. Biol. 2017, 17, 1. [Google Scholar] [CrossRef]
- Su, T.; Han, M.; Min, J.; Chen, P.; Mao, Y.; Huang, Q.; Tong, Q.; Liu, Q.; Fang, Y. Genome-wide survey of invertase encoding genes and functional characterization of an extracellular fungal pathogen-responsive invertase in Glycine max. Int. J. Mol. Sci. 2018, 19, 2395. [Google Scholar] [CrossRef]
- Qin, G.; Zhu, Z.; Wang, W.; Cai, J.; Chen, Y.; Li, L.; Tian, S. A tomato vacuolar invertase inhibitor mediates sucrose metabolism and influences fruit ripening. Plant Physiol. 2016, 172, 1596–1611. [Google Scholar] [CrossRef]
- Bahaji, A.; Baroja-Fernández, E.; Ricarte-Bermejo, A.; Sánchez-López, Á.M.; Muñoz, F.J.; Romero, J.M.; Ruiz, M.T.; Baslam, M.; Almagro, G.; Sesma, M.T.; et al. Characterization of multiple sps knockout mutants reveals redundant functions of the four arabidopsis sucrose phosphate synthase isoforms in plant viability, and strongly indicates that enhanced respiration and accelerated starch turnover can alleviate the blockage of sucrose biosynthesis. Plant. Sci. 2015, 238, 135–147. [Google Scholar]
- Anur, R.M.; Mufithah, N.; Sawitri, W.D.; Sakakibara, H.; Sugiharto, B. Overexpression of sucrose phosphate synthase enhanced sucrose content and biomass production in transgenic sugarcane. Plants (Basel) 2020, 9, 200. [Google Scholar] [CrossRef]
- Haigler, C.H.; Singh, B.; Zhang, D.; Hwang, S.; Wu, C.; Cai, W.X.; Hozain, M.; Kang, W.; Kiedaisch, B.; Strauss, R.E.; et al. Transgenic cotton over-producing spinach sucrose phosphate synthase showed enhanced leaf sucrose synthesis and improved fiber quality under controlled environmental conditions. Plant. Mol. Biol. 2007, 63, 815–832. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-Y.; Canam, T.; Kang, K.-Y.; Ellis, D.D.; Mansfield, S.D. Over-expression of an Arabidopsis family A sucrose phosphate synthase (SPS) gene alters plant growth and fibre development. Transgenic Res. 2008, 17, 181–192. [Google Scholar] [CrossRef]
- Park, J.-Y.; Canam, T.; Kang, K.-Y.; Unda, F.; Mansfield, S.D. Sucrose phosphate synthase expression influences poplar phenology. Tree Physiol. 2009, 29, 937–946. [Google Scholar] [CrossRef]
- Baxter, C.J.; Foyer, C.H.; Turner, J.; Rolfe, S.A.; Quick, W.P. Elevated sucrose-phosphate synthase activity in transgenic tobacco sustains photosynthesis in older leaves and alters development. J. Exp. Bot. 2003, 54, 1813–1820. [Google Scholar] [CrossRef]
- Hirose, T.; Hashida, Y.; Aoki, N.; Okamura, M.; Yonekura, M.; Ohto, C.; Terao, T.; Ohsugi, R. Analysis of gene-disruption mutants of a sucrose phosphate synthase gene in rice, OsSPS1, shows the importance of sucrose synthesis in pollen germination. Plant. Sci. 2014, 225, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Ma, L.; Zhao, C.; Hao, H.; Gong, B.; Yu, X.; Wang, X. Antisense repression of sucrose phosphate synthase in transgenic muskmelon alters plant growth and fruit development. Biochem. Biophys. Res. Commun. 2010, 393, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Barratt, D.H.P.; Derbyshire, P.; Findlay, K.; Pike, M.; Wellner, N.; Lunn, J.; Feil, R.; Simpson, C.; Maule, A.J.; Smith, A.M. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA 2009, 106, 13124–13129. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Qu, Z.; Zhang, L.; Zhao, S.; Bi, Z.; Ji, X.; Wang, X.; Wei, H. Overexpression of poplar xylem sucrose synthase in tobacco leads to a thickened cell wall and increased height. PLoS ONE 2015, 10, e0120669. [Google Scholar] [CrossRef] [PubMed]
- Goren, S.; Lugassi, N.; Stein, O.; Yeselson, Y.; Schaffer, A.A.; David-Schwartz, R.; Granot, D. Suppression of sucrose synthase affects auxin signaling and leaf morphology in tomato. PLoS ONE 2017, 12, e0182334. [Google Scholar] [CrossRef] [PubMed]
- Gordon, A.J.; Minchin, F.R.; James, C.L.; Komina, O. Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol. 1999, 120, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Hua, L.-N.; Liu, X.-F.; Li, Y.-Z.; Shen, Y.-Y.; Guo, J.-X. Sucrose synthase FaSS1 plays an important role in the regulation of strawberry fruit ripening. Plant. Growth Regul. 2017, 81, 175–181. [Google Scholar] [CrossRef]
- Martín, M.L.; Lechner, L.; Zabaleta, E.J.; Salerno, G.L. A mitochondrial alkaline/neutral invertase isoform (A/N-InvC) functions in developmental energy-demanding processes in Arabidopsis. Planta 2013, 237, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Zhang, B.; Mao, C.; Li, J.; Wu, Y.; Wu, P.; Wu, Z. OsCYT-INV1 for alkaline/neutral invertase is involved in root cell development and reproductivity in rice (Oryza sativa L.). Planta 2008, 228, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Zanor, M.I.; Osorio, S.; Nunes-Nesi, A.; Carrari, F.; Lohse, M.; Usadel, B.; Kühn, C.; Bleiss, W.; Giavalisco, P.; Willmitzer, L.; et al. RNA interference of lin5 in tomato confirms its role in controlling brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiol. 2009, 150, 1204–1218. [Google Scholar] [CrossRef]
- Takehara, K.; Murata, K.; Yamaguchi, T.; Yamaguchi, K.; Chaya, G.; Kido, S.; Iwasaki, Y.; Ogiwara, H.; Ebitani, T.; Miura, K. Thermo-responsive allele of sucrose synthase 3 (Sus3) provides high-temperature tolerance during the ripening stage in rice (Oryza sativa L.). Breed. Sci. 2018, 68, 336–342. [Google Scholar] [CrossRef]
- Wang, H.; Sui, X.; Guo, J.; Wang, Z.; Cheng, J.; Ma, S.; Li, X.; Zhang, Z. Antisense suppression of cucumber (Cucumis sativus L.) sucrose synthase 3 (CsSUS3) reduces hypoxic stress tolerance. Plant. Cell Environ. 2014, 37, 795–810. [Google Scholar] [CrossRef]
- Qian, W.; Xiao, B.; Wang, L.; Hao, X.; Yue, C.; Cao, H.; Wang, Y.; Li, N.; Yu, Y.; Zeng, J.; et al. CsINV5, a tea vacuolar invertase gene enhances cold tolerance in transgenic Arabidopsis. BMC Plant. Biol. 2018, 18, 228. [Google Scholar] [CrossRef]
- Liu, Y.-H.; Offler, C.E.; Ruan, Y.-L. Cell wall invertase promotes fruit set under heat stress by suppressing ros-independent cell death. Plant Physiol. 2016, 172, 163–180. [Google Scholar] [CrossRef]
- Xu, X.-x.; Hu, Q.; Yang, W.-n.; Jin, Y. The roles of cell wall invertase inhibitor in regulating chilling tolerance in tomato. BMC Plant. Biol. 2017, 17, 195. [Google Scholar] [CrossRef]
- Wang, F.; Smith, A.G.; Brenner, M.L. Isolation and sequencing of tomato fruit sucrose synthase cDNA. Plant Physiol. 1993, 103, 1463–1464. [Google Scholar] [CrossRef] [PubMed]
- D’Aoust, M.-A.; Yelle, S.; Nguyen-Quoc, B. Antisense inhibition of tomato fruit sucrose synthase decreases fruit setting and the sucrose unloading capacity of young fruit. Plant. Cell 1999, 11, 2407–2418. [Google Scholar] [CrossRef]
- Chengappa, S.; Loader, N.; Shields, R. Cloning, expression, and mapping of a second tomato sucrose synthase gene, Sus3. Plant Physiol. 1998, 118, 1533. [Google Scholar]
- Goren, S.; Huber, S.C.; Granot, D. Comparison of a novel tomato sucrose synthase, SlSUS4, with previously described SlSUS isoforms reveals distinct sequence features and differential expression patterns in association with stem maturation. Planta 2011, 233, 1011–1023. [Google Scholar] [CrossRef]
- Godt, D.E.; Roitsch, T. Regulation and tissue-specific distribution of mRNAs for three extracellular invertase isoenzymes of tomato suggests an important function in establishing and maintaining sink metabolism. Plant Physiol. 1997, 115, 273–282. [Google Scholar] [CrossRef]
- Klann, E.; Yelle, S.; Bennett, A.B. Tomato fruit acid invertase complementary dna nucleotide and deduced amino acid sequences. Plant Physiol. 1992, 99, 351–353. [Google Scholar] [CrossRef]
- Leskow, C.C.; Conte, M.; del Pozo, T.; Bermúdez, L.; Lira, B.S.; Gramegna, G.; Baroli, I.; Burgos, E.; Zavallo, D.; Kamenetzky, L.; et al. The cytosolic invertase NI6 affects vegetative growth, flowering, fruit set and yield in tomato. J. Exp. Bot. 2020, 72, 2525–2543. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E.; Ashton, A.R.; Hatch, M.D.; Heldt, H.W. Purification, molecular cloning, and sequence analysis of sucrose-6F-phosphate phosphohydrolase from plants. Proc. Natl. Acad. Sci. USA 2000, 97, 12914–12919. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E.; MacRae, E. New complexities in the synthesis of sucrose. Curr. Opin. Plant. Biol. 2003, 6, 208–214. [Google Scholar] [CrossRef]
- Lammens, W.; Roy, K.L.; Schroeven, L.; Laere, A.V.; Rabijns, A.; Ende, W.V.d.E. Structural insights into glycoside hydrolase family 32 and 68 enzymes: Functional implications. J. Exp. Bot. 2009, 60, 727–740. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant. Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhao, J.; Hu, B.; Li, J.; Qin, Y.; Chen, L.; Qin, Y.; Hu, G. Identification and expression profile analysis of the sucrose phosphate synthase gene family in Litchi chinensis Sonn. PeerJ 2018, 6, e4379. [Google Scholar] [CrossRef]
- Mitchell-Olds, T.; Clauss, M.J. Plant evolutionary genomics. Curr. Opin.Plant. Biol. 2002, 5, 74–79. [Google Scholar] [CrossRef]
- Jiang, S.-Y.; Chi, Y.-H.; Wang, J.-Z.; Zhou, J.-X.; Cheng, Y.-S.; Zhang, B.-L.; Ma, A.; Vanitha, J.; Ramachandran, S. Sucrose metabolism gene families and their biological functions. Sci. Rep. 2015, 5, 17583. [Google Scholar] [CrossRef] [PubMed]
- Bocock, P.N.; Morse, A.M.; Dervinis, C.; Davis, J.M. Evolution and diversity of invertase genes in Populus trichocarpa. Planta 2008, 227, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Sturm, A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999, 121, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.-B.; Yao, Y.; He, H.; Qin, Y.-L.; Liu, Z.-J.; Liu, W.-X.; Qi, Z.-Q.; Yang, L.-J.; Cao, Z.-M.; Yang, Y. Genome-wide identification, expression, and functional analysis of the alkaline/neutral invertase gene family in pepper. Int. J. Mol. Sci. 2018, 19, 224. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.G.; Kodama, R.; Wang, H.; Ichii, M.; Taketa, S.; Yoshida, H. Analysis of the rice SHORT-ROOT5 gene revealed functional diversification of plant neutral/alkaline invertase family. Plant. Sci. 2009, 176, 627–634. [Google Scholar] [CrossRef]
- Palmer, W.M.; Ru, L.; Jin, Y.; Patrick, J.W.; Ruan, Y.-L. Tomato ovary-to-fruit transition is characterized by a spatial shift of mRNAs for cell wall invertase and its inhibitor with the encoded proteins localized to sieve elements. Mol. Plant. 2015, 8, 315–328. [Google Scholar] [CrossRef]
- Proels, R.K.; González, M.-C.; Roitsch, T. Gibberellin-dependent induction of tomato extracellular invertase Lin7 is required for pollen development. Funct. Plant. Biol. 2006, 33, 547–554. [Google Scholar] [CrossRef]
- Nguyen-Quoc, B.; Foyer, C.H. A role for ‘futile cycles’ involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. J. Exp. Bot. 2001, 52, 881–889. [Google Scholar] [CrossRef]
- Hu, T.; Hu, L.; Zhang, X.; Zhang, P.; Zhao, Z.; Fu, J. Differential responses of CO2 assimilation, carbohydrate allocation and gene expression to nacl stress in perennial ryegrass with different salt tolerance. PLoS ONE 2013, 8, e66090. [Google Scholar]
- Solís-Guzmán, M.G.; Argüello-Astorga, G.; López-Bucio, J.; Ruiz-Herrera, L.F.; López-Meza, J.E.; Sánchez-Calderón, L.; Carreón-Abud, Y.; Martínez-Trujillo, M. Arabidopsis thaliana sucrose phosphate synthase (SPS) genes are expressed differentially in organs and tissues, and their transcription is regulated by osmotic stress. Gene Expr. Patterns 2017, 25, 92–101. [Google Scholar] [CrossRef]
- Dahro, B.; Wang, F.; Peng, T.; Liu, J.-H. PtrA/NINV, an alkaline/neutral invertase gene of Poncirus trifoliata, confers enhanced tolerance to multiple abiotic stresses by modulating ROS levels and maintaining photosynthetic efficiency. BMC Plant. Biol. 2016, 16, 76. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Large-scale plant protein subcellular location prediction. J. Cell. Biochem. 2007, 100, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.G.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2000, 30, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Bodén, M.; Buske, F.A.; Frith, M.C.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.-f.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The water culture method for growing plants without soil. Calif. Agric. Exp. Stn. Circ. 1950, 347, 32. [Google Scholar]
- Expósito-Rodríguez, M.; Borges, A.A.; Borges-Pérez, A.; Pérez, J.A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant. Biol. 2008, 8, 131. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004, 136, 2621–2632. [Google Scholar] [CrossRef] [PubMed]



| Gene | Gene Structure | Protein Characteristics | Gene Name in the Literature | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Locus | Chr | Sequence Length (bp) | CDS Length (bp) | Exon No. | Intron No. | Length (Aa) | IP | MW (KDa) | Subcellular Localization | Phosphorylation Site No. | (References) |
| SPS1 | Solyc07g007790 | 7 | 8511 | 3165 | 13 | 12 | 1054 | 6.05 | 118.5 | PM | 24 | SPSA1 [17] |
| SPS2 | Solyc08g042000 | 8 | 7283 | 3147 | 13 | 12 | 1048 | 6.26 | 117.9 | PM | 21 | SPSA2 [17] |
| SPS3 | Solyc09g092130 | 9 | 8270 | 3195 | 12 | 11 | 1064 | 6.13 | 119.6 | PM | 15 | SPSB [17] |
| SPS4 | Solyc11g045110 | 11 | 7534 | 3090 | 14 | 13 | 1029 | 6.59 | 116.2 | PM | 21 | SPSC [17] |
| SUS1 | Solyc12g009300 | 12 | 5834 | 2418 | 13 | 12 | 805 | 5.94 | 92.5 | PM | 9 | SUS1 [39]; TOMMSSF [40] |
| SUS3 | Solyc07g042550 | 7 | 5663 | 2418 | 13 | 12 | 805 | 5.96 | 92.5 | PM | 13 | SUS3 [41] |
| SUS4 | Solyc09g098590 | 9 | 6464 | 2439 | 15 | 14 | 812 | 5.91 | 92.9 | PM | 18 | SUS4 [42] |
| SUS5 | Solyc07g042520 | 7 | 3624 | 2412 | 11 | 10 | 803 | 5.97 | 91.6 | PM | 13 | SS5 [17] |
| SUS6 | Solyc03g098290 | 3 | 4379 | 2676 | 15 | 14 | 891 | 5.87 | 100.7 | PM | 20 | SS6 [17] |
| SUS7 | Solyc02g081300 | 2 | 4757 | 2550 | 14 | 13 | 849 | 6.94 | 96.3 | PM | 14 | SS7 [17] |
| INVCW1 | Solyc03g121680 | 3 | 2716 | 1716 | 6 | 5 | 571 | 6.8 | 64.2 | CW | 13 | |
| INVCW2 | Solyc06g064620 | 6 | 2834 | 1596 | 8 | 7 | 531 | 4.96 | 59.7 | CW | 8 | |
| INVCW3 | Solyc09g010080 | 9 | 3863 | 1755 | 6 | 5 | 584 | 9.2 | 67.2 | CW * | 8 | LIN5 [43] |
| INVCW4 | Solyc09g010090 | 9 | 3530 | 1752 | 6 | 5 | 583 | 6.93 | 66.2 | CW | 14 | LIN7 [43] |
| INVCW5 | Solyc10g083290 | 10 | 5513 | 1749 | 6 | 5 | 582 | 9.23 | 65.9 | CW | 9 | LIN6 [43] |
| INVCW6 | Solyc10g083300 | 10 | 4528 | 1560 | 6 | 5 | 519 | 8.69 | 58.8 | CW | 8 | LIN8 [43] |
| INVCW7 | Solyc10g085360 | 10 | 4459 | 1737 | 6 | 5 | 578 | 8.7 | 65.9 | CW | 5 | |
| INVCW8 | Solyc10g085640 | 10 | 3498 | 1731 | 6 | 5 | 576 | 7.26 | 65.7 | CW | 5 | |
| INVCW9 | Solyc10g085650 | 10 | 3659 | 1737 | 6 | 5 | 578 | 6.39 | 66.6 | CW | 10 | |
| INVVR1 | Solyc03g083910 | 3 | 4187 | 1911 | 7 | 6 | 636 | 5.54 | 70.1 | Vac | 8 | VI [44] |
| INVVR2 | Solyc08g079080 | 8 | 4194 | 1959 | 7 | 6 | 652 | 6.21 | 72.8 | Vac | 9 | LIN9 [17] |
| INVAN1 | Solyc01g058010 | 1 | 5694 | 2010 | 7 | 6 | 669 | 5.84 | 76.0 | Mito | 10 | NI1 [17] |
| INVAN2 | Solyc01g100810 | 1 | 5852 | 1962 | 6 | 5 | 653 | 8.18 | 74.5 | Mito | 13 | NI2 [4,17] |
| INVAN3 | Solyc01g111100 | 1 | 5189 | 1707 | 4 | 3 | 568 | 6.45 | 64.9 | Cyt | 13 | CIN2 [4] |
| INVAN4 | Solyc04g081440 | 4 | 4713 | 1713 | 4 | 3 | 570 | 5.97 | 65.2 | Cytosol * | 13 | CIN3 [4]; NI6 [45] |
| INVAN5 | Solyc06g065210 | 6 | 5788 | 1656 | 4 | 3 | 551 | 6.16 | 62.8 | Cyt | 17 | NI3 [17]; CIN4 [4] |
| INVAN6 | Solyc11g007270 | 11 | 6087 | 1968 | 6 | 5 | 655 | 5.84 | 73.5 | Chl | 12 | CIN5 [4] |
| INVAN7 | Solyc11g020610 | 11 | 6175 | 1659 | 4 | 3 | 552 | 6.06 | 62.6 | Cyt | 16 | NI4 [17]; CIN6 [4] |
| INVAN8 | Solyc11g067050 | 11 | 4291 | 2019 | 6 | 5 | 672 | 6.89 | 76.2 | Chl | 8 | NI5 [17]; CIN7 [4] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, Y.; Yang, L.; Zhu, H.; Zhou, J.; Sun, H.; Gong, H. Structure and Expression Analysis of Sucrose Phosphate Synthase, Sucrose Synthase and Invertase Gene Families in Solanum lycopersicum. Int. J. Mol. Sci. 2021, 22, 4698. https://doi.org/10.3390/ijms22094698
Duan Y, Yang L, Zhu H, Zhou J, Sun H, Gong H. Structure and Expression Analysis of Sucrose Phosphate Synthase, Sucrose Synthase and Invertase Gene Families in Solanum lycopersicum. International Journal of Molecular Sciences. 2021; 22(9):4698. https://doi.org/10.3390/ijms22094698
Chicago/Turabian StyleDuan, Yaoke, Lan Yang, Haijia Zhu, Jie Zhou, Hao Sun, and Haijun Gong. 2021. "Structure and Expression Analysis of Sucrose Phosphate Synthase, Sucrose Synthase and Invertase Gene Families in Solanum lycopersicum" International Journal of Molecular Sciences 22, no. 9: 4698. https://doi.org/10.3390/ijms22094698
APA StyleDuan, Y., Yang, L., Zhu, H., Zhou, J., Sun, H., & Gong, H. (2021). Structure and Expression Analysis of Sucrose Phosphate Synthase, Sucrose Synthase and Invertase Gene Families in Solanum lycopersicum. International Journal of Molecular Sciences, 22(9), 4698. https://doi.org/10.3390/ijms22094698







