Copy Number Variation and Rearrangements Assessment in Cancer: Comparison of Droplet Digital PCR with the Current Approaches
Abstract
:1. Introduction
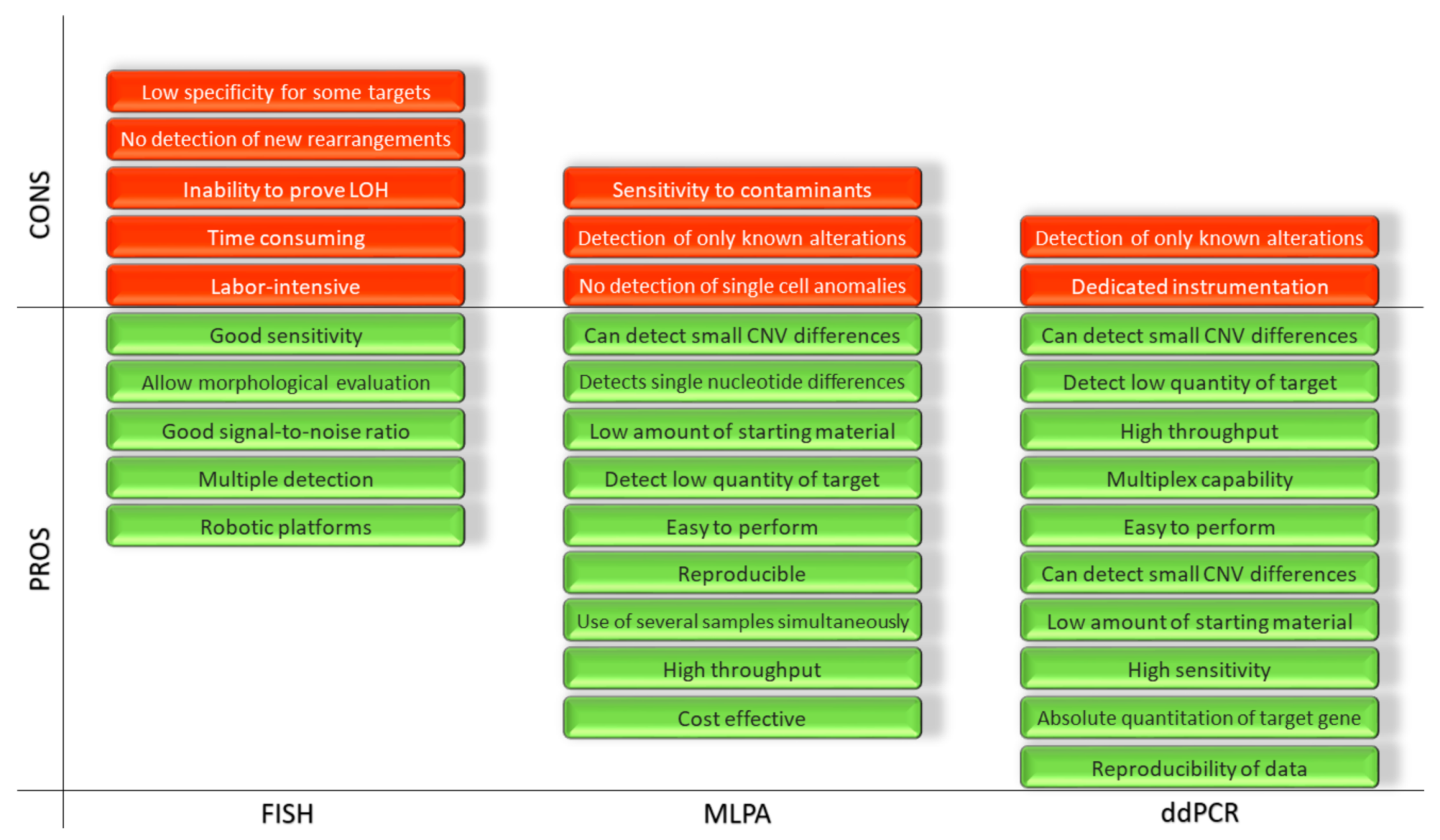
2. Fluorescence in Situ Hybridization (FISH)
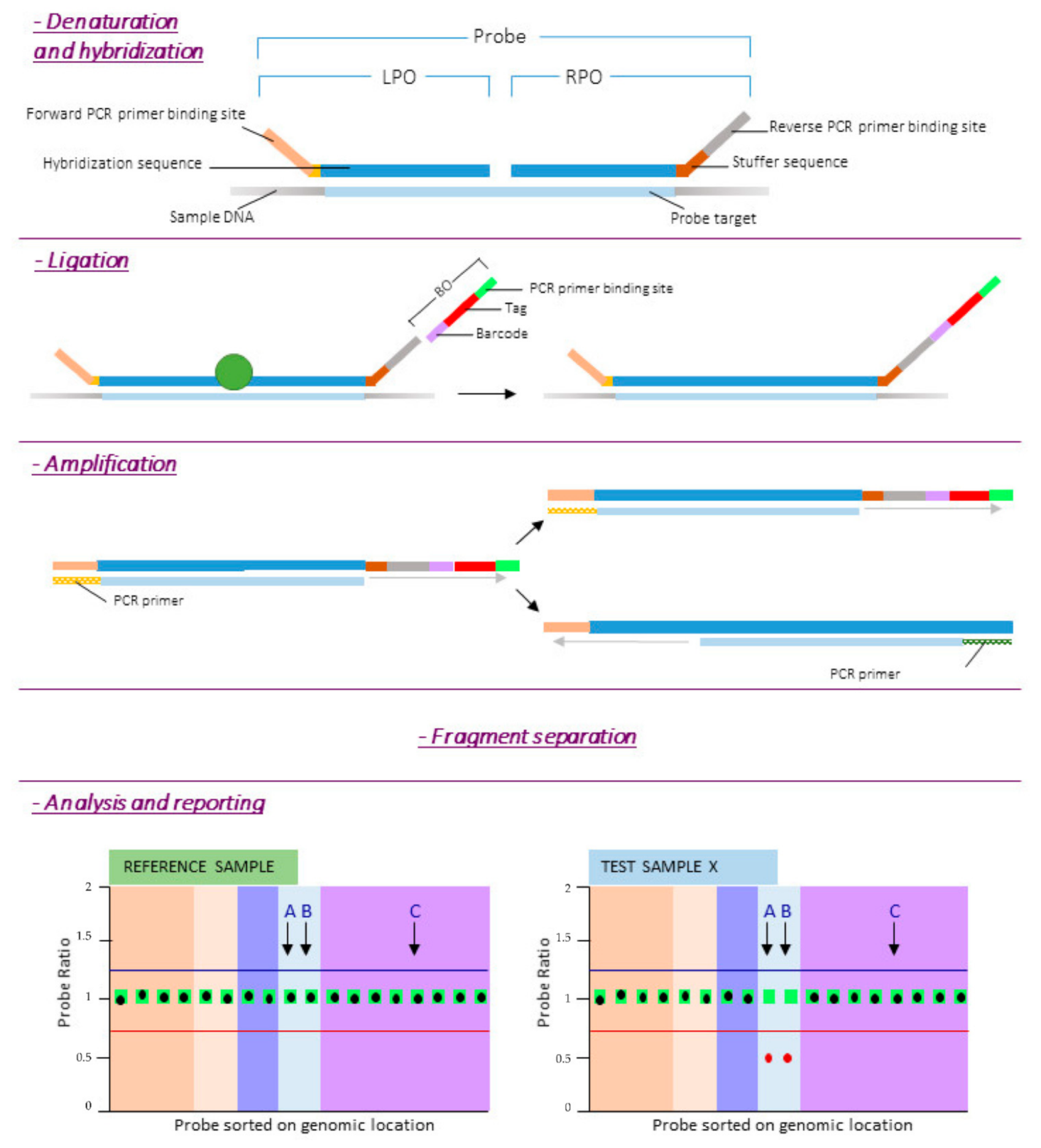
3. Multiplex Ligation-Dependent Probe Amplification (MLPA)
- -
- It is a cost effective way to check for rearrangements, duplications and deletions;
- -
- It can be applied to a large number of targets (high throughput);
- -
- It can be performed on a large number of samples simultaneously;
- -
- It is reproducible, easy to perform, and it is capable of detecting a low quantity of the target;
- -
- It requires only 50 ng of human DNA, can distinguish sequences differing by a single nucleotide and can detect small copy number differences.
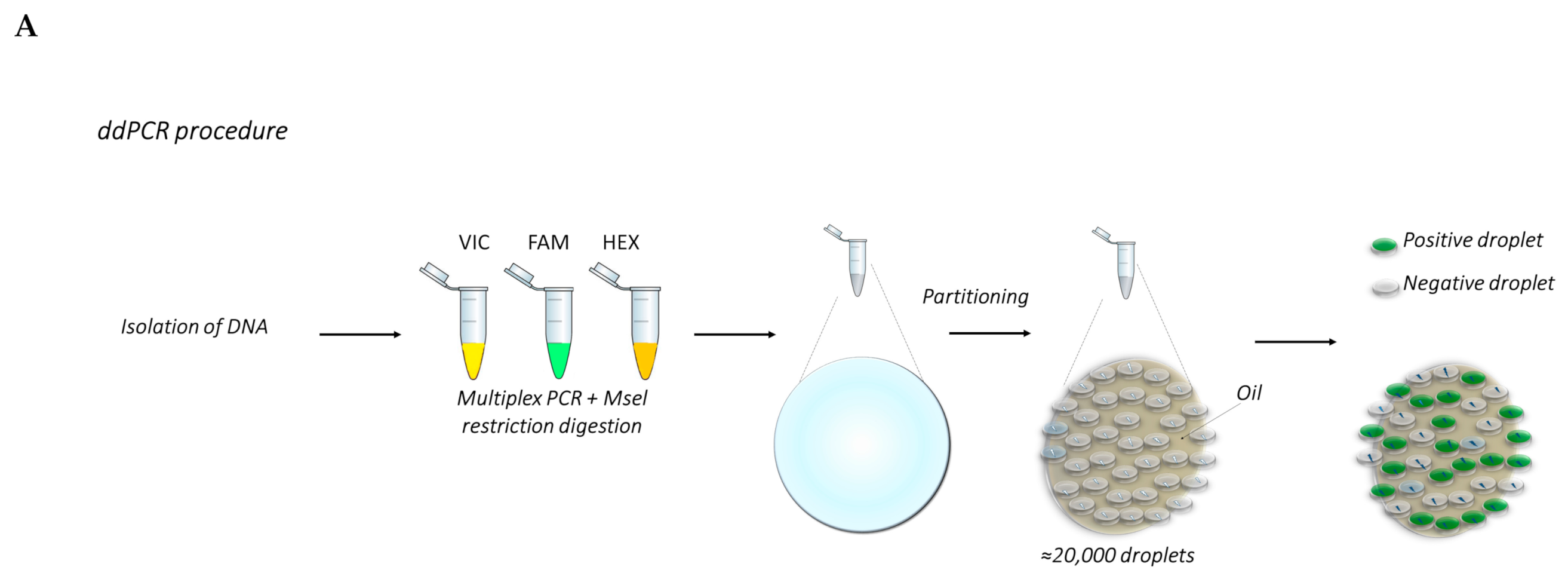
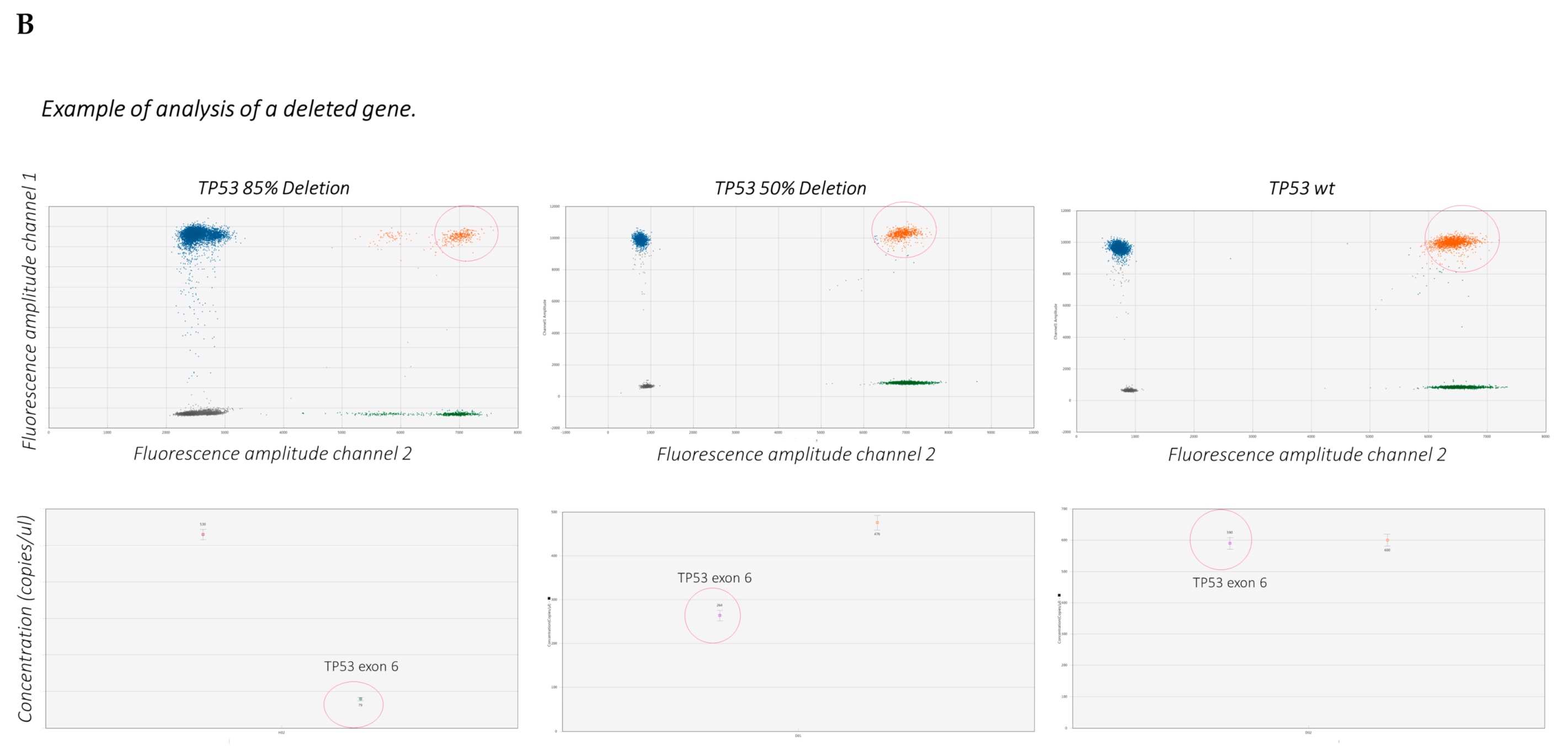
4. Droplet Digital PCR (ddPCR)
- -
- Reproducibility of the data, given by the absolute quantitation of the target genes;
- -
- Sensitivity of the quantitation, obtained through the amplification into an emulsion matrix;
- -
- Very low amount of starting material requested for any of the possible applications, due to the high sensitivity of the technique.
- -
- The few disadvantages known up to now are:
- -
- ddPCR requires special and dedicated instrumentation;
- -
- ddPCR detects only known mutations.
- -
- The procedure is represented in Figure 3.
5. Protocols and Graphical Representation of the Described Techniques
5.1. FISH Protocol
5.1.1. Slide Preparation
- Starting material is either formalin-fixed, paraffin-embedded tissue, needle aspirates or cell slides;
- Incubate with 200 µL RNase for 1 h at 37 °C
- Wash slides in 2× saline-sodium citate buffer (SSC) for 5 min, repeat.
- Rinse slides in 10 mM HCl.
- Incubate with 200 µL pepsin for 10 min at 37 °C.
- Rinse slides in deionized H2O.
- Wash slides in 2× SSC for 5 min, repeat.
- Stabilize slides in paraformaldehyde for 10 min.
- Wash slides in 2× SSC for 5 min, repeat.
- Dehydrate slides in an ethanol series: 70%, 80% and 95%; 2 min each.
- Air dry.
5.1.2. Hybridization
- Prepare 30 µL hybridization solution per slide containing the specific probes for a target of interest. Heat to 70 °C for 10 min and place on ice.
- Place 30 µL of hybridization solution on each slide and cover with a plastic cover slip.
- Denature slide at 65–70 °C for 5 min on heat block.
- Gradually decrease temperature to 37 °C.
- Hybridize at 37 °C overnight in humidity chamber.
5.1.3. Detection
- Wash slides in 2× SSC to remove coverslip.
- Wash slides in wash buffer at 40 °C for 5 min, repeat.
- Wash slides in 0.1× SSC at 40 °C for 5–15 min.
- Wash slides in 2× SSC at 40 °C for 5–15 min.
- Cool slides to room temperature.
- Equilibrate slides in detection buffer for 5 min.
- Block in blocking buffer for 20–30 min.
- Incubate with 50 µL antibody or detection compound for 30–60 min (e.g., 5 µg/mL Streptavidin-Cy3 in blocking buffer).
- Wash slides in 2× SSC for 5 min, repeat twice.
- Counterstain with DAPI solution for 10 min.
- Rinse briefly and mount in antifade mounting medium.
- Analyze with a fluorescence microscope.
5.2. MLPA Protocol
- DNA denaturation:
- -
- Incubate 100 ng DNA sample in 5 μL of Tris-EDTA (pH 8) for 5 min at 98 °C.
- Probes hybridization to sample DNA:
- -
- Cool down the samples to room temperature;
- -
- Add 3 μL hybridization master mix;
- -
- In a thermal cycler incubate 1 min at 95 °C followed by 16 h at 60 °C.
- Ligation of hybridized probes:
- -
- Lower thermocycler temperature to 54 °C;
- -
- Add 32 μL Ligase-65 master mix, incubate 15 min at 54 °C;
- -
- Heat inactivates the ligase enzyme: 5 min at 98 °C.
- PCR amplification of ligated probes:
- -
- Cool down the samples to room temperature;
- -
- Add 10 μL polymerase master mix at room temperature;
- -
- Start PCR amplification.
- Fragment separation by capillary electrophoresis and data analysis.
5.3. ddPCR Protocol
- Prepare the samples by diluting genomic DNA at the desired concentration before setting up the reaction mix (e.g., dilute the DNA at 25 ng/microliter);
- Prepare master mixes containing the specific reaction buffer for probes, each of the sampling and reference probes, 25 ng DNA/sample and water to a final volume of 20 μL/sample. The master mixes shall be sufficient for three-four technical replicates each target deletion;
- Mix thoroughly and allow reaction tubes to equilibrate at room temperature for about 3 min;
- Once the reaction mixtures are ready, load 20 μL of each reaction mix into a sample well of a cartridge, followed by 70 μL of droplet generation oil for probes into the oil wells;
- Put the cartridge in the automated droplet generator;
- After droplet generation, carefully transfer droplets into a clean 96-well PCR plate. Seal the plate with an aluminum foil in the PCR plate sealer.
- Proceed to thermal cycling and PCR amplification, followed by the acquisition of droplets in the QX100 or QX200 droplet reader.
- Design the experimental template through QuantaSoft™ Software (plate layout). After that the droplets acquisition is complete, data are analyzed with the proper setup for any specific application (CNV, gene expression, mutation detection, etc.).
6. Time Required for the Report, Sensitivity and Specificity
7. Applications in Cancer Research and Molecular Testing
7.1. Chronic Lymphocytic Leukemia
7.2. Glioma and Glioblastoma
7.3. Breast Cancer, Lung Cancer and Colorectal Cancer
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, Z.; Wang, L.; Tang, G.; Medeiros, L.J. Fluorescence in situ hybridization (fish) for detecting anaplastic lymphoma kinase (alk) rearrangement in lung cancer: Clinically relevant technical aspects. Int. J. Mol. Sci. 2019, 20, 3939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sussman, R.T.; Oran, A.R.; Paolillo, C.; Lieberman, D.; Morrissette, J.J.D.; Rosenbaum, J.N. Validation of a next-generation sequencing assay targeting rna for the multiplexed detection of fusion transcripts and oncogenic isoforms. Arch. Pathol. Lab. Med. 2020, 144, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.R.; Chien, H.P.; Chen, K.S.; Hwang, C.C.; Chen, H.Y.; Yeh, K.Y.; Hsieh, T.Y.; Chang, L.C.; Hsu, Y.C.; Lu, R.J.; et al. Amplification of her2 and top2a and deletion of top2a genes in a series of taiwanese breast cancer. Medicine 2017, 96, e5582. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, S.; Shi, X.; Luo, Y.; Pang, J.; Wang, C.; Mao, F.; Liang, Z.; Zeng, X. Her2 double-equivocal breast cancer in chinese patients: A high concordance of her2 status between different blocks from the same tumor. Breast Cancer Res. Treat. 2019, 178, 275–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nevisi, F.; Yaghmaie, M.; Pashaiefar, H.; Alimoghaddam, K.; Iravani, M.; Javadi, G.; Ghavamzadeh, A. Correlation of her2, mdm2, c-myc, c-met, and tp53 copy number alterations in circulating tumor cells with tissue in gastric cancer patients: A pilot study. Iran Biomed. J. 2020, 24, 47–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pessoa, I.A.; Amorim, C.K.; Ferreira, W.A.S.; Sagica, F.; Brito, J.R.; Othman, M.; Meyer, B.; Liehr, T.; de Oliveira, E.H.C. Detection and correlation of single and concomitant tp53, pten, and cdkn2a alterations in gliomas. Int. J. Mol. Sci. 2019, 20, 2658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raponi, S.; Del Giudice, I.; Ilari, C.; Cafforio, L.; Messina, M.; Cappelli, L.V.; Bonina, S.; Piciocchi, A.; Marinelli, M.; Peragine, N.; et al. Biallelic birc3 inactivation in chronic lymphocytic leukaemia patients with 11q deletion identifies a subgroup with very aggressive disease. Br. J. Haematol. 2019, 185, 156–159. [Google Scholar] [CrossRef]
- Eijk-Van Os, P.G.; Schouten, J.P. Multiplex ligation-dependent probe amplification (mlpa(r)) for the detection of copy number variation in genomic sequences. Methods Mol. Biol. 2011, 688, 97–126. [Google Scholar]
- Abel, H.J.; Al-Kateb, H.; Cottrell, C.E.; Bredemeyer, A.J.; Pritchard, C.C.; Grossmann, A.H.; Wallander, M.L.; Pfeifer, J.D.; Lockwood, C.M.; Duncavage, E.J. Detection of gene rearrangements in targeted clinical next-generation sequencing. J. Mol. Diagn. 2014, 16, 405–417. [Google Scholar] [CrossRef] [Green Version]
- Baert-Desurmont, S.; Coutant, S.; Charbonnier, F.; Macquere, P.; Lecoquierre, F.; Schwartz, M.; Blanluet, M.; Vezain, M.; Lanos, R.; Quenez, O.; et al. Optimization of the diagnosis of inherited colorectal cancer using ngs and capture of exonic and intronic sequences of panel genes. Eur. J. Hum. Genet. 2018, 26, 1597–1602. [Google Scholar] [CrossRef] [Green Version]
- Yohe, S.; Thyagarajan, B. Review of clinical next-generation sequencing. Arch. Pathol. Lab. Med. 2017, 141, 1544–1557. [Google Scholar] [CrossRef] [Green Version]
- Mundle, S.D.; Sokolova, I. Clinical implications of advanced molecular cytogenetics in cancer. Expert Rev. Mol. Diagn. 2004, 4, 71–81. [Google Scholar] [CrossRef]
- Sunil, P.; Ramachandran, C.; Gokul, S.; Jaisanghar, N. Fluorescence in-situ hybridization technique as a diagnostic and prognostic tool in oral squamous cell carcinoma. J. Oral. Maxillofac. Pathol. 2013, 17, 61–64. [Google Scholar] [CrossRef]
- Cui, C.; Shu, W.; Li, P. Fluorescence in situ hybridization: Cell-based genetic diagnostic and research applications. Front. Cell Dev. Biol. 2016, 4, 89. [Google Scholar] [CrossRef] [Green Version]
- Halling, K.C.; Kipp, B.R. Fluorescence in situ hybridization in diagnostic cytology. Hum. Pathol. 2007, 38, 1137–1144. [Google Scholar] [CrossRef]
- Rudkin, G.T.; Stollar, B.D. High resolution detection of DNA-rna hybrids in situ by indirect immunofluorescence. Nature 1977, 265, 472–473. [Google Scholar] [CrossRef]
- Bauman, J.G.; Wiegant, J.; Borst, P.; van Duijn, P. A new method for fluorescence microscopical localization of specific DNA sequences by in situ hybridization of fluorochromelabelled rna. Exp. Cell Res. 1980, 128, 485–490. [Google Scholar] [CrossRef]
- Singer, R.H.; Ward, D.C. Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinated nucleotide analog. Proc. Natl. Acad. Sci. USA 1982, 79, 7331–7335. [Google Scholar] [CrossRef] [Green Version]
- Langer, P.R.; Waldrop, A.A.; Ward, D.C. Enzymatic synthesis of biotin-labeled polynucleotides: Novel nucleic acid affinity probes. Proc. Natl. Acad. Sci. USA 1981, 78, 6633–6637. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef]
- Nilsson, M.; Malmgren, H.; Samiotaki, M.; Kwiatkowski, M.; Chowdhary, B.P.; Landegren, U. Padlock probes: Circularizing oligonucleotides for localized DNA detection. Science 1994, 265, 2085–2088. [Google Scholar] [CrossRef]
- Femino, A.M.; Fay, F.S.; Fogarty, K.; Singer, R.H. Visualization of single rna transcripts in situ. Science 1998, 280, 585–590. [Google Scholar] [CrossRef] [Green Version]
- Silahtaroglu, A.; Pfundheller, H.; Koshkin, A.; Tommerup, N.; Kauppinen, S. Lna-modified oligonucleotides are highly efficient as fish probes. Cytogenet. Genome Res. 2004, 107, 32–37. [Google Scholar] [CrossRef]
- Larsson, C.; Grundberg, I.; Soderberg, O.; Nilsson, M. In situ detection and genotyping of individual mrna molecules. Nat. Methods 2010, 7, 395–397. [Google Scholar] [CrossRef] [PubMed]
- Speicher, M.R.; Gwyn Ballard, S.; Ward, D.C. Karyotyping human chromosomes by combinatorial multi-fluor fish. Nat. Genet. 1996, 12, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Levsky, J.M.; Shenoy, S.M.; Pezo, R.C.; Singer, R.H. Single-cell gene expression profiling. Science 2002, 297, 836–840. [Google Scholar] [CrossRef] [PubMed]
- Sieben, V.J.; Debes-Marun, C.S.; Pilarski, L.M.; Backhouse, C.J. An integrated microfluidic chip for chromosome enumeration using fluorescence in situ hybridization. Lab Chip 2008, 8, 2151–2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.O.; Matsuno, H.; Ikeda, S.; Nakamura, A.; Yanagisawa, H.; Hayashi, Y.; Okamoto, A. A quick and simple fish protocol with hybridization-sensitive fluorescent linear oligodeoxynucleotide probes. RNA 2012, 18, 166–175. [Google Scholar] [CrossRef] [Green Version]
- Sigma-Aldrich. Available online: Https://www.Sigmaaldrich.Com/technical-documents/protocols/biology/fish-procedure.Html (accessed on 1 April 2021).
- Schouten, J.P.; McElgunn, C.J.; Waaijer, R.; Zwijnenburg, D.; Diepvens, F.; Pals, G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002, 30, e57. [Google Scholar] [CrossRef] [Green Version]
- Den Dunnen, J.T.; White, S.J. Mlpa and maph: Sensitive detection of deletions and duplications. Curr. Protoc. Hum. Genet. 2006, 7, 7–14. [Google Scholar] [CrossRef]
- Lips, E.H.; Laddach, N.; Savola, S.P.; Vollebergh, M.A.; Oonk, A.M.; Imholz, A.L.; Wessels, L.F.; Wesseling, J.; Nederlof, P.M.; Rodenhuis, S. Quantitative copy number analysis by multiplex ligation-dependent probe amplification (mlpa) of brca1-associated breast cancer regions identifies brcaness. Breast Cancer Res. 2011, 13, R107. [Google Scholar] [CrossRef] [Green Version]
- Schouten, J.; van Vught, P.; Galjaard, R.J. Multiplex ligation-dependent probe amplification (mlpa) for prenatal diagnosis of common aneuploidies. Methods Mol. Biol. 2019, 1885, 161–170. [Google Scholar]
- MRC Holland. Available online: https://www.Mrcholland.com/ (accessed on 1 April 2021).
- Manoj, P. Droplet digital pcr technology promises new applications and research areas. Mitochondrial DNA A DNA Mapp Seq. Anal. 2016, 27, 742–746. [Google Scholar] [CrossRef]
- Kanagal-Shamanna, R. Digital pcr: Principles and applications. Methods Mol. Biol. 2016, 1392, 43–50. [Google Scholar]
- Vogelstein, B.; Kinzler, K.W. Digital pcr. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [Green Version]
- Morley, A.A. Digital pcr: A brief history. Biomol. Detect. Quantif. 2014, 1, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Taly, V.; Pekin, D.; Benhaim, L.; Kotsopoulos, S.K.; Le Corre, D.; Li, X.; Atochin, I.; Link, D.R.; Griffiths, A.D.; Pallier, K.; et al. Multiplex picodroplet digital pcr to detect kras mutations in circulating DNA from the plasma of colorectal cancer patients. Clin. Chem. 2013, 59, 1722–1731. [Google Scholar] [CrossRef]
- Nakano, M.; Komatsu, J.; Matsuura, S.; Takashima, K.; Katsura, S.; Mizuno, A. Single-molecule pcr using water-in-oil emulsion. J. Biotechnol. 2003, 102, 117–124. [Google Scholar] [CrossRef]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The digital miqe guidelines: Minimum information for publication of quantitative digital pcr experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef]
- Huggett, J.F. The digital miqe guidelines update: Minimum information for publication of quantitative digital pcr experiments for 2020. Clin. Chem. 2020, 66, 1012–1029. [Google Scholar] [CrossRef]
- Frazzi, R.; Bizzarri, V.; Albertazzi, L.; Cusenza, V.Y.; Coppolecchia, L.; Luminari, S.; Ilariucci, F. Droplet digital pcr is a sensitive tool for the detection of tp53 deletions and point mutations in chronic lymphocytic leukaemia. Br. J. Haematol. 2020, 189, e49–e52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiktor, A.E.; Van Dyke, D.L.; Stupca, P.J.; Ketterling, R.P.; Thorland, E.C.; Shearer, B.M.; Fink, S.R.; Stockero, K.J.; Majorowicz, J.R.; Dewald, G.W. Preclinical validation of fluorescence in situ hybridization assays for clinical practice. Genet. Med. 2006, 8, 16–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, J.; Smith, J.L.; Dowling, P.K. Fluorescence in situ hybridization probe validation for clinical use. Methods Mol. Biol. 2017, 1541, 101–118. [Google Scholar] [PubMed]
- Abdool, A.; Donahue, A.C.; Wohlgemuth, J.G.; Yeh, C.H. Detection, analysis and clinical validation of chromosomal aberrations by multiplex ligation-dependent probe amplification in chronic leukemia. PLoS ONE 2010, 5, e15407. [Google Scholar] [CrossRef]
- Blakemore, S.J.; Clifford, R.; Parker, H.; Antoniou, P.; Stec-Dziedzic, E.; Larrayoz, M.; Davis, Z.; Kadalyayil, L.; Colins, A.; Robbe, P.; et al. Clinical significance of tp53, birc3, atm and mapk-erk genes in chronic lymphocytic leukaemia: Data from the randomised uk lrf cll4 trial. Leukemia 2020, 34, 1760–1774. [Google Scholar] [CrossRef]
- Homig-Holzel, C.; Savola, S. Multiplex ligation-dependent probe amplification (mlpa) in tumor diagnostics and prognostics. Diagn. Mol. Pathol. 2012, 21, 189–206. [Google Scholar] [CrossRef]
- Al Zaabi, E.A.; Fernandez, L.A.; Sadek, I.A.; Riddell, D.C.; Greer, W.L. Multiplex ligation-dependent probe amplification versus multiprobe fluorescence in situ hybridization to detect genomic aberrations in chronic lymphocytic leukemia: A tertiary center experience. J. Mol. Diagn. 2010, 12, 197–203. [Google Scholar] [CrossRef]
- Alhourani, E.; Rincic, M.; Othman, M.A.; Pohle, B.; Schlie, C.; Glaser, A.; Liehr, T. Comprehensive chronic lymphocytic leukemia diagnostics by combined multiplex ligation dependent probe amplification (mlpa) and interphase fluorescence in situ hybridization (ifish). Mol. Cytogenet. 2014, 7, 79. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, V.K.; Naseem, S.; Varma, N.; Lad, D.P.; Malhotra, P. Genomic alterations in chronic lymphocytic leukemia and their correlation with clinico-hematological parameters and disease progression. Blood Res. 2020, 55, 131–138. [Google Scholar] [CrossRef]
- Krober, A.; Bloehdorn, J.; Hafner, S.; Buhler, A.; Seiler, T.; Kienle, D.; Winkler, D.; Bangerter, M.; Schlenk, R.F.; Benner, A.; et al. Additional genetic high-risk features such as 11q deletion, 17p deletion, and v3-21 usage characterize discordance of zap-70 and vh mutation status in chronic lymphocytic leukemia. J. Clin. Oncol. 2006, 24, 969–975. [Google Scholar] [CrossRef] [Green Version]
- Rose-Zerilli, M.J.; Forster, J.; Parker, H.; Parker, A.; Rodriguez, A.E.; Chaplin, T.; Gardiner, A.; Steele, A.J.; Collins, A.; Young, B.D.; et al. Atm mutation rather than birc3 deletion and/or mutation predicts reduced survival in 11q-deleted chronic lymphocytic leukemia: Data from the uk lrf cll4 trial. Haematologica 2014, 99, 736–742. [Google Scholar] [CrossRef]
- Stankovic, T.; Skowronska, A. The role of atm mutations and 11q deletions in disease progression in chronic lymphocytic leukemia. Leuk. Lymphoma 2014, 55, 1227–1239. [Google Scholar] [CrossRef]
- Frazzi, R. Birc3 and birc5: Multi-faceted inhibitors in cancer. Cell Biosci. 2021, 11, 8. [Google Scholar] [CrossRef]
- Veronese, L.; Tournilhac, O.; Combes, P.; Prie, N.; Pierre-Eymard, E.; Guieze, R.; Veyrat-Masson, R.; Bay, J.O.; Vago, P.; Tchirkov, A. Contribution of mlpa to routine diagnostic testing of recurrent genomic aberrations in chronic lymphocytic leukemia. Cancer Genet. 2013, 206, 19–25. [Google Scholar] [CrossRef]
- Tausch, E.; Stilgenbauer, S. Birc3 mutations in chronic lymphocytic leukemia—Uncommon and unfavorable. Haematologica 2020, 105, 255–256. [Google Scholar] [CrossRef]
- Chan, T.S.; Lee, Y.S.; Del Giudice, I.; Marinelli, M.; Ilari, C.; Cafforio, L.; Guarini, A.; Tan, D.; Phipps, C.; Goh, Y.T.; et al. Clinicopathological features and outcome of chronic lymphocytic leukaemia in Chinese patients. Oncotarget 2017, 8, 25455–25468. [Google Scholar] [CrossRef] [Green Version]
- Balgobind, B.V.; Hollink, I.H.; Reinhardt, D.; van Wering, E.R.; de Graaf, S.S.; Baruchel, A.; Stary, J.; Beverloo, H.B.; de Greef, G.E.; Pieters, R.; et al. Low frequency of mll-partial tandem duplications in paediatric acute myeloid leukaemia using mlpa as a novel DNA screenings technique. Eur. J. Cancer 2010, 46, 1892–1899. [Google Scholar] [CrossRef]
- Amin, N.A.; Seymour, E.; Saiya-Cork, K.; Parkin, B.; Shedden, K.; Malek, S.N. A quantitative analysis of subclonal and clonal gene mutations before and after therapy in chronic lymphocytic leukemia. Clin. Cancer Res. 2016, 22, 4525–4535. [Google Scholar] [CrossRef] [Green Version]
- Dolecek, T.A.; Propp, J.M.; Stroup, N.E.; Kruchko, C. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the united states in 2005–2009. Neuro Oncol. 2012, 14. [Google Scholar] [CrossRef]
- Gressot, L.V.; Doucette, T.; Yang, Y.; Fuller, G.N.; Manyam, G.; Rao, A.; Latha, K.; Rao, G. Analysis of the inhibitors of apoptosis identifies birc3 as a facilitator of malignant progression in glioma. Oncotarget 2017, 8, 12695–12704. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, D.S.; Wright, R.D.; Kesari, S.; Lemieux, M.E.; Tran, M.A.; Jain, M.; Zawel, L.; Kung, A.L. Resistance of human glioblastoma multiforme cells to growth factor inhibitors is overcome by blockade of inhibitor of apoptosis proteins. J. Clin. Investig. 2008, 118, 3109–3122. [Google Scholar] [CrossRef] [Green Version]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in pdgfra, idh1, egfr, and nf1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [Green Version]
- Yip, S.; Butterfield, Y.S.; Morozova, O.; Chittaranjan, S.; Blough, M.D.; An, J.; Birol, I.; Chesnelong, C.; Chiu, R.; Chuah, E.; et al. Concurrent cic mutations, idh mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J. Pathol. 2012, 226, 7–16. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Acquaviva, J.; Ramachandran, P.; Boskovitz, A.; Woolfenden, S.; Pfannl, R.; Bronson, R.T.; Chen, J.W.; Weissleder, R.; Housman, D.E.; et al. Oncogenic egfr signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 2712–2716. [Google Scholar] [CrossRef] [Green Version]
- Okada, Y.; Hurwitz, E.E.; Esposito, J.M.; Brower, M.A.; Nutt, C.L.; Louis, D.N. Selection pressures of tp53 mutation and microenvironmental location influence epidermal growth factor receptor gene amplification in human glioblastomas. Cancer Res. 2003, 63, 413–416. [Google Scholar]
- Kraus, J.A.; Glesmann, N.; Beck, M.; Krex, D.; Klockgether, T.; Schackert, G.; Schlegel, U. Molecular analysis of the pten, tp53 and cdkn2a tumor suppressor genes in long-term survivors of glioblastoma multiforme. J. Neurooncol. 2000, 48, 89–94. [Google Scholar] [CrossRef]
- Ichimura, K.; Bolin, M.B.; Goike, H.M.; Schmidt, E.E.; Moshref, A.; Collins, V.P. Deregulation of the p14arf/mdm2/p53 pathway is a prerequisite for human astrocytic gliomas with g1-s transition control gene abnormalities. Cancer Res. 2000, 60, 417–424. [Google Scholar]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. Idh1 and idh2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Nakamura, J.L. The epidermal growth factor receptor in malignant gliomas: Pathogenesis and therapeutic implications. Expert Opin. Ther. Targets 2007, 11, 463–472. [Google Scholar] [CrossRef]
- Schober, R.; Bilzer, T.; Waha, A.; Reifenberger, G.; Wechsler, W.; von Deimling, A.; Wiestler, O.D.; Westphal, M.; Kemshead, J.T.; Vega, F.; et al. The epidermal growth factor receptor in glioblastoma: Genomic amplification, protein expression, and patient survival data in a therapeutic trial. Clin. Neuropathol. 1995, 14, 169–174. [Google Scholar] [PubMed]
- Li, L.; Dutra, A.; Pak, E.; Labrie, J.E., 3rd; Gerstein, R.M.; Pandolfi, P.P.; Recht, L.D.; Ross, A.H. Egfrviii expression and pten loss synergistically induce chromosomal instability and glial tumors. Neuro Oncol. 2009, 11, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Lassman, A.B.; Aldape, K.D.; Ansell, P.J.; Bain, E.; Curran, W.J.; Eoli, M.; French, P.J.; Kinoshita, M.; Looman, J.; Mehta, M.; et al. Epidermal growth factor receptor (egfr) amplification rates observed in screening patients for randomized trials in glioblastoma. J. Neurooncol. 2019, 144, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Hidalgo, L.; San-Miguel, T.; Megias, J.; Monleon, D.; Navarro, L.; Roldan, P.; Cerda-Nicolas, M.; Lopez-Gines, C. Somatic copy number alterations are associated with egfr amplification and shortened survival in patients with primary glioblastoma. Neoplasia 2020, 22, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Saadeh, F.S.; Mahfouz, R.; Assi, H.I. Egfr as a clinical marker in glioblastomas and other gliomas. Int. J. Biol. Markers 2018, 33, 22–32. [Google Scholar] [CrossRef] [Green Version]
- Korshunov, A.; Sycheva, R.; Golanov, A. Molecular stratification of diagnostically challenging high-grade gliomas composed of small cells: The utility of fluorescence in situ hybridization. Clin Cancer Res. 2004, 10, 7820–7826. [Google Scholar] [CrossRef] [Green Version]
- Jeuken, J.; Cornelissen, S.; Boots-Sprenger, S.; Gijsen, S.; Wesseling, P. Multiplex ligation-dependent probe amplification: A diagnostic tool for simultaneous identification of different genetic markers in glial tumors. J. Mol. Diagn. 2006, 8, 433–443. [Google Scholar] [CrossRef] [Green Version]
- Trabelsi, S.; Chabchoub, I.; Ksira, I.; Karmeni, N.; Mama, N.; Kanoun, S.; Burford, A.; Jury, A.; Mackay, A.; Popov, S.; et al. Molecular diagnostic and prognostic subtyping of gliomas in tunisian population. Mol. Neurobiol. 2017, 54, 2381–2394. [Google Scholar] [CrossRef]
- Weller, M.; Felsberg, J.; Hartmann, C.; Berger, H.; Steinbach, J.P.; Schramm, J.; Westphal, M.; Schackert, G.; Simon, M.; Tonn, J.C.; et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: A prospective translational study of the german glioma network. J. Clin. Oncol. 2009, 27, 5743–5750. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.S.; Perry, A.; Borell, T.J.; Lee, H.K.; O’Fallon, J.; Hosek, S.M.; Kimmel, D.; Yates, A.; Burger, P.C.; Scheithauer, B.W.; et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J. Clin. Oncol. 2000, 18, 636–645. [Google Scholar] [CrossRef]
- Sievert, A.J.; Jackson, E.M.; Gai, X.; Hakonarson, H.; Judkins, A.R.; Resnick, A.C.; Sutton, L.N.; Storm, P.B.; Shaikh, T.H.; Biegel, J.A. Duplication of 7q34 in pediatric low-grade astrocytomas detected by high-density single-nucleotide polymorphism-based genotype arrays results in a novel braf fusion gene. Brain Pathol. 2009, 19, 449–458. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, C.; Walker, E.; Mohamed, N.; Zhang, C.; Jacob, K.; Shirinian, M.; Alon, N.; Kahn, D.; Fried, I.; Scheinemann, K.; et al. Braf-kiaa1549 fusion predicts better clinical outcome in pediatric low-grade astrocytoma. Clin. Cancer Res. 2011, 17, 4790–4798. [Google Scholar] [CrossRef] [Green Version]
- Ghasimi, S.; Wibom, C.; Dahlin, A.M.; Brannstrom, T.; Golovleva, I.; Andersson, U.; Melin, B. Genetic risk variants in the cdkn2a/b, rtel1 and egfr genes are associated with somatic biomarkers in glioma. J. Neurooncol. 2016, 127, 483–492. [Google Scholar] [CrossRef] [Green Version]
- Trevisan, P.; Graziadio, C.; Rodrigues, D.B.K.; Rosa, R.F.M.; Soares, F.P.; Provenzi, V.O.; de Oliveira, C.A.V.; Paskulin, G.A.; Varella-Garcia, M.; Zen, P.R.G. Clinical and molecular characterization of adult glioblastomas in southern brazil. J. Neuropathol. Exp. Neurol. 2019, 78, 297–304. [Google Scholar] [CrossRef] [Green Version]
- Broniscer, A.; Hwang, S.N.; Chamdine, O.; Lin, T.; Pounds, S.; Onar-Thomas, A.; Chi, L.; Shurtleff, S.; Allen, S.; Gajjar, A.; et al. Bithalamic gliomas may be molecularly distinct from their unilateral high-grade counterparts. Brain Pathol. 2018, 28, 112–120. [Google Scholar] [CrossRef]
- Purkait, S.; Mallick, S.; Sharma, V.; Kumar, A.; Pathak, P.; Jha, P.; Biswas, A.; Julka, P.K.; Gupta, D.; Suri, A.; et al. A simplified approach for molecular classification of glioblastomas (gbms): Experience from a tertiary care center in india. Brain Tumor Pathol. 2016, 33, 183–190. [Google Scholar] [CrossRef]
- Holtkamp, N.; Ziegenhagen, N.; Malzer, E.; Hartmann, C.; Giese, A.; von Deimling, A. Characterization of the amplicon on chromosomal segment 4q12 in glioblastoma multiforme. Neuro Oncol. 2007, 9, 291–297. [Google Scholar] [CrossRef] [Green Version]
- Gielen, G.H.; Gessi, M.; Buttarelli, F.R.; Baldi, C.; Hammes, J.; zur Muehlen, A.; Doerner, E.; Denkhaus, D.; Warmuth-Metz, M.; Giangaspero, F.; et al. Genetic analysis of diffuse high-grade astrocytomas in infancy defines a novel molecular entity. Brain Pathol. 2015, 25, 409–417. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, Y.Y.; Li, J.F.; Guo, C.C.; Chen, F.R.; Su, H.K.; Zhao, H.F.; Long, Y.K.; Shao, J.Y.; To, S.; et al. Idh1 mutation detection by droplet digital pcr in glioma. Oncotarget 2015, 6, 39651–39660. [Google Scholar] [CrossRef] [Green Version]
- Ge, J.; Liu, M.Y.; Li, L.; Deng, Q.; Liu, F.; Luo, Y.; Wang, L.; Yao, G.; Zhu, D.; Lu, H.; et al. Detection of idh1 and tert promoter mutations with droplet digital pcr in diffuse gliomas. Int. J. Clin. Exp. Pathol. 2020, 13, 230–238. [Google Scholar]
- Harat, M.; Blok, M.; Harat, A.; Soszynska, K. The impact of adjuvant radiotherapy on molecular prognostic markers in gliomas. Onco Targets Ther. 2019, 12, 2215–2224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokogami, K.; Yamasaki, K.; Matsumoto, F.; Yamashita, S.; Saito, K.; Tacheva, A.; Mizuguchi, A.; Watanabe, T.; Ohta, H.; Takeshima, H. Impact of pcr-based molecular analysis in daily diagnosis for the patient with gliomas. Brain Tumor Pathol. 2018, 35, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Fina, F.; Barets, D.; Colin, C.; Bouvier, C.; Padovani, L.; Nanni-Metellus, I.; Ouafik, L.; Scavarda, D.; Korshunov, A.; Jones, D.T.; et al. Droplet digital pcr is a powerful technique to demonstrate frequent fgfr1 duplication in dysembryoplastic neuroepithelial tumors. Oncotarget 2017, 8, 2104–2113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tse, C.; Gauchez, A.S.; Jacot, W.; Lamy, P.J. Her2 shedding and serum her2 extracellular domain: Biology and clinical utility in breast cancer. Cancer Treat. Rev. 2012, 38, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Yeh, I.T.; Martin, M.A.; Robetorye, R.S.; Bolla, A.R.; McCaskill, C.; Shah, R.K.; Gorre, M.E.; Mohammed, M.S.; Gunn, S.R. Clinical validation of an array cgh test for her2 status in breast cancer reveals that polysomy 17 is a rare event. Mod. Pathol. 2009, 22, 1169–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vranic, S.; Teruya, B.; Repertinger, S.; Ulmer, P.; Hagenkord, J.; Gatalica, Z. Assessment of her2 gene status in breast carcinomas with polysomy of chromosome 17. Cancer 2011, 117, 48–53. [Google Scholar] [CrossRef]
- Marchio, C.; Lambros, M.B.; Gugliotta, P.; Di Cantogno, L.V.; Botta, C.; Pasini, B.; Tan, D.S.; Mackay, A.; Fenwick, K.; Tamber, N.; et al. Does chromosome 17 centromere copy number predict polysomy in breast cancer? A fluorescence in situ hybridization and microarray-based cgh analysis. J. Pathol. 2009, 219, 16–24. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of american pathologists clinical practice guideline focused update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef] [Green Version]
- Moerland, E.; van Hezik, R.L.; van der Aa, T.C.; van Beek, M.W.; van den Brule, A.J. Detection of her2 amplification in breast carcinomas: Comparison of multiplex ligation-dependent probe amplification (mlpa) and fluorescence in situ hybridization (fish) combined with automated spot counting. Cell Oncol. 2006, 28, 151–159. [Google Scholar]
- Soosanabadi, M.; Mirfakhraie, R.; Atanesyan, L.; Biglarian, A.; Aghakhani Moghadam, F.; Rahimi, M.; Behjati, F.; Keyhani, E. Application of multiplex ligation-dependent probe amplification in determining the copy number alterations of her gene family members in invasive ductal breast carcinoma. Rep. Biochem. Mol. Biol. 2019, 8, 91–101. [Google Scholar]
- Tantiwetrueangdet, A.; Panvichian, R.; Wongwaisayawan, S.; Sueangoen, N.; Lertsithichai, P. Droplet digital pcr using her2/eif2c1 ratio for detection of her2 amplification in breast cancer tissues. Med. Oncol. 2018, 35, 149. [Google Scholar] [CrossRef] [Green Version]
- Faraz, M.; Tellstrom, A.; Ardnor, C.E.; Grankvist, K.; Huminiecki, L.; Tavelin, B.; Henriksson, R.; Hedman, H.; Ljuslinder, I. Lrig1 gene copy number analysis by ddpcr and correlations to clinical factors in breast cancer. BMC Cancer 2020, 20, 459. [Google Scholar] [CrossRef]
- Veenstra, C.; Karlsson, E.; Mirwani, S.M.; Nordenskjold, B.; Fornander, T.; Perez-Tenorio, G.; Stal, O. The effects of ptpn2 loss on cell signalling and clinical outcome in relation to breast cancer subtype. J. Cancer Res. Clin. Oncol. 2019, 145, 1845–1856. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Zheng, J.; Wu, C.; Liu, S.; Chen, Y.; Liu, X.; Du, J.; Wang, J. Breast cancer subtype classification using 4-plex droplet digital pcr. Clin. Chem. 2019, 65, 1051–1059. [Google Scholar] [CrossRef]
- Medford, A.J.; Gillani, R.N.; Park, B.H. Detection of cancer DNA in early stage and metastatic breast cancer patients. Methods Mol. Biol. 2018, 1768, 209–227. [Google Scholar]
- Otsuji, K.; Sasaki, T.; Tanabe, M.; Seto, Y. Droplet-digital pcr reveals frequent mutations in tert promoter region in breast fibroadenomas and phyllodes tumours, irrespective of the presence of med12 mutations. Br. J. Cancer 2021, 124, 466–473. [Google Scholar] [CrossRef]
- Del Re, M.; Bertolini, I.; Crucitta, S.; Fontanelli, L.; Rofi, E.; De Angelis, C.; Diodati, L.; Cavallero, D.; Gianfilippo, G.; Salvadori, B.; et al. Overexpression of tk1 and cdk9 in plasma-derived exosomes is associated with clinical resistance to cdk4/6 inhibitors in metastatic breast cancer patients. Breast Cancer Res. Treat. 2019, 178, 57–62. [Google Scholar] [CrossRef]
- Ooi, A.; Inokuchi, M.; Horike, S.I.; Kawashima, H.; Ishikawa, S.; Ikeda, H.; Nakamura, R.; Oyama, T.; Dobashi, Y. Amplicons in breast cancers analyzed by multiplex ligation-dependent probe amplification and fluorescence in situ hybridization. Hum. Pathol. 2019, 85, 33–43. [Google Scholar] [CrossRef]
- Hirai, N.; Sasaki, T.; Okumura, S.; Sado, M.; Akiyama, N.; Kitada, M.; Takei, H.; Ohsaki, Y. Novel alk-specific mrna in situ hybridization assay for non-small-cell lung carcinoma. Transl. Lung Cancer Res. 2020, 9, 257–268. [Google Scholar] [CrossRef]
- Badawy, O.M.; Loay, I. Fish analysis of top2a and her-2 aberrations in female breast carcinoma on archived material: Egyptian nci experience. Appl. Immunohistochem Mol. Morphol. 2019, 27, 216–222. [Google Scholar] [CrossRef]
- Gogas, H.; Kotoula, V.; Alexopoulou, Z.; Christodoulou, C.; Kostopoulos, I.; Bobos, M.; Raptou, G.; Charalambous, E.; Tsolaki, E.; Xanthakis, I.; et al. Myc copy gain, chromosomal instability and pi3k activation as potential markers of unfavourable outcome in trastuzumab-treated patients with metastatic breast cancer. J. Transl. Med. 2016, 14, 136. [Google Scholar] [CrossRef] [Green Version]
- Brody, H. Colorectal cancer. Nature 2015, 521, S1. [Google Scholar] [CrossRef]
- Villalobos, P.; Wistuba, I.I. Lung cancer biomarkers. Hematol. Oncol. Clin. N. Am. 2017, 31, 13–29. [Google Scholar] [CrossRef] [Green Version]
- Ciardiello, F.; Tortora, G. Egfr antagonists in cancer treatment. N. Engl. J. Med. 2008, 358, 1160–1174. [Google Scholar] [CrossRef] [Green Version]
- Birkman, E.M.; Avoranta, T.; Algars, A.; Korkeila, E.; Lintunen, M.; Lahtinen, L.; Kuopio, T.; Ristamaki, R.; Carpen, O.; Sundstrom, J. Egfr gene copy number decreases during anti-egfr antibody therapy in colorectal cancer. Hum. Pathol. 2018, 82, 163–171. [Google Scholar] [CrossRef]
- Lievre, A.; Bachet, J.B.; Le Corre, D.; Boige, V.; Landi, B.; Emile, J.F.; Cote, J.F.; Tomasic, G.; Penna, C.; Ducreux, M.; et al. Kras mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006, 66, 3992–3995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amado, R.G.; Wolf, M.; Peeters, M.; Van Cutsem, E.; Siena, S.; Freeman, D.J.; Juan, T.; Sikorski, R.; Suggs, S.; Radinsky, R.; et al. Wild-type kras is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 1626–1634. [Google Scholar] [CrossRef] [PubMed]
- Linardou, H.; Dahabreh, I.J.; Kanaloupiti, D.; Siannis, F.; Bafaloukos, D.; Kosmidis, P.; Papadimitriou, C.A.; Murray, S. Assessment of somatic k-ras mutations as a mechanism associated with resistance to egfr-targeted agents: A systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008, 9, 962–972. [Google Scholar] [CrossRef]
- Allegra, C.J.; Rumble, R.B.; Hamilton, S.R.; Mangu, P.B.; Roach, N.; Hantel, A.; Schilsky, R.L. Extended ras gene mutation testing in metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy: American society of clinical oncology provisional clinical opinion update 2015. J. Clin. Oncol. 2016, 34, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Atreya, C.E.; Corcoran, R.B.; Kopetz, S. Expanded ras: Refining the patient population. J. Clin. Oncol. 2015, 33, 682–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorich, M.J.; Wiese, M.D.; Rowland, A.; Kichenadasse, G.; McKinnon, R.A.; Karapetis, C.S. Extended ras mutations and anti-egfr monoclonal antibody survival benefit in metastatic colorectal cancer: A meta-analysis of randomized, controlled trials. Ann. Oncol. 2015, 26, 13–21. [Google Scholar] [CrossRef]
- Al-Kuraya, K.; Novotny, H.; Bavi, P.; Siraj, A.K.; Uddin, S.; Ezzat, A.; Sanea, N.A.; Al-Dayel, F.; Al-Mana, H.; Sheikh, S.S.; et al. Her2, top2a, ccnd1, egfr and c-myc oncogene amplification in colorectal cancer. J. Clin. Pathol. 2007, 60, 768–772. [Google Scholar] [CrossRef]
- Sartore-Bianchi, A.; Fieuws, S.; Veronese, S.; Moroni, M.; Personeni, N.; Frattini, M.; Torri, V.; Cappuzzo, F.; Vander Borght, S.; Martin, V.; et al. Standardisation of egfr fish in colorectal cancer: Results of an international interlaboratory reproducibility ring study. J. Clin. Pathol. 2012, 65, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Spindler, K.L.; Lindebjerg, J.; Nielsen, J.N.; Olsen, D.A.; Bisgard, C.; Brandslund, I.; Jakobsen, A. Epidermal growth factor receptor analyses in colorectal cancer: A comparison of methods. Int. J. Oncol. 2006, 29, 1159–1165. [Google Scholar] [CrossRef] [Green Version]
- Afrasanie, V.A.; Marinca, M.V.; Alexa-Stratulat, T.; Gafton, B.; Paduraru, M.; Adavidoaiei, A.M.; Miron, L.; Rusu, C. Kras, nras, braf, her2 and microsatellite instability in metastatic colorectal cancer—Practical implications for the clinician. Radiol. Oncol. 2019, 53, 265–274. [Google Scholar] [CrossRef] [Green Version]
- Bos, J.L.; Fearon, E.R.; Hamilton, S.R.; Verlaan-de Vries, M.; van Boom, J.H.; van der Eb, A.J.; Vogelstein, B. Prevalence of ras gene mutations in human colorectal cancers. Nature 1987, 327, 293–297. [Google Scholar] [CrossRef]
- Finkelstein, S.D.; Sayegh, R.; Christensen, S.; Swalsky, P.A. Genotypic classification of colorectal adenocarcinoma. Biologic behavior correlates with k-ras-2 mutation type. Cancer 1993, 71, 3827–3838. [Google Scholar] [CrossRef]
- Boughdady, I.S.; Kinsella, A.R.; Haboubi, N.Y.; Schofield, P.F. K-ras gene mutations in adenomas and carcinomas of the colon. Surg. Oncol. 1992, 1, 275–282. [Google Scholar] [CrossRef]
- Orhan, T.; Nielsen, P.B.; Hviid, T.V.F.; Rosen, A.W.; Gogenur, I. Expression of circadian clock genes in human colorectal cancer tissues using droplet digital pcr. Cancer Investig. 2019, 37, 90–98. [Google Scholar] [CrossRef]
- Bidshahri, R.; Attali, D.; Fakhfakh, K.; McNeil, K.; Karsan, A.; Won, J.R.; Wolber, R.; Bryan, J.; Hughesman, C.; Haynes, C. Quantitative detection and resolution of braf v600 status in colorectal cancer using droplet digital pcr and a novel wild-type negative assay. J. Mol. Diagn. 2016, 18, 190–204. [Google Scholar] [CrossRef] [Green Version]
- Vanova, B.; Kalman, M.; Jasek, K.; Kasubova, I.; Burjanivova, T.; Farkasova, A.; Kruzliak, P.; Busselberg, D.; Plank, L.; Lasabova, Z. Droplet digital pcr revealed high concordance between primary tumors and lymph node metastases in multiplex screening of kras mutations in colorectal cancer. Clin. Exp. Med. 2019, 19, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Alcaide, M.; Cheung, M.; Bushell, K.; Arthur, S.E.; Wong, H.L.; Karasinska, J.; Renouf, D.; Schaeffer, D.F.; McNamara, S.; Tertre, M.C.D.; et al. A novel multiplex droplet digital pcr assay to identify and quantify kras mutations in clinical specimens. J. Mol. Diagn. 2019, 21, 214–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.T.; Gopalan, V.; Islam, F.; Wahab, R.; Mamoori, A.; Lu, C.T.; Smith, R.A.; Lam, A.K. Gaec1 mutations and copy number aberration is associated with biological aggressiveness of colorectal cancer. Eur. J. Cell Biol. 2018, 97, 230–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herring, E.; Kanaoka, S.; Tremblay, E.; Beaulieu, J.F. Droplet digital pcr for quantification of itga6 in a stool mrna assay for the detection of colorectal cancers. World J. Gastroenterol. 2017, 23, 2891–2898. [Google Scholar] [CrossRef] [PubMed]
- Lagerstedt-Robinson, K.; Rohlin, A.; Aravidis, C.; Melin, B.; Nordling, M.; Stenmark-Askmalm, M.; Lindblom, A.; Nilbert, M. Mismatch repair gene mutation spectrum in the swedish lynch syndrome population. Oncol. Rep. 2016, 36, 2823–2835. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, A.J.; Capo-Chichi, J.M.; Spence, T.; Grenier, S.; Stockley, T.; Kamel-Reid, S.; Serra, S.; Sabatini, P.; Chetty, R. Heterogenous loss of mismatch repair (mmr) protein expression: A challenge for immunohistochemical interpretation and microsatellite instability (msi) evaluation. J. Pathol. Clin. Res. 2019, 5, 115–129. [Google Scholar] [CrossRef]
- Dong, Z.; Kong, L.; Wan, Z.; Zhu, F.; Zhong, M.; Lv, Y.; Zhao, P.; Shi, H. Somatic mutation profiling and her2 status in kras-positive chinese colorectal cancer patients. Sci. Rep. 2019, 9, 16894. [Google Scholar] [CrossRef]
- Khan, S.A.; Zeng, Z.; Shia, J.; Paty, P.B. Egfr gene amplification and kras mutation predict response to combination targeted therapy in metastatic colorectal cancer. Pathol. Oncol. Res. 2017, 23, 673–677. [Google Scholar] [CrossRef]
- Fiedler, D.; Heselmeyer-Haddad, K.; Hirsch, D.; Hernandez, L.S.; Torres, I.; Wangsa, D.; Hu, Y.; Zapata, L.; Rueschoff, J.; Belle, S.; et al. Single-cell genetic analysis of clonal dynamics in colorectal adenomas indicates cdx2 gain as a predictor of recurrence. Int. J. Cancer 2019, 144, 1561–1573. [Google Scholar] [CrossRef]
- Pao, W.; Girard, N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011, 12, 175–180. [Google Scholar] [CrossRef]
- Wang, R.; Pan, Y.; Li, C.; Hu, H.; Zhang, Y.; Li, H.; Luo, X.; Zhang, J.; Fang, Z.; Li, Y.; et al. The use of quantitative real-time reverse transcriptase pcr for 5′ and 3′ portions of alk transcripts to detect alk rearrangements in lung cancers. Clin. Cancer Res. 2012, 18, 4725–4732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soda, M.; Choi, Y.L.; Enomoto, M.; Takada, S.; Yamashita, Y.; Ishikawa, S.; Fujiwara, S.; Watanabe, H.; Kurashina, K.; Hatanaka, H.; et al. Identification of the transforming eml4-alk fusion gene in non-small-cell lung cancer. Nature 2007, 448, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Kwak, E.L.; Bang, Y.J.; Camidge, D.R.; Shaw, A.T.; Solomon, B.; Maki, R.G.; Ou, S.H.; Dezube, B.J.; Janne, P.A.; Costa, D.B.; et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010, 363, 1693–1703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Yoo, S.B.; Choe, J.Y.; Paik, J.H.; Xu, X.; Nitta, H.; Zhang, W.; Grogan, T.M.; Lee, C.T.; Jheon, S.; et al. Detection of alk gene rearrangement in non-small cell lung cancer: A comparison of fluorescence in situ hybridization and chromogenic in situ hybridization with correlation of alk protein expression. J. Thorac. Oncol. 2011, 6, 1359–1366. [Google Scholar] [CrossRef] [Green Version]
- Inamura, K.; Takeuchi, K.; Togashi, Y.; Hatano, S.; Ninomiya, H.; Motoi, N.; Mun, M.Y.; Sakao, Y.; Okumura, S.; Nakagawa, K.; et al. Eml4-alk lung cancers are characterized by rare other mutations, a ttf-1 cell lineage, an acinar histology, and young onset. Mod. Pathol. 2009, 22, 508–515. [Google Scholar] [CrossRef] [Green Version]
- Shaw, A.T.; Yeap, B.Y.; Mino-Kenudson, M.; Digumarthy, S.R.; Costa, D.B.; Heist, R.S.; Solomon, B.; Stubbs, H.; Admane, S.; McDermott, U.; et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor eml4-alk. J. Clin. Oncol. 2009, 27, 4247–4253. [Google Scholar] [CrossRef] [Green Version]
- Rodig, S.J.; Mino-Kenudson, M.; Dacic, S.; Yeap, B.Y.; Shaw, A.; Barletta, J.A.; Stubbs, H.; Law, K.; Lindeman, N.; Mark, E.; et al. Unique clinicopathologic features characterize alk-rearranged lung adenocarcinoma in the western population. Clin. Cancer Res. 2009, 15, 5216–5223. [Google Scholar] [CrossRef] [Green Version]
- Jokoji, R.; Yamasaki, T.; Minami, S.; Komuta, K.; Sakamaki, Y.; Takeuchi, K.; Tsujimoto, M. Combination of morphological feature analysis and immunohistochemistry is useful for screening of eml4-alk-positive lung adenocarcinoma. J. Clin. Pathol. 2010, 63, 1066–1070. [Google Scholar] [CrossRef]
- Yi, E.S.; Boland, J.M.; Maleszewski, J.J.; Roden, A.C.; Oliveira, A.M.; Aubry, M.C.; Erickson-Johnson, M.R.; Caron, B.L.; Li, Y.; Tang, H.; et al. Correlation of ihc and fish for alk gene rearrangement in non-small cell lung carcinoma: Ihc score algorithm for fish. J. Thorac. Oncol. 2011, 6, 459–465. [Google Scholar] [CrossRef] [Green Version]
- Paik, J.H.; Choe, G.; Kim, H.; Choe, J.Y.; Lee, H.J.; Lee, C.T.; Lee, J.S.; Jheon, S.; Chung, J.H. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: Correlation with fluorescence in situ hybridization. J. Thorac. Oncol. 2011, 6, 466–472. [Google Scholar] [CrossRef] [Green Version]
- Just, P.A.; Cazes, A.; Audebourg, A.; Cessot, A.; Pallier, K.; Danel, C.; Vacher-Lavenu, M.C.; Laurent-Puig, P.; Terris, B.; Blons, H. Histologic subtypes, immunohistochemistry, fish or molecular screening for the accurate diagnosis of alk-rearrangement in lung cancer: A comprehensive study of caucasian non-smokers. Lung Cancer 2012, 76, 309–315. [Google Scholar] [CrossRef]
- McLeer-Florin, A.; Moro-Sibilot, D.; Melis, A.; Salameire, D.; Lefebvre, C.; Ceccaldi, F.; de Fraipont, F.; Brambilla, E.; Lantuejoul, S. Dual ihc and fish testing for alk gene rearrangement in lung adenocarcinomas in a routine practice: A french study. J. Thorac. Oncol. 2012, 7, 348–354. [Google Scholar] [CrossRef] [Green Version]
- Park, H.S.; Lee, J.K.; Kim, D.W.; Kulig, K.; Kim, T.M.; Lee, S.H.; Jeon, Y.K.; Chung, D.H.; Heo, D.S. Immunohistochemical screening for anaplastic lymphoma kinase (alk) rearrangement in advanced non-small cell lung cancer patients. Lung Cancer 2012, 77, 288–292. [Google Scholar] [CrossRef]
- Conklin, C.M.; Craddock, K.J.; Have, C.; Laskin, J.; Couture, C.; Ionescu, D.N. Immunohistochemistry is a reliable screening tool for identification of alk rearrangement in non-small-cell lung carcinoma and is antibody dependent. J. Thorac. Oncol. 2013, 8, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Selinger, C.I.; Rogers, T.M.; Russell, P.A.; O’Toole, S.; Yip, P.; Wright, G.M.; Wainer, Z.; Horvath, L.G.; Boyer, M.; McCaughan, B.; et al. Testing for alk rearrangement in lung adenocarcinoma: A multicenter comparison of immunohistochemistry and fluorescent in situ hybridization. Mod. Pathol. 2013, 26, 1545–1553. [Google Scholar] [CrossRef] [Green Version]
- Conde, E.; Suarez-Gauthier, A.; Benito, A.; Garrido, P.; Garcia-Campelo, R.; Biscuola, M.; Paz-Ares, L.; Hardisson, D.; de Castro, J.; Camacho, M.C.; et al. Accurate identification of alk positive lung carcinoma patients: Novel fda-cleared automated fluorescence in situ hybridization scanning system and ultrasensitive immunohistochemistry. PLoS ONE 2014, 9, e107200. [Google Scholar] [CrossRef] [Green Version]
- Wynes, M.W.; Sholl, L.M.; Dietel, M.; Schuuring, E.; Tsao, M.S.; Yatabe, Y.; Tubbs, R.R.; Hirsch, F.R. An international interpretation study using the alk ihc antibody d5f3 and a sensitive detection kit demonstrates high concordance between alk ihc and alk fish and between evaluators. J. Thorac. Oncol. 2014, 9, 631–638. [Google Scholar] [CrossRef] [Green Version]
- Soda, M.; Isobe, K.; Inoue, A.; Maemondo, M.; Oizumi, S.; Fujita, Y.; Gemma, A.; Yamashita, Y.; Ueno, T.; Takeuchi, K.; et al. A prospective pcr-based screening for the eml4-alk oncogene in non-small cell lung cancer. Clin. Cancer Res. 2012, 18, 5682–5689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Pan, Y.; Wang, R.; Sun, Y.; Hu, H.; Shen, X.; Lu, Y.; Shen, L.; Zhu, X.; Chen, H. Alk-rearranged lung cancer in chinese: A comprehensive assessment of clinicopathology, ihc, fish and rt-pcr. PLoS ONE 2013, 8, e69016. [Google Scholar] [CrossRef] [Green Version]
- Nakamichi, S.; Seike, M.; Miyanaga, A.; Chiba, M.; Matsuda, K.; Kobayashi, K.; Takahashi, A.; Takeuchi, S.; Minegishi, Y.; Kubota, K.; et al. Rt-pcr for detecting alk translocations in cytology samples from lung cancer patients. Anticancer Res. 2017, 37, 3295–3299. [Google Scholar]
- Pekar-Zlotin, M.; Hirsch, F.R.; Soussan-Gutman, L.; Ilouze, M.; Dvir, A.; Boyle, T.; Wynes, M.; Miller, V.A.; Lipson, D.; Palmer, G.A.; et al. Fluorescence in situ hybridization, immunohistochemistry, and next-generation sequencing for detection of eml4-alk rearrangement in lung cancer. Oncologist 2015, 20, 316–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, S.M.; Hensing, T.; Schrock, A.B.; Allen, J.; Sanford, E.; Gowen, K.; Kulkarni, A.; He, J.; Suh, J.H.; Lipson, D.; et al. Comprehensive genomic profiling identifies a subset of crizotinib-responsive alk-rearranged non-small cell lung cancer not detected by fluorescence in situ hybridization. Oncologist 2016, 21, 762–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Wu, S.; Huang, F.; Shen, M.; Jiang, H.; Yu, Y.; Yu, Q.; Yang, Y.; Zhao, Y.; Zhou, Y.; et al. Analytical and clinical validation of a novel amplicon-based ngs assay for the evaluation of circulating tumor DNA in metastatic colorectal cancer patients. Clin. Chem. Lab. Med. 2019, 57, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yang, X.; He, Y.; Ma, Q.; Lin, L.; Fu, P.; Xiao, H. Droplet digital pcr for absolute quantification of eml4-alk gene rearrangement in lung adenocarcinoma. J. Mol. Diagn. 2015, 17, 515–520. [Google Scholar] [CrossRef] [Green Version]
- Lund, H.L.; Hughesman, C.B.; Fakhfakh, K.; McNeil, K.; Clemens, S.; Hocken, K.; Pettersson, R.; Karsan, A.; Foster, L.J.; Haynes, C. Initial diagnosis of alk-positive non-small-cell lung cancer based on analysis of alk status utilizing droplet digital pcr. Anal. Chem. 2016, 88, 4879–4885. [Google Scholar] [CrossRef]
- Junca, A.; Tachon, G.; Evrard, C.; Villalva, C.; Frouin, E.; Karayan-Tapon, L.; Tougeron, D. Detection of colorectal cancer and advanced adenoma by liquid biopsy (decalib study): The ddpcr challenge. Cancers 2020, 12, 1482. [Google Scholar] [CrossRef]
- Ferrara, R.; Auger, N.; Auclin, E.; Besse, B. Clinical and translational implications of ret rearrangements in non-small cell lung cancer. J. Thorac. Oncol. 2018, 13, 27–45. [Google Scholar] [CrossRef] [Green Version]
- Luk, P.P.; Selinger, C.I.; Mahar, A.; Cooper, W.A. Biomarkers for alk and ros1 in lung cancer: Immunohistochemistry and fluorescent in situ hybridization. Arch. Pathol. Lab. Med. 2018, 142, 922–928. [Google Scholar] [CrossRef] [Green Version]
- Piton, N.; Ruminy, P.; Gravet, C.; Marchand, V.; Colasse, E.; Lamy, A.; Naoures Mear, C.L.; Bibeau, F.; Marguet, F.; Guisier, F.; et al. Ligation-dependent rt-pcr: A new specific and low-cost technique to detect alk, ros, and ret rearrangements in lung adenocarcinoma. Lab. Investig. 2018, 98, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Sung, J.S.; Chong, H.Y.; Kwon, N.J.; Kim, H.M.; Lee, J.W.; Kim, B.; Lee, S.B.; Park, C.W.; Choi, J.Y.; Chang, W.J.; et al. Detection of somatic variants and egfr mutations in cell-free DNA from non-small cell lung cancer patients by ultra-deep sequencing using the ion ampliseq cancer hotspot panel and droplet digital polymerase chain reaction. Oncotarget 2017, 8, 106901–106912. [Google Scholar] [CrossRef] [Green Version]
- Yamaura, T.; Muto, S.; Mine, H.; Takagi, H.; Watanabe, M.; Ozaki, Y.; Inoue, T.; Fukuhara, M.; Okabe, N.; Matsumura, Y.; et al. Genetic alterations in epidermal growth factor receptor-tyrosine kinase inhibitor-naive non-small cell lung carcinoma. Oncol. Lett. 2020, 19, 4169–4176. [Google Scholar]
- Li, C.; He, Q.; Liang, H.; Cheng, B.; Li, J.; Xiong, S.; Zhao, Y.; Guo, M.; Liu, Z.; He, J.; et al. Diagnostic accuracy of droplet digital pcr and amplification refractory mutation system pcr for detecting egfr mutation in cell-free DNA of lung cancer: A meta-analysis. Front. Oncol. 2020, 10, 290. [Google Scholar] [CrossRef] [Green Version]

| Cancer Type/Subtype | FISH | MLPA | ddPCR | References |
|---|---|---|---|---|
| Chronic Lymphocytic leukaemia |
|
|
| [7,43,47,48,49,50,51,52,53,54,55,56,57,58,59,60] |
| Celebral cancer (Gliomas, glioblastoma) |
|
|
| [61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95] |
| Breast cancer |
|
|
| [96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113] |
| Colorectal cancer |
|
|
| [114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141] |
| Lung cancer |
|
|
| [1,9,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cusenza, V.Y.; Bisagni, A.; Rinaldini, M.; Cattani, C.; Frazzi, R. Copy Number Variation and Rearrangements Assessment in Cancer: Comparison of Droplet Digital PCR with the Current Approaches. Int. J. Mol. Sci. 2021, 22, 4732. https://doi.org/10.3390/ijms22094732
Cusenza VY, Bisagni A, Rinaldini M, Cattani C, Frazzi R. Copy Number Variation and Rearrangements Assessment in Cancer: Comparison of Droplet Digital PCR with the Current Approaches. International Journal of Molecular Sciences. 2021; 22(9):4732. https://doi.org/10.3390/ijms22094732
Chicago/Turabian StyleCusenza, Vincenza Ylenia, Alessandra Bisagni, Monia Rinaldini, Chiara Cattani, and Raffaele Frazzi. 2021. "Copy Number Variation and Rearrangements Assessment in Cancer: Comparison of Droplet Digital PCR with the Current Approaches" International Journal of Molecular Sciences 22, no. 9: 4732. https://doi.org/10.3390/ijms22094732
APA StyleCusenza, V. Y., Bisagni, A., Rinaldini, M., Cattani, C., & Frazzi, R. (2021). Copy Number Variation and Rearrangements Assessment in Cancer: Comparison of Droplet Digital PCR with the Current Approaches. International Journal of Molecular Sciences, 22(9), 4732. https://doi.org/10.3390/ijms22094732






