Fluorescence Correlation Spectroscopy Analysis of Effect of Molecular Crowding on Self-Assembly of β-Annulus Peptide into Artificial Viral Capsid
Abstract
1. Introduction
2. Results
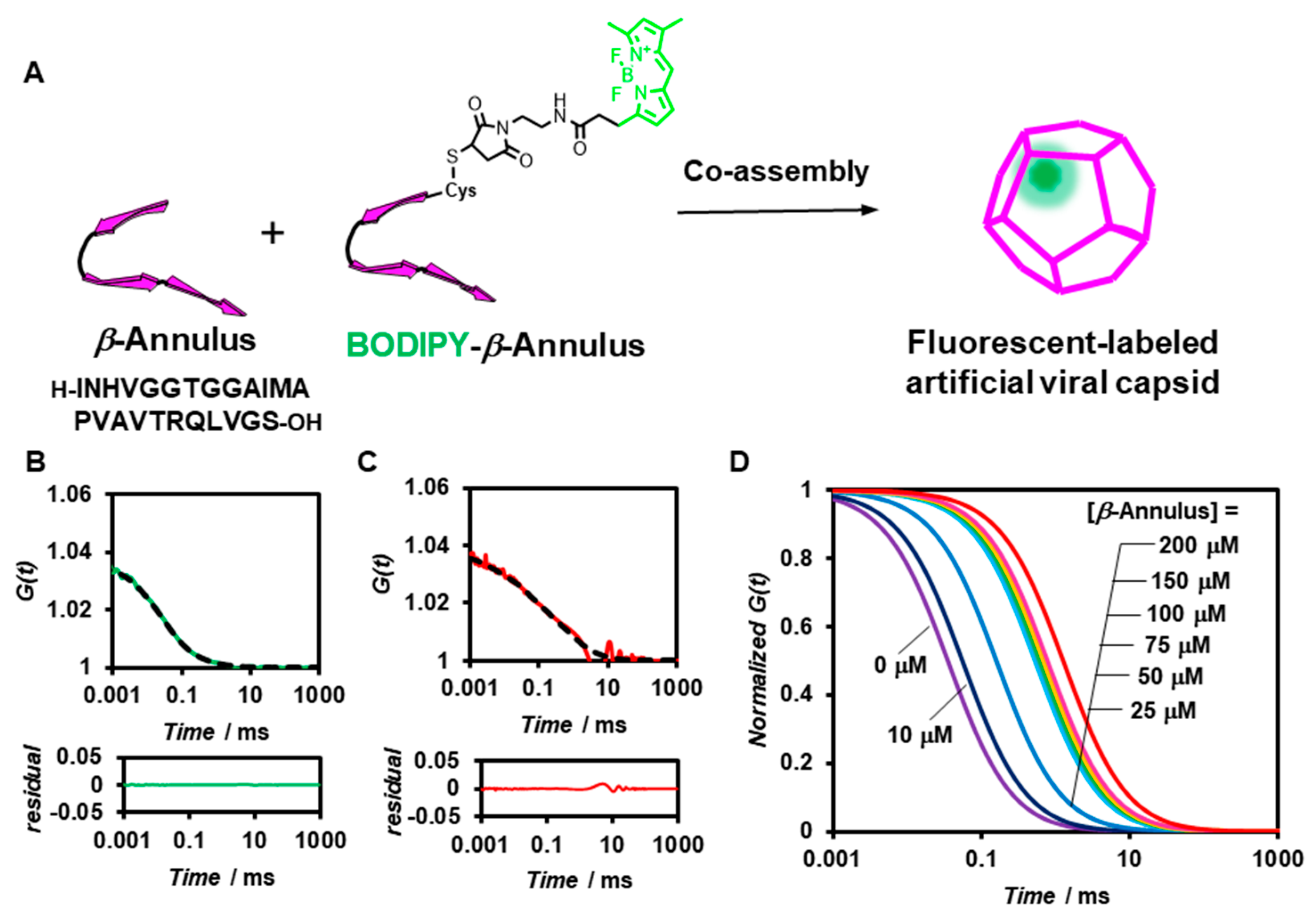
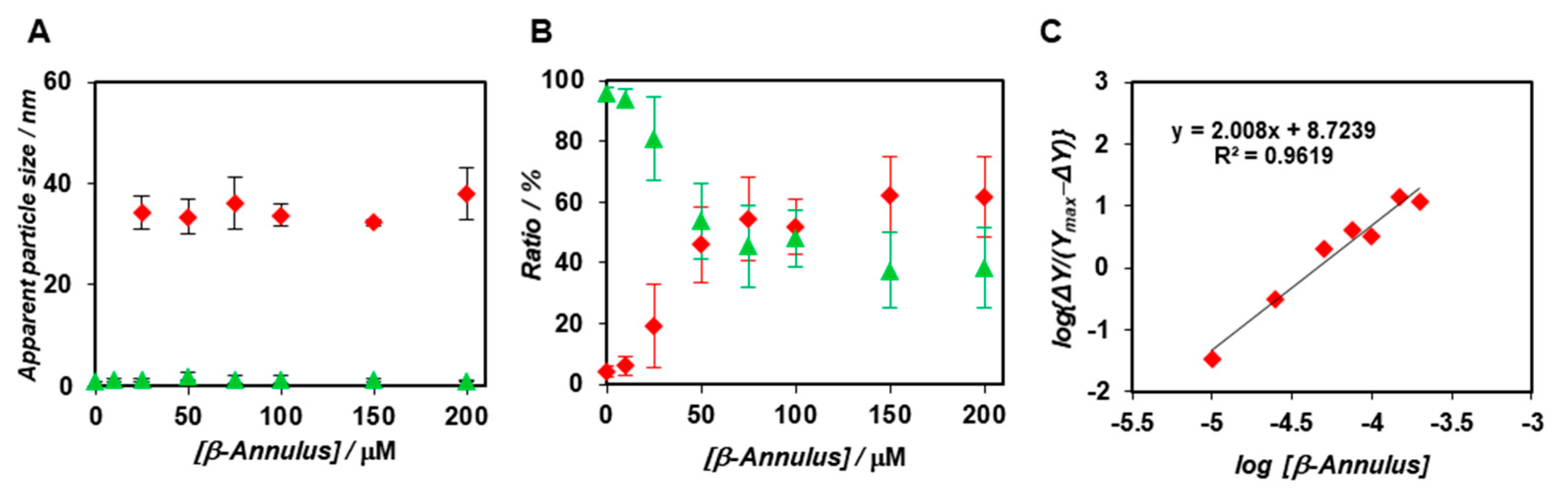
2.1. FCS Analyses of Artificial Viral Capsid Formation in Diluted Aqueous Solution
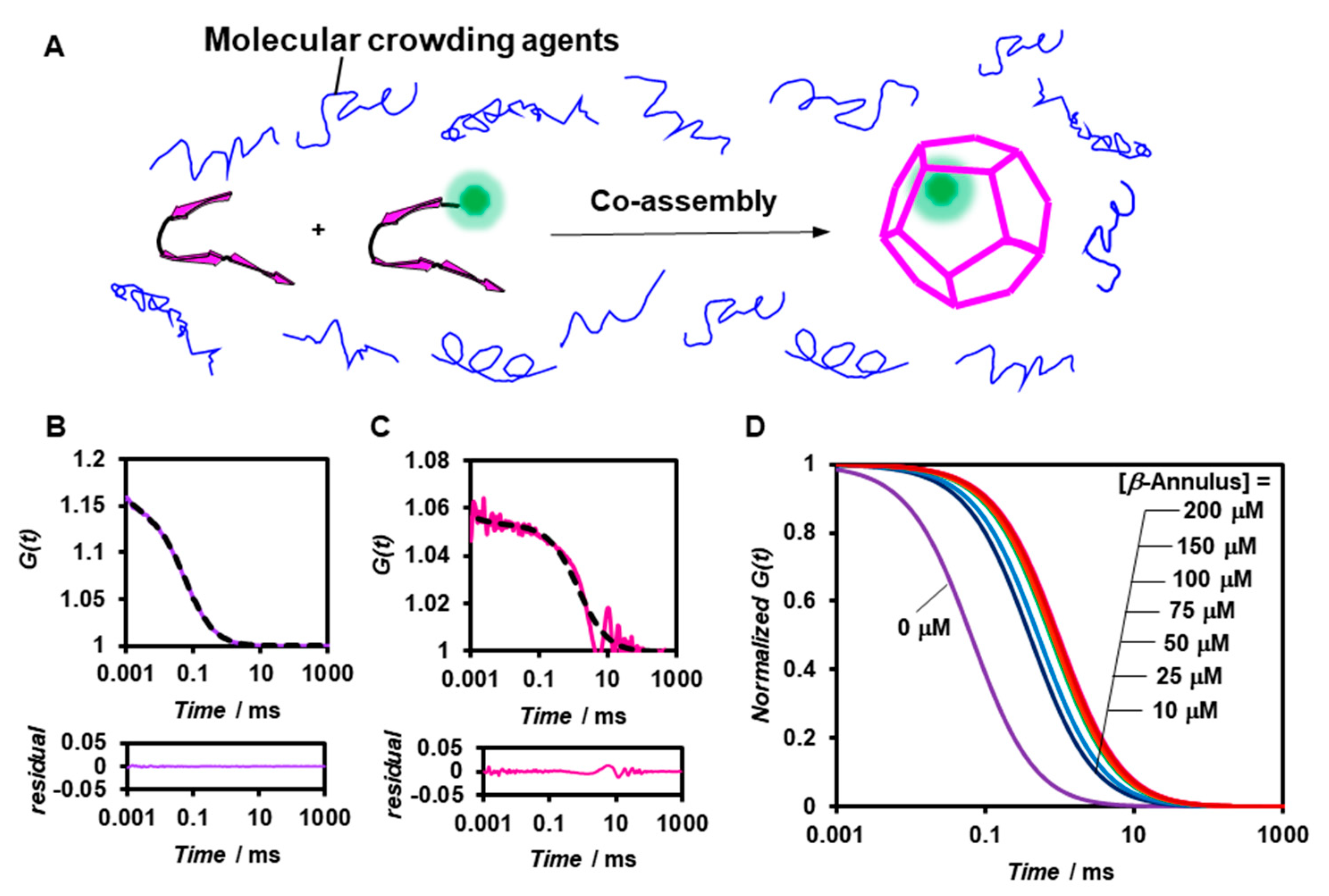
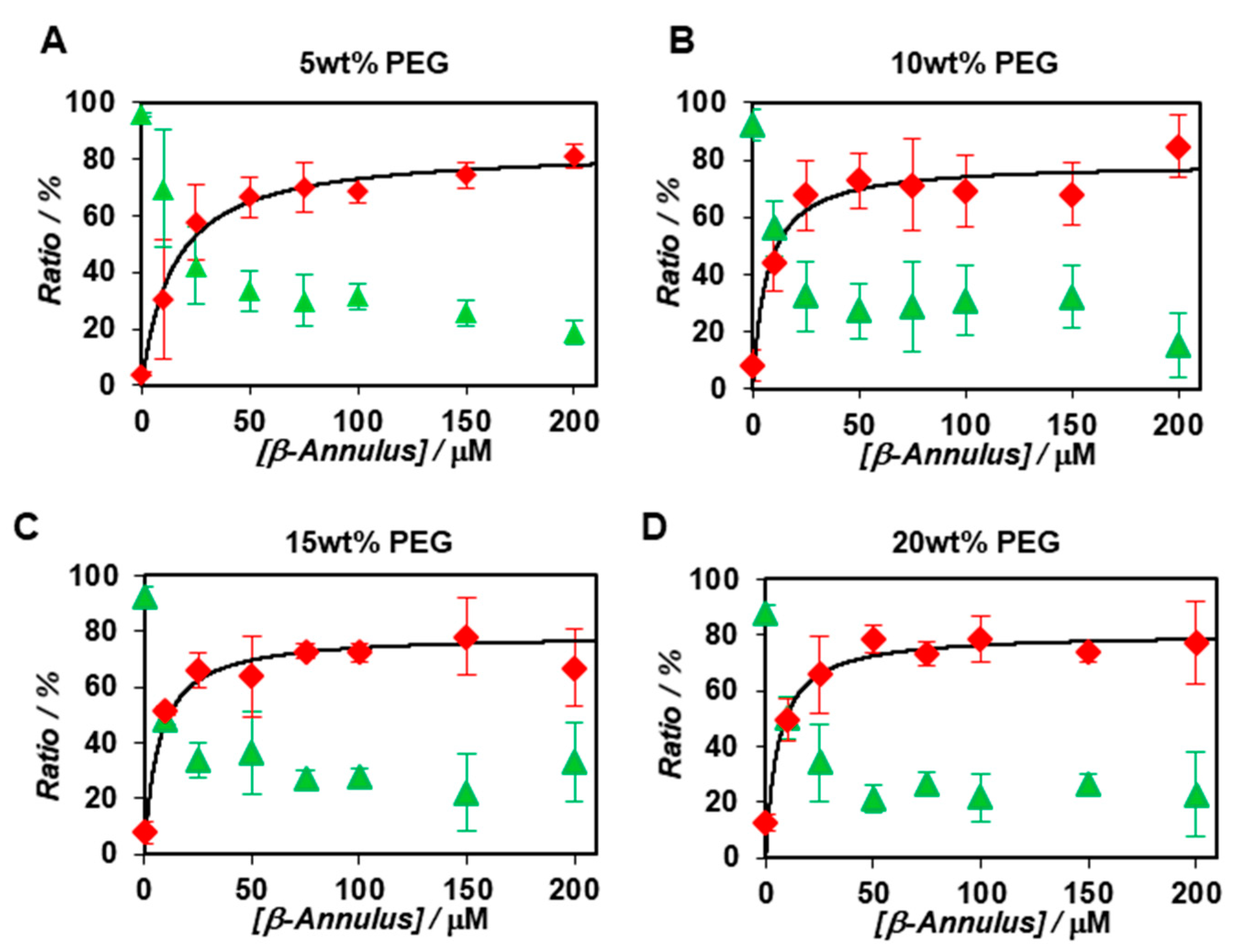
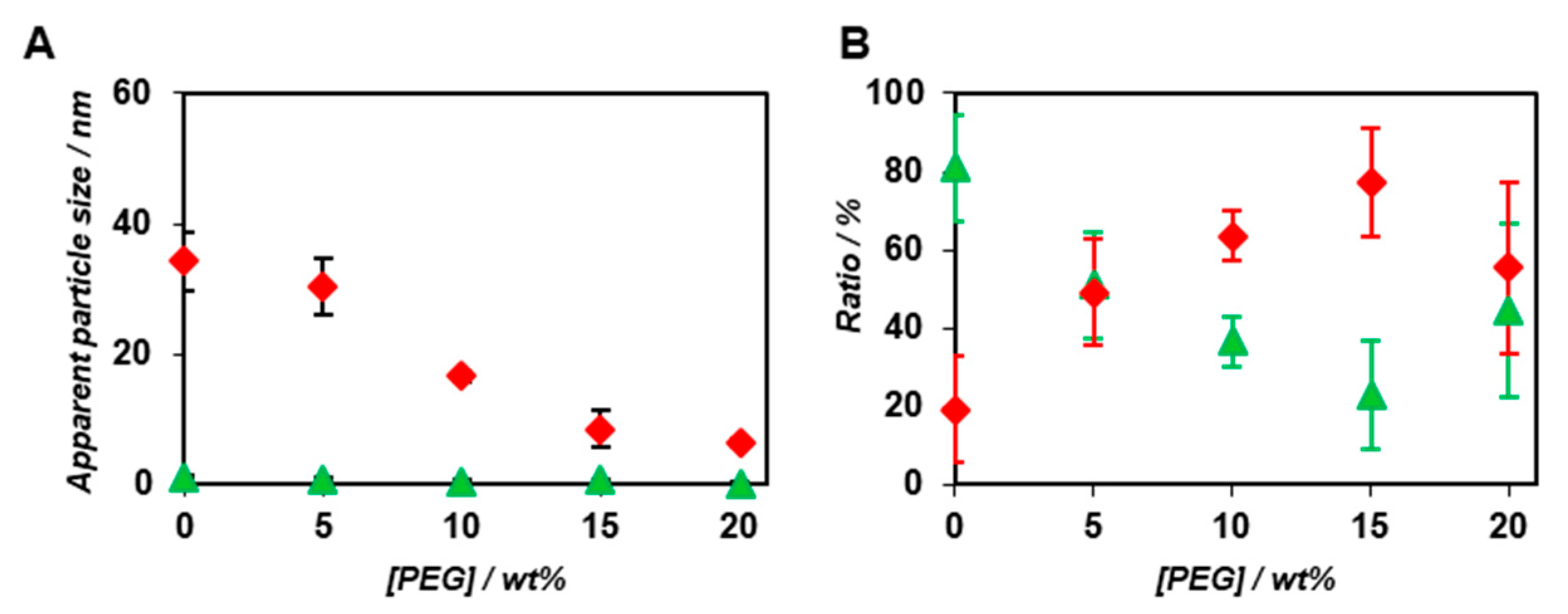
2.2. FCS Analyses of Artificial Viral Capsid Formation under Molecular Crowding Conditions.
3. Discussion
4. Materials and Methods
4.1. General
4.2. Synthesis of Cys-β-Annulus Peptide.
4.3. Synthesis of BODIPY-β-Annulus Peptide
4.4. Co-Assembly of BODIPY-β-Annulus Peptides and β-Annulus Peptides in Solutions Containing PEG
4.5. Fluorescence Correlation Spectroscopy (FCS)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fulton, A.B. How crowded is the cytoplasm? Cell 1982, 30, 345–347. [Google Scholar] [CrossRef]
- Nakano, S.; Miyoshi, D.; Sugimoto, N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem. Rev. 2014, 114, 2733–2758. [Google Scholar] [CrossRef]
- Minton, A.P. Implications of macromolecular crowding for protein assembly. Curr. Opin. Struct. Biol. 2000, 10, 34–39. [Google Scholar] [CrossRef]
- Minton, A.P. The Influence of Macromolecular Crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 2001, 276, 10577–10580. [Google Scholar] [CrossRef]
- Ma, Q.; Fan, J.-B.; Zhou, Z.; Zhou, B.-R.; Meng, S.-R.; Hu, J.-Y.; Chen, J.; Liang, Y. The contrasting effect of macromolecular crowding on amyloid fibril formation. PLoS ONE 2012, 7, e36288. [Google Scholar] [CrossRef]
- Junker, N.O.; Vaghefikia, F.; Albarghash, A.; Höfig, H.; Kempe, D.; Walter, J.; Otten, J.; Pohl, M.; Katranidis, A.; Wiegand, S.; et al. Impact of molecular crowding on translational mobility and conformational properties of biological macromolecules. J. Phys. Chem. B 2019, 123, 4477–4486. [Google Scholar] [CrossRef]
- Sasahara, K.; McPhie, P.; Minton, A.P. Effect of dextran on protein stability and conformation attributed to macromolecular crowding. J. Mol. Biol. 2003, 326, 1227–1237. [Google Scholar] [CrossRef]
- Toluriki, N.; Kinjo, M.; Negi, S.; Hoshino, M.; Goto, Y.; Urabe, I.; Yomo, T. Protein folding by the effects of macromolecular crowding. Protein Sci. 2004, 13, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Khudyakov, Y.; Pumpens, P. (Eds.) Viral Nanotechnology; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- van Rijn, P.; Schirhagl, R. Viruses, artificial viruses and virus-based structures for biomedical applications. Adv. Healthc. Mater. 2016, 5, 1386–1400. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.L.; Twyman, R.M.; Fiering, S.; Steinmetz, N.F. Virus-based nanoparticles as platform technologies for modern vaccines. WIREs Nanomed. Nnobiotech. 2016, 8, 554–578. [Google Scholar] [CrossRef]
- Wen, A.M.; Steinmetz, N.F. Design of virus-based nanomaterials for medicine, biotechnology, and energy. Chem. Soc. Rev. 2016, 45, 4074–4126. [Google Scholar] [CrossRef]
- Perlmutter, J.D.; Hagan, M.F. Mechanisms of virus assembly. Annu. Rev. Phys. Chem. 2015, 66, 217–239. [Google Scholar] [CrossRef]
- Bruinsma, R.F.; Wuite, G.J.L.; Roos, W.H. Physics of viral dynamics. Nat. Rev. Phys. 2021, 3, 76–91. [Google Scholar] [CrossRef]
- Uetrecht, C.; Barbu, I.; Shoemaker, G.; Duijn, E.; Heck, A. Interrogating viral capsid assembly with ion mobility–mass spectrometry. Nat. Chem. 2011, 3, 126–132. [Google Scholar] [CrossRef]
- Medrano, M.; Fuertes, M.Á.; Valbuena, A.; Carrillo, P.J.P.; Rodríguez-Huete, A.; Mateu, M.G. Imaging and quantitation of a succession of transient intermediates reveal the reversible self-assembly pathway of a simple icosahedral virus capsid. J. Am. Chem. Soc. 2016, 138, 15385–15396. [Google Scholar] [CrossRef]
- Li, H.; Sun, J.; Xiao, S.; Zhang, L.; Zhou, D. Triterpenoid-mediated inhibition of virus-host interaction: Is now the time for discovering viral entry/release Inhibitors from nature? J. Med. Chem. 2020, 63, 15371–15388. [Google Scholar] [CrossRef]
- Alamo, M.; Rivas, G.; Mateu, M.G. Effect of macromolecular crowding agents on human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Virol. 2005, 79, 14271–14281. [Google Scholar] [CrossRef]
- Matsuurua, K. Rational design of self-assembled proteins and peptides for nano- and micro-sized architectures. RSC Adv. 2014, 4, 2942–2953. [Google Scholar] [CrossRef]
- Lou, S.; Wang, X.; Yu, Z.; Shi, L. Peptide tectonics: Encoded structural complementarity dictates programmable self-assembly. Adv. Sci. 2019, 6, 1802043. [Google Scholar] [CrossRef]
- De Santis, E.; Alkassem, H.; Lamarre, B.; Faruqui, N.; Bella, A.; Noble, J.E.; Micale, N.; Ray, S.; Burns, J.R.; Yon, A.R.; et al. Antimicrobial peptide capsids of de novo design. Nat. Commun. 2017, 8, 2263. [Google Scholar] [CrossRef]
- Kepiro, I.E.; Marzuoli, I.; Hammond, K.; Ba, X.; Lewis, H.; Shaw, M.; Gunnoo, S.B.; De Santis, E.; Łapińska, U.; Pagliara, S.; et al. Engineering chirally blind protein pseudocapsids into antibacterial persisters. ACS Nano 2020, 14, 1609–1622. [Google Scholar] [CrossRef]
- Matsuura, K. Synthetic approaches to construct viral capsid-like spherical nanomaterials. Chem. Commun. 2018, 54, 8944–8959. [Google Scholar] [CrossRef]
- Matsuura, K.; Watanabe, K.; Sakurai, K.; Matsuzaki, T.; Kimizuka, N. Self-assembled synthetic viral capsids from a 24-mer viral peptide fragment. Angew. Chem. Int. Ed. 2010, 49, 9662–9665. [Google Scholar] [CrossRef]
- Matsuura, K.; Watanabe, K.; Matsushita, Y.; Kimizuka, N. Guest-binding behavior of peptide nanocapsules self-assembled from viral peptide fragments. Polymer J. 2013, 45, 529–534. [Google Scholar] [CrossRef]
- Nakamura, Y.; Inaba, H.; Matsuura, K. Construction of artificial viral capsids encapsulating short DNAs via disulfide bonds and controlled release of DNAs by reduction. Chem. Lett. 2019, 48, 544–546. [Google Scholar] [CrossRef]
- Fujita, S.; Matsuura, K. Encapsulation of CdTe quantum dots into synthetic viral capsids. Chem. Lett. 2016, 45, 922–924. [Google Scholar] [CrossRef]
- Matsuura, K.; Ueno, G.; Fujita, S. Self-assembled artificial viral capsid decorated with gold nanoparticles. Polymer J. 2015, 47, 146–151. [Google Scholar] [CrossRef]
- Fujita, S.; Matsuura, K. Self-assembled artificial viral capsids bearing coiled-coils at the surface. Org. Biomol. Chem. 2017, 15, 5070–5077. [Google Scholar] [CrossRef]
- Nakamura, Y.; Yamada, S.; Nishikawa, S.; Matsuura, K. DNA-modified artificial viral capsids self-assembled from DNA-conjugated β-annulus peptide. J. Pept. Sci. 2017, 23, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Honjo, T. Artificial viral capsid dressed up with human serum albumin. Bioconjug. Chem. 2019, 30, 1636–1641. [Google Scholar] [CrossRef]
- Matsuura, K.; Ota, J.; Fujita, S.; Shiomi, Y.; Inaba, H. Construction of ribonuclease-decorated artificial virus-like capsid by peptide self-assembly. J. Org. Chem. 2020, 85, 1668–1673. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, H.; Inaba, H.; Inoue, F.; Sasaki, Y.; Akiyoshi, K.; Matsuura, K. Enveloped artificial viral capsids self-assembled from anionic β-annulus peptide and cationic lipid bilayer. Chem. Commun. 2020, 56, 7092–7095. [Google Scholar] [CrossRef] [PubMed]
- Al-Soufi, W.; Reija, B.; Felekyan, S.; Seidel, C.A.M.; Novo, M. Dynamics of supramolecular association monitored by fluorescence correlation spectroscopy. ChemPhysChem 2008, 9, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Comas-Garcia, M.; Garmann, R.F.; Singaram, W.S.; Ben-Shaul, A.; Knobler, C.M.; Gebart, W.M. Characterization of viral capsid protein self-assembly around short single-stranded RNA. J. Phys. Chem. B 2014, 118, 7510–7519. [Google Scholar] [CrossRef]
| Solvent | Kd/μM | ΔG/kJ mol−1 |
|---|---|---|
| 10 mM Tris-HCl buffer (pH 7.0) | 45.2 ± 10.6 | −24.8 ± 0.739 |
| 5 wt% PEG2000 | 14.4 ± 7.81 | −27.6 ± 0.899 |
| 10 wt% PEG2000 | 6.69 ± 3.25 | −29.5 ± 0.952 |
| 15 wt% PEG2000 | 4.24 ± 3.22 | −30.7 ± 1.27 |
| 20 wt% PEG2000 | 5.66 ± 2.15 | −29.9 ± 0.710 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kobayashi, R.; Inaba, H.; Matsuura, K. Fluorescence Correlation Spectroscopy Analysis of Effect of Molecular Crowding on Self-Assembly of β-Annulus Peptide into Artificial Viral Capsid. Int. J. Mol. Sci. 2021, 22, 4754. https://doi.org/10.3390/ijms22094754
Kobayashi R, Inaba H, Matsuura K. Fluorescence Correlation Spectroscopy Analysis of Effect of Molecular Crowding on Self-Assembly of β-Annulus Peptide into Artificial Viral Capsid. International Journal of Molecular Sciences. 2021; 22(9):4754. https://doi.org/10.3390/ijms22094754
Chicago/Turabian StyleKobayashi, Risako, Hiroshi Inaba, and Kazunori Matsuura. 2021. "Fluorescence Correlation Spectroscopy Analysis of Effect of Molecular Crowding on Self-Assembly of β-Annulus Peptide into Artificial Viral Capsid" International Journal of Molecular Sciences 22, no. 9: 4754. https://doi.org/10.3390/ijms22094754
APA StyleKobayashi, R., Inaba, H., & Matsuura, K. (2021). Fluorescence Correlation Spectroscopy Analysis of Effect of Molecular Crowding on Self-Assembly of β-Annulus Peptide into Artificial Viral Capsid. International Journal of Molecular Sciences, 22(9), 4754. https://doi.org/10.3390/ijms22094754







