Pancreatic Cancer and Therapy: Role and Regulation of Cancer Stem Cells
Abstract
1. Introduction
1.1. Characteristic Features of Cancer Stem Cells
1.2. Cancer Stem Cells and PC Heterogeneity
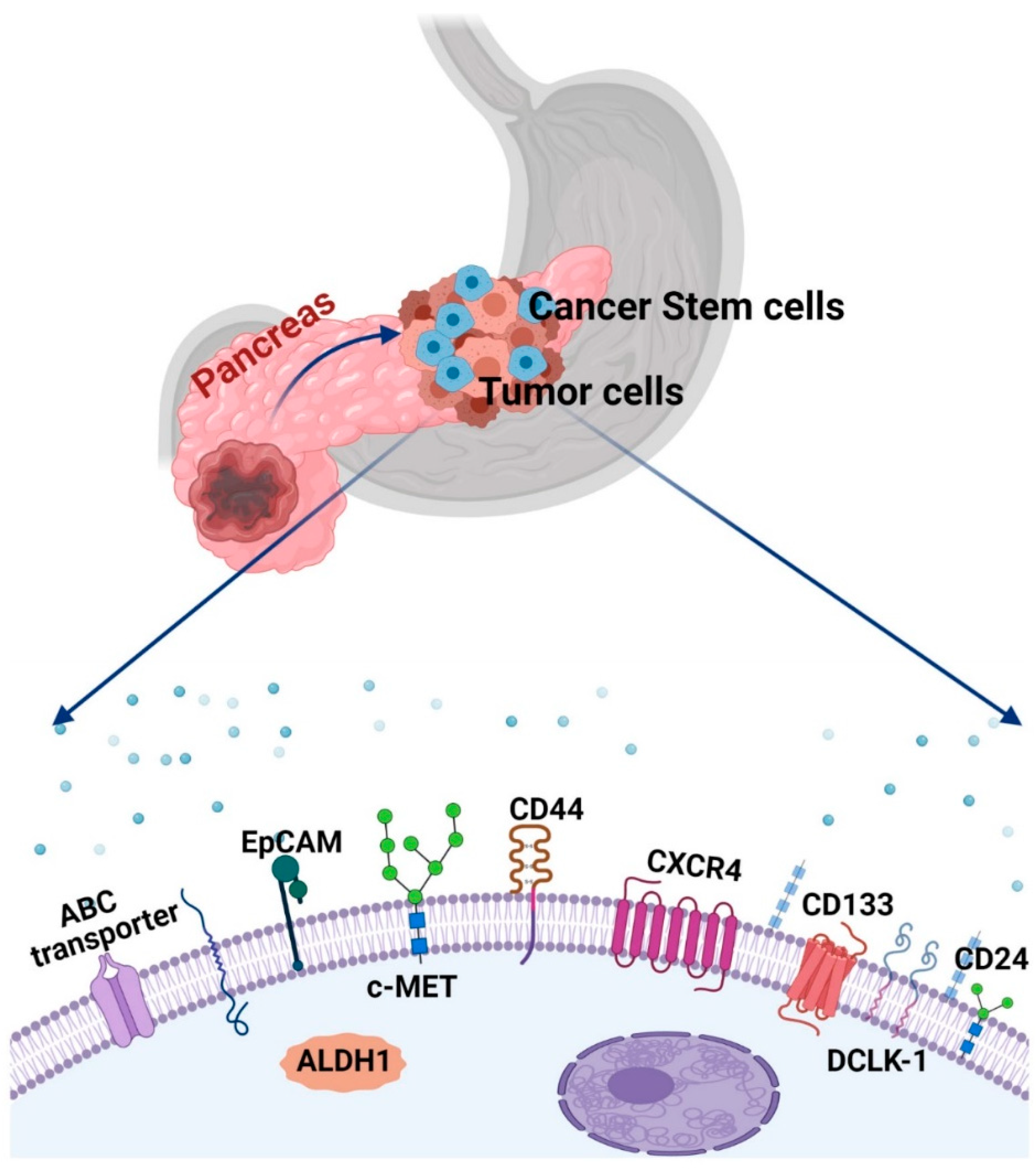
1.3. Strategies for Identification and Isolation of PaCSCs
2. Targeting Major Signaling Pathways to Regulate CSCs for PC Therapy
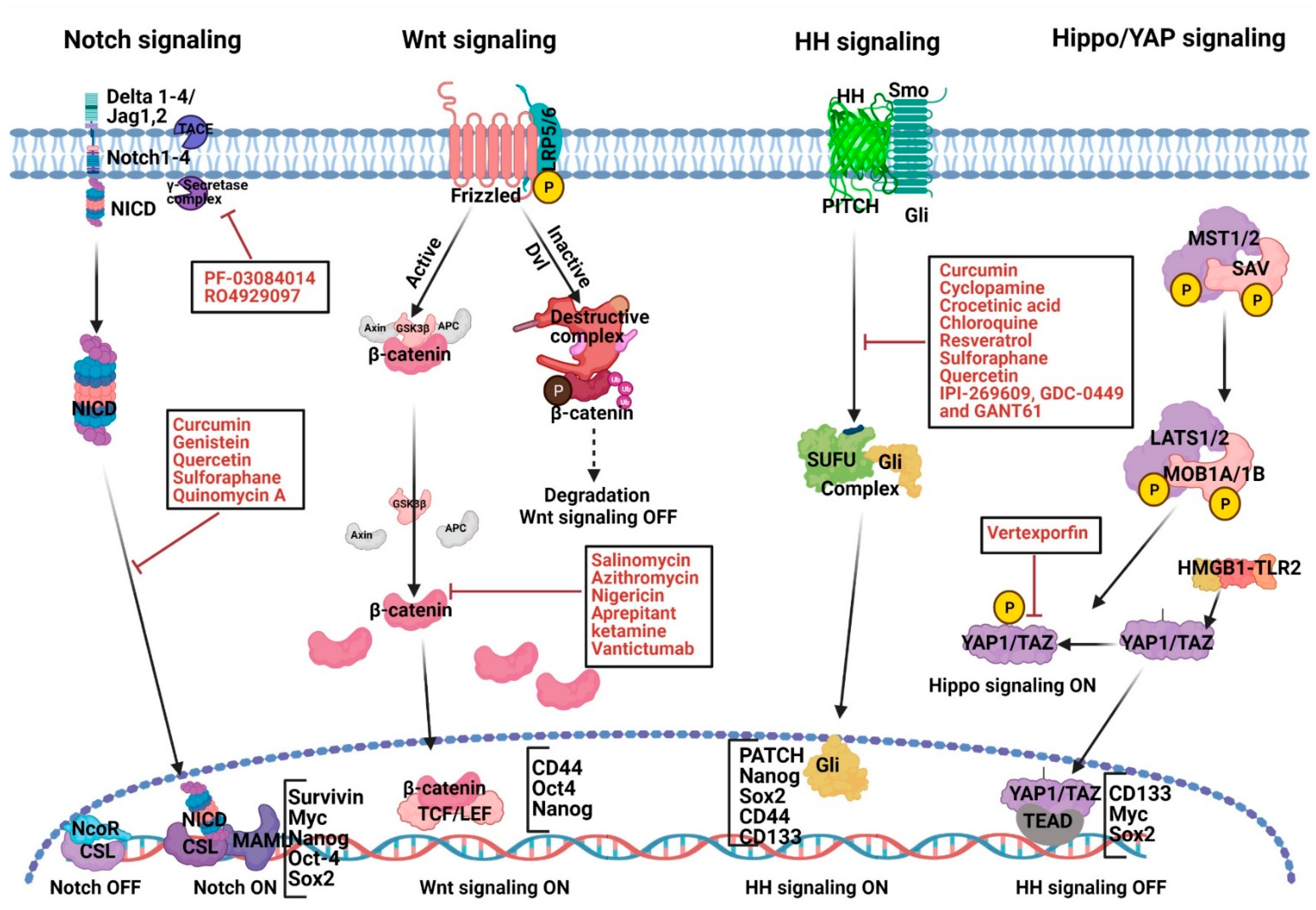
2.1. Abnormal Notch Signaling Activation and Therapeutic Strategies in PC Development
2.2. Abnormal Wnt Signaling Activation and Therapeutic Strategies in PC Development
2.3. Abnormal HH Signaling Activation, PaCSCs and Therapeutic Strategies
2.4. Targeting Hippo Signaling
2.5. Targeting JAK-STAT Pathway
2.6. Targeting PI3K/Akt/mTOR Signaling
2.7. Targeting MAPK-ERK Pathway
2.8. Targeting CXCR4 Signaling
2.9. Targeting NODAL/ACTIVIN Signaling
2.10. Targeting MicroRNAs to Regulate CSCs for PC Therapy
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Global Cancer Observatory. Available online: https://gco.iarc.fr/ (accessed on 17 March 2021).
- Survival Rates for Pancreatic Cancer. Available online: https://www.cancer.org/cancer/pancreatic-cancer/detection-diagnosis-staging/survival-rates.html (accessed on 15 March 2021).
- Khalaf, N.; El-Serag, H.B.; Abrams, H.R.; Thrift, A.P. Burden of Pancreatic Cancer: From Epidemiology to Practice. Clin. Gastroenterol. Hepatol. 2020, 19, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lok, V.; Ngai, C.H.; Zhang, L.; Yuan, J.; Lao, X.Q.; Ng, K.; Chong, C.; Zheng, Z.-J.; Wong, M.C.S. Worldwide Burden of, Risk Factors for, and Trends in Pancreatic Cancer. Gastroenterology 2021, 160, 744–754. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.-M.; Gingras, M.-C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic Analyses Identify Molecular Subtypes of Pancreatic Cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.H.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual Microdissection Identifies Distinct Tumor- and Stroma-Specific Subtypes of Pancreatic Ductal Adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Korc, M. Role of Growth Factors in Pancreatic Cancer. Surg. Oncol. Clin. N. Am. 1998, 7, 25–41. [Google Scholar] [CrossRef]
- Neoptolemos, J.P.; Stocken, D.D.; Friess, H.; Bassi, C.; Dunn, J.A.; Hickey, H.; Beger, H.; Fernandez-Cruz, L.; Dervenis, C.; Lacaine, F.; et al. A Randomized Trial of Chemoradiotherapy and Chemotherapy after Resection of Pancreatic Cancer. N. Engl. J. Med. 2004, 350, 1200–1210. [Google Scholar] [CrossRef]
- Yue, Q.; Gao, G.; Zou, G.; Yu, H.; Zheng, X. Natural Products as Adjunctive Treatment for Pancreatic Cancer: Recent Trends and Advancements. Biomed. Res. Int. 2017, 2017, 1–13. [Google Scholar] [CrossRef]
- Zong, Y.; Peng, Z.; Wang, X.; Lu, M.; Shen, L.; Zhou, J. Efficacy and Safety of Nab-Paclitaxel Plus S-1 versus Nab-Paclitaxel Plus Gemcitabine for First-Line Chemotherapy in Advanced Pancreatic Ductal Adenocarcinoma. Cancer Manag. Res. 2020, 12, 12657–12666. [Google Scholar] [CrossRef]
- Mortezaee, K. Enriched Cancer Stem Cells, Dense Stroma, and Cold Immunity: Interrelated Events in Pancreatic Cancer. J. Biochem. Mol. Toxicol. N/A 2021, e22708. [Google Scholar] [CrossRef]
- Das, P.K.; Pillai, S.; Md Rakib, A.; Khanam, J.A.; Gopalan, V.; Lam, A.K.Y.; Islam, F. Plasticity of Cancer Stem Cell: Origin and Role in Disease Progression and Therapy Resistance. Stem Cell Rev. Rep. 2020, 16, 397–412. [Google Scholar] [CrossRef]
- Garcia-Mayea, Y.; Mir, C.; Masson, F.; Paciucci, R.; LLeonart, M.E. Insights into New Mechanisms and Models of Cancer Stem Cell Multidrug Resistance. Semin. Cancer Biol. 2020, 60, 166–180. [Google Scholar] [CrossRef]
- Marcu, L.G. Cancer stem cells as therapeutic targets of pancreatic cancer. Fundam. Clin. Pharmacol. 2020, 34, 200–201. [Google Scholar] [CrossRef] [PubMed]
- Valle, S.; Martin-Hijano, L.; Alcalá, S.; Alonso-Nocelo, M.; Sainz, B., Jr. The ever-evolving concept of the cancer stem cell in pancreatic cancer. Cancers 2018, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Tang, D. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012, 22, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Eun, K.; Ham, S.W.; Kim, H. Cancer stem cell heterogeneity: Origin and new perspectives on CSC targeting. BMB Rep. 2017, 50, 117. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.J.; Dosch, J.; Simeone, D.M. Pancreatic Cancer Stem Cells. JCO 2008, 26, 2806–2812. [Google Scholar] [CrossRef] [PubMed]
- Nimmakayala, R.K.; Batra, S.K.; Ponnusamy, M.P. Unraveling the Journey of Cancer Stem Cells from Origin to Metastasis. Biochim. Biophys. Acta (BBA) Rev. Cancer 2019, 1871, 50–63. [Google Scholar] [CrossRef]
- Steinbichler, T.B.; Savic, D.; Dudás, J.; Kvitsaridze, I.; Skvortsov, S.; Riechelmann, H.; Skvortsova, I.-I. Cancer Stem Cells and Their Unique Role in Metastatic Spread. Semin. Cancer Biol. 2020, 60, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of Pancreatic Cancer Stem Cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.J.; Chiu, T.-J.; Lin, Y.C.; Weng, C.-C.; Weng, Y.-T.; Hsiao, C.-C.; Cheng, K. Inactivation of APC Induces CD34 Upregulation to Promote Epithelial-Mesenchymal Transition and Cancer Stem Cell Traits in Pancreatic Cancer. Int. J. Mol. Sci. 2020, 21, 4473. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.P.; Wen, J.; Bang, S.; Park, S.; Song, S.Y. CD44-Positive Cells Are Responsible for Gemcitabine Resistance in Pancreatic Cancer Cells. Int. J. Cancer 2009, 125, 2323–2331. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, H.; Gu, J.; Zheng, L. Prognostic Value of Cancer Stem Cell Marker CD133 Expression in Pancreatic Ductal Adenocarcinoma (PDAC): A Systematic Review and Meta-Analysis. Int. J. Clin. Exp. Pathol. 2015, 8, 12084–12092. [Google Scholar]
- Nomura, A.; Banerjee, S.; Chugh, R.; Dudeja, V.; Yamamoto, M.; Vickers, S.M.; Saluja, A.K. CD133 Initiates Tumors, Induces Epithelial-Mesenchymal Transition and Increases Metastasis in Pancreatic Cancer. Oncotarget 2015, 6, 8313–8322. [Google Scholar] [CrossRef]
- Wei, H.-J.; Yin, T.; Zhu, Z.; Shi, P.-F.; Tian, Y.; Wang, C.-Y. Expression of CD44, CD24 and ESA in Pancreatic Adenocarcinoma Cell Lines Varies with Local Microenvironment. Hepatobiliary Pancreat. Dis. Int. 2011, 10, 428–434. [Google Scholar] [CrossRef]
- Kahlert, C.; Bergmann, F.; Beck, J.; Welsch, T.; Mogler, C.; Herpel, E.; Dutta, S.; Niemietz, T.; Koch, M.; Weitz, J. Low Expression of Aldehyde Deyhdrogenase 1A1 (ALDH1A1) Is a Prognostic Marker for Poor Survival in Pancreatic Cancer. BMC Cancer 2011, 11, 275. [Google Scholar] [CrossRef]
- Maréchal, R.; Demetter, P.; Nagy, N.; Berton, A.; Decaestecker, C.; Polus, M.; Closset, J.; Devière, J.; Salmon, I.; Van Laethem, J.-L. High Expression of CXCR4 May Predict Poor Survival in Resected Pancreatic Adenocarcinoma. Br. J. Cancer 2009, 100, 1444–1451. [Google Scholar] [CrossRef]
- Bailey, J.M.; Alsina, J.; Rasheed, Z.A.; McAllister, F.M.; Fu, Y.; Plentz, R.; Zhang, H.; Pasricha, P.J.; Bardeesy, N.; Matsui, W.; et al. DCLK1 Marks a Morphologically Distinct Subpopulation of Cells With Stem Cell Properties in Preinvasive Pancreatic Cancer. Gastroenterology 2014, 146, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Tanaka, S.; Akiyama, Y.; Shimada, S.; Adikrisna, R.; Matsumura, S.; Aihara, A.; Mitsunori, Y.; Ban, D.; Ochiai, T.; et al. Dominant Expression of DCLK1 in Human Pancreatic Cancer Stem Cells Accelerates Tumor Invasion and Metastasis. PLoS ONE 2016, 11, e0146564. [Google Scholar] [CrossRef] [PubMed]
- Ohara, Y.; Oda, T.; Sugano, M.; Hashimoto, S.; Enomoto, T.; Yamada, K.; Akashi, Y.; Miyamoto, R.; Kobayashi, A.; Fukunaga, K.; et al. Histological and Prognostic Importance of CD44+/CD24+/EpCAM+ Expression in Clinical Pancreatic Cancer. Cancer Sci. 2013, 104, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; He, J.; Liu, Y.; Simeone, D.M.; Lubman, D.M. Identification of Glycoprotein Markers for Pancreatic Cancer CD24+CD44+ Stem-like Cells Using Nano-LC–MS/MS and Tissue Microarray. J. Proteome Res. 2012, 11, 2272–2281. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.; D’Alterio, C.; Camerlingo, R.; Tirino, V.; Consales, C.; Riccio, A.; Ieranò, C.; Cecere, S.C.; Losito, N.S.; Greggi, S.; et al. Identification of a Distinct Population of CD133 + CXCR4 + Cancer Stem Cells in Ovarian Cancer. Sci. Rep. 2015, 5, 10357. [Google Scholar] [CrossRef] [PubMed]
- Sleightholm, R.L.; Neilsen, B.K.; Li, J.; Steele, M.M.; Singh, R.K.; Hollingsworth, M.A.; Oupicky, D. Emerging Roles of the CXCL12/CXCR4 Axis in Pancreatic Cancer Progression and Therapy. Pharmacol. Ther. 2017, 179, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, J.; Hynes, M.; Dosch, J.; Sarkar, B.; Welling, T.H.; di Magliano, M.; Simeone, D.M. C-Met Is a Marker of Pancreatic Cancer Stem Cells and Therapeutic Target. Gastroenterology 2011, 141, 2218–2227. [Google Scholar] [CrossRef] [PubMed]
- Herreros-Villanueva, M.; Zubia-Olascoaga, A.; Bujanda, L. C-Met in Pancreatic Cancer Stem Cells: Therapeutic Implications. World J. Gastroenterol 2012, 18, 5321–5323. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Yang, X.; Lassus, H.; Liang, S.; Kaur, S.; Ye, Q.; Li, C.; Wang, L.-P.; Roby, K.F.; Orsulic, S.; et al. Distinct Expression Levels and Patterns of Stem Cell Marker, Aldehyde Dehydrogenase Isoform 1 (ALDH1), in Human Epithelial Cancers. PLoS ONE 2010, 5, e10277. [Google Scholar] [CrossRef] [PubMed]
- Skoda, J.; Hermanova, M.; Loja, T.; Nemec, P.; Neradil, J.; Karasek, P.; Veselska, R. Co-Expression of Cancer Stem Cell Markers Corresponds to a Pro-Tumorigenic Expression Profile in Pancreatic Adenocarcinoma. PLoS ONE 2016, 11, e0159255. [Google Scholar]
- Gupta, V.K.; Sharma, N.S.; Kesh, K.; Dauer, P.; Nomura, A.; Giri, B.; Dudeja, V.; Banerjee, S.; Bhattacharya, S.; Saluja, A.; et al. Metastasis and Chemoresistance in CD133 Expressing Pancreatic Cancer Cells Are Dependent on Their Lipid Raft Integrity. Cancer Lett. 2018, 439, 101–112. [Google Scholar] [CrossRef]
- Ren, C.; Chen, H.; Han, C.; Wang, D.; Fu, D. Increased Plasma MicroRNA and CD133/CK18-positive Cancer Cells in the Pleural Fluid of a Pancreatic Cancer Patient with Liver and Pleural Metastases and Correlation with Chemoresistance. Oncol. Lett. 2012, 4, 691–694. [Google Scholar] [CrossRef]
- Mueller, M.; Hermann, P.C.; Witthauer, J.; Rubio–Viqueira, B.; Leicht, S.F.; Huber, S.; Ellwart, J.W.; Mustafa, M.; Bartenstein, P.; D’Haese, J.G.; et al. Combined Targeted Treatment to Eliminate Tumorigenic Cancer Stem Cells in Human Pancreatic Cancer. Gastroenterology 2009, 137, 1102–1113. [Google Scholar] [CrossRef]
- Kise, K.; Kinugasa-Katayama, Y.; Takakura, N. Tumor Microenvironment for Cancer Stem Cells. Adv. Drug. Deliv. Rev. 2016, 99, 197–205. [Google Scholar] [CrossRef]
- Bocci, F.; Gearhart-Serna, L.; Boareto, M.; Ribeiro, M.; Ben-Jacob, E.; Devi, G.R.; Levine, H.; Onuchic, J.N.; Jolly, M.K. Toward Understanding Cancer Stem Cell Heterogeneity in the Tumor Microenvironment. Proc. Natl. Acad. Sci. USA 2019, 116, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Begum, A.; McMillan, R.H.; Chang, Y.-T.; Penchev, V.R.; Maitra, A.; Goggins, M.G.; Eshelman, J.R.; Wolfgang, C.L.; Rasheed, Z.A. Direct Interactions With Cancer-Associated Fibroblasts Lead to Enhanced Pancreatic Cancer Stem Cell Function. Pancreas 2019, 48, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, X.; Contino, G.; Liesa, M.; Sahin, E.; Ying, H.; Bause, A.; Li, Y.; Stommel, J.M.; Dell’Antonio, G.; et al. Pancreatic Cancers Require Autophagy for Tumor Growth. Genes Dev. 2011, 25, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Bian, Q.; Zhang, Y.-J.; Shao, C.-H.; Li, G.; Liu, A.-A.; Jing, W.; Liu, R.; Zhou, Y.-Q.; Jin, G.; et al. Downregulation of ASPP2 in Pancreatic Cancer Cells Contributes to Increased Resistance to Gemcitabine through Autophagy Activation. Mol. Cancer 2015, 14, 177. [Google Scholar] [CrossRef]
- Yuen, C.A.; Asuthkar, S.; Guda, M.R.; Tsung, A.J.; Velpula, K.K. Cancer Stem Cell Molecular Reprogramming of the Warburg Effect in Glioblastomas: A New Target Gleaned from an Old Concept. CNS Oncol. 2016, 5, 101–108. [Google Scholar] [CrossRef]
- Fabian, A.; Stegner, S.; Miarka, L.; Zimmermann, J.; Lenk, L.; Rahn, S.; Buttlar, J.; Viol, F.; Knaack, H.; Esser, D.; et al. Metastasis of Pancreatic Cancer: An Uninflamed Liver Micromilieu Controls Cell Growth and Cancer Stem Cell Properties by Oxidative Phosphorylation in Pancreatic Ductal Epithelial Cells. Cancer Lett. 2019, 453, 95–106. [Google Scholar] [CrossRef]
- Cros, J.; Raffenne, J.; Couvelard, A.; Poté, N. Tumor Heterogeneity in Pancreatic Adenocarcinoma. PAT 2018, 85, 64–71. [Google Scholar] [CrossRef]
- di Magliano, M.P.; Logsdon, C.D. Roles for KRAS in Pancreatic Tumor Development and Progression. Gastroenterology 2013, 144, 1220–1229. [Google Scholar] [CrossRef]
- Kulemann, B.; Liss, A.S.; Warshaw, A.L.; Seifert, S.; Bronsert, P.; Glatz, T.; Pitman, M.B.; Hoeppner, J. KRAS Mutations in Pancreatic Circulating Tumor Cells: A Pilot Study. Tumor Biol. 2016, 37, 7547–7554. [Google Scholar] [CrossRef]
- Guo, S.; Shi, X.; Shen, J.; Gao, S.; Wang, H.; Shen, S.; Pan, Y.; Li, B.; Xu, X.; Shao, Z.; et al. Preoperative Detection of KRAS G12D Mutation in CtDNA Is a Powerful Predictor for Early Recurrence of Resectable PDAC Patients. Br. J. Cancer 2020, 122, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.P.; Timpson, P.; Karim, S.A.; Ridgway, R.A.; Athineos, D.; Doyle, B.; Jamieson, N.B.; Oien, K.A.; Lowy, A.M.; Brunton, V.G.; et al. Mutant P53 Drives Metastasis and Overcomes Growth Arrest/Senescence in Pancreatic Cancer. Proc. Natl. Acad. Sci. USA 2010, 107, 246–251. [Google Scholar] [CrossRef]
- Haugk, B. Pancreatic Intraepithelial Neoplasia—Can We Detect Early Pancreatic Cancer? Histopathology 2010, 57, 503–514. [Google Scholar] [CrossRef]
- Bailey, J.M.; Hendley, A.M.; Lafaro, K.J.; Pruski, M.A.; Jones, N.C.; Alsina, J.; Younes, M.; Maitra, A.; McAllister, F.; Iacobuzio-Donahue, C.A.; et al. P53 Mutations Cooperate with Oncogenic Kras to Promote Adenocarcinoma from Pancreatic Ductal Cells. Oncogene 2016, 35, 4282–4288. [Google Scholar] [CrossRef]
- Bünger, S.; Barow, M.; Thorns, C.; Freitag-Wolf, S.; Danner, S.; Tiede, S.; Pries, R.; Görg, S.; Bruch, H.-P.; Roblick, U.J.; et al. Pancreatic Carcinoma Cell Lines Reflect Frequency and Variability of Cancer Stem Cell Markers in Clinical Tissue. ESR 2012, 49, 88–98. [Google Scholar] [CrossRef]
- Sheikh, A.; Hussain, S.A.; Ghori, Q.; Naeem, N.; Fazil, A.; Giri, S.; Sathian, B.; Mainali, P.; Al Tamimi, D.M. The Spectrum of Genetic Mutations in Breast Cancer. Asian Pac. J. Cancer Prev. 2015, 16, 2177–2185. [Google Scholar] [CrossRef]
- Pellagatti, A.; Roy, S.; Di Genua, C.; Burns, A.; McGraw, K.; Valletta, S.; Larrayoz, M.J.; Fernandez-Mercado, M.; Mason, J.; Killick, S.; et al. Targeted Resequencing Analysis of 31 Genes Commonly Mutated in Myeloid Disorders in Serial Samples from Myelodysplastic Syndrome Patients Showing Disease Progression. Leukemia 2016, 30, 248–250. [Google Scholar] [CrossRef]
- Maddipati, R.; Stanger, B.Z. Pancreatic Cancer Metastases Harbor Evidence of Polyclonality. Cancer Discov. 2015, 5, 1086–1097. [Google Scholar] [CrossRef]
- Ball, C.R.; Oppel, F.; Ehrenberg, K.R.; Dubash, T.D.; Dieter, S.M.; Hoffmann, C.M.; Abel, U.; Herbst, F.; Koch, M.; Werner, J.; et al. Succession of Transiently Active Tumor-Initiating Cell Clones in Human Pancreatic Cancer Xenografts. Embo Mol. Med. 2017, 9, 918–932. [Google Scholar] [CrossRef]
- Seth, S.; Li, C.-Y.; Ho, I.-L.; Corti, D.; Loponte, S.; Sapio, L.; Del Poggetto, E.; Yen, E.-Y.; Robinson, F.S.; Peoples, M.; et al. Pre-Existing Functional Heterogeneity of Tumorigenic Compartment as the Origin of Chemoresistance in Pancreatic Tumors. Cell Rep. 2019, 26, 1518–1532. [Google Scholar] [CrossRef]
- Saikrishna, L.; Kasa, P.; Momin, S.; Bhaskar, L.V.K.S. Perspectives and Molecular Understanding of Pancreatic Cancer Stem Cells. In Exploring Pancreatic Metabolism and Malignancy; Nagaraju, G.P., BM Reddy, A., Eds.; Springer: Singapore, 2019; pp. 157–172. ISBN 978-981-329-393-9. [Google Scholar]
- Ikenaga, N.; Ohuchida, K.; Mizumoto, K.; Yu, J.; Kayashima, T.; Hayashi, A.; Nakata, K.; Tanaka, M. Characterization of CD24 Expression in Intraductal Papillary Mucinous Neoplasms and Ductal Carcinoma of the Pancreas. Hum. Pathol. 2010, 41, 1466–1474. [Google Scholar] [CrossRef]
- Hermann, P.C.; Huber, S.L.; Herrler, T.; Aicher, A.; Ellwart, J.W.; Guba, M.; Bruns, C.J.; Heeschen, C. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar] [CrossRef]
- D’Alterio, C.; Cindolo, L.; Portella, L.; Polimeno, M.; Consales, C.; Riccio, A.; Cioffi, M.; Franco, R.; Chiodini, P.; Cartenì, G.; et al. Differential Role of CD133 and CXCR4 in Renal Cell Carcinoma. Cell Cycle 2010, 9, 4492–4500. [Google Scholar] [CrossRef]
- Bertolini, G.; D’Amico, L.; Moro, M.; Landoni, E.; Perego, P.; Miceli, R.; Gatti, L.; Andriani, F.; Wong, D.; Caserini, R.; et al. Microenvironment-Modulated Metastatic CD133+/CXCR4+/EpCAM− Lung Cancer–Initiating Cells Sustain Tumor Dissemination and Correlate with Poor Prognosis. Cancer Res. 2015, 75, 3636–3649. [Google Scholar] [CrossRef]
- Sun, Y.; Yoshida, T.; Okabe, M.; Zhou, K.; Wang, F.; Soko, C.; Saito, S.; Nikaido, T. Isolation of Stem-Like Cancer Cells in Primary Endometrial Cancer Using Cell Surface Markers CD133 and CXCR4. Transl. Oncol. 2017, 10, 976–987. [Google Scholar] [CrossRef] [PubMed]
- Sancho, P.; Alcala, S.; Usachov, V.; Hermann, P.C.; Sainz, B. The Ever-Changing Landscape of Pancreatic Cancer Stem Cells. Pancreatology 2016, 16, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Shuto, T.; Suzuki, S.; Sato, T.; Koga, T.; Suico, M.A.; Kusuhara, H.; Sugiyama, Y.; Cyr, D.M.; Kai, H. Posttranslational Negative Regulation of Glycosylated and Non-Glycosylated BCRP Expression by Derlin-1. Biochem. Biophys. Res. Commun. 2011, 404, 853–858. [Google Scholar] [CrossRef]
- Murtaugh, L.C.; Stanger, B.Z.; Kwan, K.M.; Melton, D.A. Notch Signaling Controls Multiple Steps of Pancreatic Differentiation. Proc. Natl. Acad. Sci. USA 2003, 100, 14920–14925. [Google Scholar] [CrossRef]
- Sharon, N.; Vanderhooft, J.; Straubhaar, J.; Mueller, J.; Chawla, R.; Zhou, Q.; Engquist, E.N.; Trapnell, C.; Gifford, D.K.; Melton, D.A. Wnt Signaling Separates the Progenitor and Endocrine Compartments during Pancreas Development. Cell Rep. 2019, 27, 2281–2291. [Google Scholar] [CrossRef]
- Hebrok, M.; Kim, S.K.; Jacques, B.S.; McMahon, A.P.; Melton, D.A. Regulation of Pancreas Development by Hedgehog Signaling. Development 2000, 127, 4905–4913. [Google Scholar] [CrossRef]
- Reichrath, J.; Reichrath, S. (Eds.) Notch Signaling in Embryology and Cancer. In Advances in Experimental Medicine and Biology; Springer Science+Business Media: Berlin/Heidelberg, Germany, 2012; ISBN 978-1-4614-0898-7. [Google Scholar]
- Mullendore, M.E.; Koorstra, J.-B.; Li, Y.-M.; Offerhaus, G.J.; Fan, X.; Henderson, C.M.; Matsui, W.; Eberhart, C.G.; Maitra, A.; Feldmann, G. Ligand-Dependent Notch Signaling Is Involved in Tumor Initiation and Tumor Maintenance in Pancreatic Cancer. Clin. Cancer Res. 2009, 15, 2291–2301. [Google Scholar] [CrossRef]
- Wang, Z.; Ahmad, A.; Li, Y.; Azmi, A.S.; Miele, L.; Sarkar, F.H. Targeting Notch to Eradicate Pancreatic Cancer Stem Cells for Cancer Therapy. Anticancer Res. 2011, 9, 1105–1113. [Google Scholar]
- Abel, E.V.; Kim, E.J.; Wu, J.; Hynes, M.; Bednar, F.; Proctor, E.; Wang, L.; Dziubinski, M.L.; Simeone, D.M. The Notch Pathway Is Important in Maintaining the Cancer Stem Cell Population in Pancreatic Cancer. PLoS ONE 2014, 9, e91983. [Google Scholar] [CrossRef] [PubMed]
- Yabuuchi, S.; Pai, S.G.; Campbell, N.R.; de Wilde, R.F.; De Oliveira, E.; Korangath, P.; Streppel, M.M.; Rasheed, Z.A.; Hidalgo, M.; Maitra, A.; et al. Notch Signaling Pathway Targeted Therapy Suppresses Tumor Progression and Metastatic Spread in Pancreatic Cancer. Cancer Lett. 2013, 335, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Song, S.Y.; Park, J.Y. Notch Pathway Activation Is Associated with Pancreatic Cancer Treatment Failure. Pancreatology 2014, 14, 48–53. [Google Scholar] [CrossRef]
- Mizuma, M.; Rasheed, Z.A.; Yabuuchi, S.; Omura, N.; Campbell, N.R.; de Wilde, R.F.; Oliveira, E.D.; Zhang, Q.; Puig, O.; Matsui, W.; et al. The Gamma Secretase Inhibitor MRK-003 Attenuates Pancreatic Cancer Growth in Preclinical Models. Mol. Cancer 2012, 11, 1999–2009. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Kumar, A.; Bharti, A.C. Anticancer Potential of Curcumin: Preclinical and Clinical Studies. Anticancer Res. 2003, 23, 363–398. [Google Scholar] [PubMed]
- Bimonte, S.; Barbieri, A.; Leongito, M.; Piccirillo, M.; Giudice, A.; Pivonello, C.; De Angelis, C.; Granata, V.; Palaia, R.; Izzo, F. Curcumin AntiCancer Studies in Pancreatic Cancer. Nutrients 2016, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Abbas Momtazi, A.; Sahebkar, A. Difluorinated Curcumin: A Promising Curcumin Analogue with Improved Anti-Tumor Activity and Pharmacokinetic Profile. Curr. Pharm. Des. 2016, 22, 4386–4397. [Google Scholar] [CrossRef]
- Xia, J.; Duan, Q.; Ahmad, A.; Bao, B.; Banerjee, S.; Shi, Y.; Ma, J.; Geng, J.; Chen, Z.; Wahidur Rahman, K.; et al. Genistein Inhibits Cell Growth and Induces Apoptosis Through Up-Regulation of MiR-34a in Pancreatic Cancer Cells. Curr. Drug Targets 2012, 13, 1750–1756. [Google Scholar] [CrossRef] [PubMed]
- Nwaeburu, C.C.; Abukiwan, A.; Zhao, Z.; Herr, I. Quercetin-Induced MiR-200b-3p Regulates the Mode of Self-Renewing Divisions in Pancreatic Cancer. Mol. Cancer 2017, 16, 23. [Google Scholar] [CrossRef]
- Pham, N.-A.; Jacobberger, J.W.; Schimmer, A.D.; Cao, P.; Gronda, M.; Hedley, D.W. The Dietary Isothiocyanate Sulforaphane Targets Pathways of Apoptosis, Cell Cycle Arrest, and Oxidative Stress in Human Pancreatic Cancer Cells and Inhibits Tumor Growth in Severe Combined Immunodeficient Mice. Mol. Cancer 2004, 3, 1239–1248. [Google Scholar]
- Appari, M.; Babu, K.R.; Kaczorowski, A.; Gross, W.; Herr, I. Sulforaphane, Quercetin and Catechins Complement Each Other in Elimination of Advanced Pancreatic Cancer by MiR-Let-7 Induction and K-Ras Inhibition. Int. J. Oncol. 2014, 45, 1391–1400. [Google Scholar] [CrossRef]
- Srivastava, R.K.; Tang, S.-N.; Zhu, W.; Meeker, D.; Shankar, S. Sulforaphane Synergizes with Quercetin to Inhibit Self-Renewal Capacity of Pancreatic Cancer Stem Cells. Front. Biosci (Elite Ed.) 2011, 3, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Kallifatidis, G.; Labsch, S.; Rausch, V.; Mattern, J.; Gladkich, J.; Moldenhauer, G.; Büchler, M.W.; Salnikov, A.V.; Herr, I. Sulforaphane Increases Drug-Mediated Cytotoxicity Toward Cancer Stem-like Cells of Pancreas and Prostate. Mol. Ther. 2011, 19, 188–195. [Google Scholar] [CrossRef]
- Seidensticker, M.J.; Behrens, J. Biochemical Interactions in the Wnt Pathway. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2000, 1495, 168–182. [Google Scholar] [CrossRef]
- Polakis, P. Wnt Signaling and Cancer. Genes Dev. 2000, 14, 1837–1851. [Google Scholar] [CrossRef]
- di Magliano, M.P.; Biankin, A.V.; Heiser, P.W.; Cano, D.A.; Gutierrez, P.J.A.; Deramaudt, T.; Segara, D.; Dawson, A.C.; Kench, J.G.; Henshall, S.M.; et al. Common Activation of Canonical Wnt Signaling in Pancreatic Adenocarcinoma. PLoS ONE 2007, 2, e1155. [Google Scholar]
- Curtin, J.C.; Lorenzi, M.V. Drug Discovery Approaches to Target Wnt Signaling in Cancer Stem Cells. Oncotarget 2010, 1, 563–577. [Google Scholar] [CrossRef]
- He, L.; Wang, F.; Dai, W.-Q.; Wu, D.; Lin, C.-L.; Wu, S.-M.; Cheng, P.; Zhang, Y.; Shen, M.; Wang, C.-F.; et al. Mechanism of Action of Salinomycin on Growth and Migration in Pancreatic Cancer Cell Lines. Pancreatology 2013, 13, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-N.; Liang, Y.; Zhou, L.-J.; Chen, S.-P.; Chen, G.; Zhang, T.-P.; Kang, T.; Zhao, Y.-P. Combination of Salinomycin and Gemcitabine Eliminates Pancreatic Cancer Cells. Cancer Lett. 2011, 313, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.; Ozsvari, B.; Lisanti, C.L.; Tanowitz, H.B.; Howell, A.; Martinez-Outschoorn, U.E.; Sotgia, F.; Lisanti, M.P. Antibiotics That Target Mitochondria Effectively Eradicate Cancer Stem Cells, across Multiple Tumor Types: Treating Cancer like an Infectious Disease. Oncotarget 2015, 6, 4569–4584. [Google Scholar] [CrossRef]
- Lu, Z.; Xu, N.; He, B.; Pan, C.; Lan, Y.; Zhou, H.; Liu, X. Inhibition of Autophagy Enhances the Selective Anti-Cancer Activity of Tigecycline to Overcome Drug Resistance in the Treatment of Chronic Myeloid Leukemia. J. Exp. Clin. Cancer Res. 2017, 36, 43. [Google Scholar] [CrossRef]
- Li, H.; Jiao, S.; Li, X.; Banu, H.; Hamal, S.; Wang, X. Therapeutic Effects of Antibiotic Drug Tigecycline against Cervical Squamous Cell Carcinoma by Inhibiting Wnt/β-Catenin Signaling. Biochem. Biophys. Res. Commun. 2015, 467, 14–20. [Google Scholar] [CrossRef]
- Dong, Z.; Abbas, M.N.; Kausar, S.; Yang, J.; Li, L.; Tan, L.; Cui, H. Biological Functions and Molecular Mechanisms of Antibiotic Tigecycline in the Treatment of Cancers. Int. J. Mol. Sci. 2019, 20, 3577. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Dong, Z.; Ren, A.; Fu, G.; Zhang, K.; Li, C.; Wang, X.; Cui, H. Antibiotic Tigecycline Inhibits Cell Proliferation, Migration and Invasion via down-Regulating CCNE2 in Pancreatic Ductal Adenocarcinoma. J. Cell. Mol. Med. 2020, 24, 4245–4260. [Google Scholar] [CrossRef] [PubMed]
- Sahib, A.K.; Loureiro, J.R.; Vasavada, M.; Anderson, C.; Kubicki, A.; Wade, B.; Joshi, S.H.; Woods, R.P.; Congdon, E.; Espinoza, R.; et al. Modulation of the Functional Connectome in Major Depressive Disorder by Ketamine Therapy. Psychol. Med. 2020, 1–10. [Google Scholar] [CrossRef]
- Hu, J.; Duan, W.; Liu, Y. Ketamine Inhibits Aerobic Glycolysis in Colorectal Cancer Cells by Blocking the NMDA Receptor-CaMK II-c-Myc Pathway. Clin. Exp. Pharmacol. Physiol. 2020, 47, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Carbone, C.; Piro, G.; Gaianigo, N.; Ligorio, F.; Santoro, R.; Merz, V.; Simionato, F.; Zecchetto, C.; Falco, G.; Conti, G.; et al. Adipocytes Sustain Pancreatic Cancer Progression through a Non-Canonical WNT Paracrine Network Inducing ROR2 Nuclear Shuttling. Int. J. Obes. 2018, 42, 334–343. [Google Scholar] [CrossRef]
- Davis, S.L.; Cardin, D.B.; Shahda, S.; Lenz, H.-J.; Dotan, E.; O’Neil, B.H.; Kapoun, A.M.; Stagg, R.J.; Berlin, J.; Messersmith, W.A.; et al. A Phase 1b Dose Escalation Study of Wnt Pathway Inhibitor Vantictumab in Combination with Nab-Paclitaxel and Gemcitabine in Patients with Previously Untreated Metastatic Pancreatic Cancer. Investig. New Drugs 2020, 38, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.M. The Hedgehog Signaling Network. Am. J. Med. Genet. Part A 2003, 123A, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Varjosalo, M.; Taipale, J. Hedgehog: Functions and Mechanisms. Genes Dev. 2008, 22, 2454–2472. [Google Scholar] [CrossRef] [PubMed]
- Thayer, S.P.; di Magliano, M.P.; Heiser, P.W.; Nielsen, C.M.; Roberts, D.J.; Lauwers, G.Y.; Qi, Y.P.; Gysin, S.; Castillo, C.F.; Yajnik, V.; et al. Hedgehog Is an Early and Late Mediator of Pancreatic Cancer Tumorigenesis. Nature 2003, 425, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Mathew, E.; Collins, M.A.; Fernandez-Barrena, M.G.; Holtz, A.M.; Yan, W.; Hogan, J.O.; Tata, Z.; Allen, B.L.; Fernandez-Zapico, M.E.; Pasca di Magliano, M. The Transcription Factor GLI1 Modulates the Inflammatory Response during Pancreatic Tissue Remodeling. J. Biol. Chem. 2014, 289, 27727–27743. [Google Scholar] [CrossRef]
- Tang, S.-N.; Fu, J.; Nall, D.; Rodova, M.; Shankar, S.; Srivastava, R.K. Inhibition of Sonic Hedgehog Pathway and Pluripotency Maintaining Factors Regulate Human Pancreatic Cancer Stem Cell Characteristics. Int. J. Cancer 2012, 131, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, Q.; Xu, Q.; Liu, H.; Lei, J.; Duan, W.; Bhat, K.; Wang, F.; Wu, E.; Wang, Z. SDF-1/CXCR4 Signaling Induces Pancreatic Cancer Cell Invasion and Epithelial–Mesenchymal Transition in Vitro through Non-Canonical Activation of Hedgehog Pathway. Cancer Lett. 2012, 322, 169–176. [Google Scholar] [CrossRef]
- Merchant, A.A.; Matsui, W. Targeting Hedgehog—A Cancer Stem Cell Pathway. Clin. Cancer Res. 2010, 16, 3130–3140. [Google Scholar] [CrossRef]
- Chen, J.K.; Taipale, J.; Cooper, M.K.; Beachy, P.A. Inhibition of Hedgehog Signaling by Direct Binding of Cyclopamine to Smoothened. Genes Dev. 2002, 16, 2743–2748. [Google Scholar] [CrossRef]
- Xu, X.-F.; Guo, C.-Y.; Liu, J.; Yang, W.-J.; Xia, Y.-J.; Xu, L.; Yu, Y.-C.; Wang, X.-P. Gli1 Maintains Cell Survival by Up-Regulating IGFBP6 and Bcl-2 through Promoter Regions in Parallel Manner in Pancreatic Cancer Cells. J. Carcinog. 2009, 8. [Google Scholar] [CrossRef]
- Huynh, D.L.; Koh, H.; Chandimali, N.; Zhang, J.J.; Kim, N.; Kang, T.Y.; Ghosh, M.; Gera, M.; Park, Y.-H.; Kwon, T.; et al. BRM270 Inhibits the Proliferation of CD44 Positive Pancreatic Ductal Adenocarcinoma Cells via Downregulation of Sonic Hedgehog Signaling. Evid. Based Complement. Altern. Med. 2019, 2019, e8620469. [Google Scholar] [CrossRef]
- Huang, F.-T.; Zhuan-Sun, Y.-X.; Zhuang, Y.-Y.; Wei, S.-L.; Tang, J.; Chen, W.-B.; Zhang, S.-N. Inhibition of Hedgehog Signaling Depresses Self-Renewal of Pancreatic Cancer Stem Cells and Reverses Chemoresistance. Int. J. Oncol. 2012, 41, 1707–1714. [Google Scholar] [CrossRef]
- Balic, A.; Sørensen, M.D.; Trabulo, S.M.; Sainz, B.; Cioffi, M.; Vieira, C.R.; Miranda-Lorenzo, I.; Hidalgo, M.; Kleeff, J.; Erkan, M.; et al. Chloroquine Targets Pancreatic Cancer Stem Cells via Inhibition of CXCR4 and Hedgehog Signaling. Mol. Cancer 2014, 13, 1758–1771. [Google Scholar] [CrossRef] [PubMed]
- Rangarajan, P.; Subramaniam, D.; Paul, S.; Kwatra, D.; Palaniyandi, K.; Islam, S.; Harihar, S.; Ramalingam, S.; Gutheil, W.; Putty, S.; et al. Crocetinic Acid Inhibits Hedgehog Signaling to Inhibit Pancreatic Cancer Stem Cells. Oncotarget 2015, 6, 27661–27673. [Google Scholar] [CrossRef]
- Sun, X.-D.; Liu, X.-E.; Huang, D.-S. Curcumin Reverses the Epithelial-Mesenchymal Transition of Pancreatic Cancer Cells by Inhibiting the Hedgehog Signaling Pathway. Oncol. Rep. 2013, 29, 2401–2407. [Google Scholar] [CrossRef]
- Cao, L.; Xiao, X.; Lei, J.; Duan, W.; Ma, Q.; Li, W. Curcumin Inhibits Hypoxia-Induced Epithelial-mesenchymal Transition in Pancreatic Cancer Cells via Suppression of the Hedgehog Signaling Pathway. Oncol. Rep. 2016, 35, 3728–3734. [Google Scholar] [CrossRef]
- Li, S.-H.; Fu, J.; Watkins, D.N.; Srivastava, R.K.; Shankar, S. Sulforaphane Regulates Self-Renewal of Pancreatic Cancer Stem Cells through the Modulation of Sonic Hedgehog–GLI Pathway. Mol. Cell Biochem. 2013, 373, 217–227. [Google Scholar] [CrossRef]
- Feldmann, G.; Fendrich, V.; McGovern, K.; Bedja, D.; Bisht, S.; Alvarez, H.; Koorstra, J.-B.M.; Habbe, N.; Karikari, C.; Mullendore, M.; et al. An Orally Bioavailable Small-Molecule Inhibitor of Hedgehog Signaling Inhibits Tumor Initiation and Metastasis in Pancreatic Cancer. Mol. Cancer 2008, 7, 2725–2735. [Google Scholar] [CrossRef]
- Singh, B.N.; Fu, J.; Srivastava, R.K.; Shankar, S. Hedgehog Signaling Antagonist GDC-0449 (Vismodegib) Inhibits Pancreatic Cancer Stem Cell Characteristics: Molecular Mechanisms. PLoS ONE 2011, 6, e27306. [Google Scholar]
- Kim, E.J.; Sahai, V.; Abel, E.V.; Griffith, K.A.; Greenson, J.K.; Takebe, N.; Khan, G.N.; Blau, J.L.; Craig, R.; Balis, U.G.; et al. Pilot Clinical Trial of Hedgehog Pathway Inhibitor GDC-0449 (Vismodegib) in Combination with Gemcitabine in Patients with Metastatic Pancreatic Adenocarcinoma. Clin. Cancer Res. 2014, 20, 5937–5945. [Google Scholar] [CrossRef]
- Misra, J.R.; Irvine, K.D. The Hippo Signaling Network and Its Biological Functions. Annu. Rev. Genet. 2018, 52, 65–87. [Google Scholar] [CrossRef]
- Pan, D. The Hippo Signaling Pathway in Development and Cancer. Dev. Cell 2010, 19, 491–505. [Google Scholar] [CrossRef]
- Ansari, D.; Ohlsson, H.; Althini, C.; Bauden, M.; Zhou, Q.; Hu, D.; Andersson, R. The Hippo Signaling Pathway in Pancreatic Cancer. Anticancer Res. 2019, 39, 3317–3321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shi, H.; Chen, H.; Gong, A.; Liu, Y.; Song, L.; Xu, X.; You, T.; Fan, X.; Wang, D.; et al. Dedifferentiation Process Driven by Radiotherapy-Induced HMGB1/TLR2/YAP/HIF-1α Signaling Enhances Pancreatic Cancer Stemness. Cell Death Dis. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Santoro, R.; Zanotto, M.; Carbone, C.; Piro, G.; Tortora, G.; Melisi, D. MEKK3 Sustains EMT and Stemness in Pancreatic Cancer by Regulating YAP and TAZ Transcriptional Activity. Anticancer Res. 2018, 38, 1937–1946. [Google Scholar]
- Chen, W.; Wang, H.; Liu, Y.; Xu, W.; Ling, C.; Li, Y.; Liu, J.; Chen, M.; Zhang, Y.; Chen, B.; et al. Linc-RoR Promotes Proliferation, Migration, and Invasion via the Hippo/YAP Pathway in Pancreatic Cancer Cells. J. Cell. Biochem. 2020, 121, 632–641. [Google Scholar] [CrossRef]
- Gruber, R.; Panayiotou, R.; Nye, E.; Spencer-Dene, B.; Stamp, G.; Behrens, A. YAP1 and TAZ Control Pancreatic Cancer Initiation in Mice by Direct Up-Regulation of JAK–STAT3 Signaling. Gastroenterology 2016, 151, 526–539. [Google Scholar] [CrossRef]
- Zhang, W.; Nandakumar, N.; Shi, Y.; Manzano, M.; Smith, A.; Graham, G.; Gupta, S.; Vietsch, E.E.; Laughlin, S.Z.; Wadhwa, M.; et al. Downstream of Mutant KRAS, the Transcription Regulator YAP Is Essential for Neoplastic Progression to Pancreatic Ductal Adenocarcinoma. Sci. Signal. 2014, 7, ra42. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, X.; Fang, L.; Lan, C.; Zheng, X.; Wang, Y.; Zhang, Y.; Han, X.; Liu, S.; Cheng, K.; et al. A Combinatorial Strategy Using YAP and Pan-RAF Inhibitors for Treating KRAS-Mutant Pancreatic Cancer. Cancer Lett. 2017, 402, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 Signalling in Cancer: New and Unexpected Biological Functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 Signalling Axis in Cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Contino, G.; Deshpande, V.; Tzatsos, A.; Conrad, C.; Benes, C.H.; Levy, D.E.; Settleman, J.; Engelman, J.A.; Bardeesy, N. STAT3 Plays a Critical Role in KRAS-Induced Pancreatic Tumorigenesis. Cancer Res. 2011, 71, 5020–5029. [Google Scholar] [CrossRef]
- Zhang, Z.; Duan, Q.; Zhao, H.; Liu, T.; Wu, H.; Shen, Q.; Wang, C.; Yin, T. Gemcitabine Treatment Promotes Pancreatic Cancer Stemness through the Nox/ROS/NF-ΚB/STAT3 Signaling Cascade. Cancer Lett. 2016, 382, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, J.; Wang, H.; Yin, G.; Liu, Y.; Lei, X.; Xiang, M. REG3A Accelerates Pancreatic Cancer Cell Growth under IL-6-Associated Inflammatory Condition: Involvement of a REG3A–JAK2/STAT3 Positive Feedback Loop. Cancer Lett. 2015, 362, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Palagani, V.; Bozko, P.; El Khatib, M.; Belahmer, H.; Giese, N.; Sipos, B.; Malek, N.P.; Plentz, R.R. Combined Inhibition of Notch and JAK/STAT Is Superior to Monotherapies and Impairs Pancreatic Cancer Progression. Carcinogenesis 2014, 35, 859–866. [Google Scholar] [CrossRef]
- Lian, J.P.; Word, B.; Taylor, S.; Hammons, G.J.; Lyn-Cook, B.D. Modulation of the Constitutive Activated STAT3 Transcription Factor in Pancreatic Cancer Prevention: Effects of Indole-3-Carbinol (I3C) and Genistein. Anticancer Res. 2004, 24, 133–137. [Google Scholar]
- Glienke, W.; Maute, L.; Wicht, J.; Bergmann, L. Curcumin Inhibits Constitutive STAT3 Phosphorylation in Human Pancreatic Cancer Cell Lines and Downregulation of Survivin/BIRC5 Gene Expression. Cancer Investig. 2009, 28, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Yue, W.; JianYu, E.; Malhotra, J.; Lu, S.; Gu, J.; Xu, F.; Tan, X.-L. In Vitro Comparative Studies of Resveratrol and Triacetylresveratrol on Cell Proliferation, Apoptosis, and STAT3 and NFκB Signaling in Pancreatic Cancer Cells. Sci. Rep. 2016, 6, 31672. [Google Scholar] [CrossRef]
- Sharma, N.; Nanta, R.; Sharma, J.; Gunewardena, S.; Singh, K.P.; Shankar, S.; Srivastava, R.K. PI3K/AKT/MTOR and Sonic Hedgehog Pathways Cooperate Together to Inhibit Human Pancreatic Cancer Stem Cell Characteristics and Tumor Growth. Oncotarget 2015, 6, 32039–32060. [Google Scholar] [CrossRef] [PubMed]
- Husain, K.; Centeno, B.A.; Coppola, D.; Trevino, J.; Sebti, S.M.; Malafa, M.P. δ-Tocotrienol, a Natural Form of Vitamin E, Inhibits Pancreatic Cancer Stem-like Cells and Prevents Pancreatic Cancer Metastasis. Oncotarget 2017, 8, 31554–31567. [Google Scholar] [CrossRef] [PubMed]
- Shin-Kang, S.; Ramsauer, V.P.; Lightner, J.; Chakraborty, K.; Stone, W.; Campbell, S.; Reddy, S.A.G.; Krishnan, K. Tocotrienols Inhibit AKT and ERK Activation and Suppress Pancreatic Cancer Cell Proliferation by Suppressing the ErbB2 Pathway. Free Radic. Biol. Med. 2011, 51, 1164–1174. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, Y.; Lian, G.; Lin, H.; Shang, C.; Zeng, L.; Chen, S.; Li, J.; Huang, C.; Huang, K.; et al. KRAS Promotes Tumor Metastasis and Chemoresistance by Repressing RKIP via the MAPK–ERK Pathway in Pancreatic Cancer. Int. J. Cancer 2018, 142, 2323–2334. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Mouzaki, M.; You, H.; Laird, J.C.; Mato, J.; Lu, S.C.; Rountree, C.B. CD133+ Liver Cancer Stem Cells from Methionine Adenosyl Transferase 1A–Deficient Mice Demonstrate Resistance to Transforming Growth Factor (TGF)-β–Induced Apoptosis. Hepatology 2009, 49, 1277–1286. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, D.; Li, Y.; Liu, Y.; Xie, X.; Wu, Y.; Zhou, Y.; Ren, J.; Zhang, J.; Zhu, H.; et al. CCL21/CCR7 Axis Contributed to CD133+ Pancreatic Cancer Stem-Like Cell Metastasis via EMT and Erk/NF-ΚB Pathway. PLoS ONE 2016, 11, e0158529. [Google Scholar] [CrossRef]
- Chai, X.; Chu, H.; Yang, X.; Meng, Y.; Shi, P.; Gou, S. Metformin Increases Sensitivity of Pancreatic Cancer Cells to Gemcitabine by Reducing CD133 + Cell Populations and Suppressing ERK/P70S6K Signaling. Sci. Rep. 2015, 5, 14404. [Google Scholar] [CrossRef]
- Mori, T.; Doi, R.; Koizumi, M.; Toyoda, E.; Ito, D.; Kami, K.; Masui, T.; Fujimoto, K.; Tamamura, H.; Hiramatsu, K.; et al. CXCR4 Antagonist Inhibits Stromal Cell-Derived Factor 1-Induced Migration and Invasion of Human Pancreatic Cancer. Mol. Cancer 2004, 3, 29–37. [Google Scholar]
- Gelmini, S.; Mangoni, M.; Serio, M.; Romagnani, P.; Lazzeri, E. The Critical Role of SDF-1/CXCR4 Axis in Cancer and Cancer Stem Cells Metastasis. J. Endocrinol. Investig. 2008, 31, 809–819. [Google Scholar] [CrossRef]
- Fitzgerald, T.L.; McCubrey, J.A. Pancreatic Cancer Stem Cells: Association with Cell Surface Markers, Prognosis, Resistance, Metastasis and Treatment. Adv. Biol. Regul. 2014, 56, 45–50. [Google Scholar] [CrossRef]
- Wang, H.; Tsang, B.K. Nodal Signalling and Apoptosis. Reproduction 2007, 133, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Lonardo, E.; Hermann, P.C.; Mueller, M.-T.; Huber, S.; Balic, A.; Miranda-Lorenzo, I.; Zagorac, S.; Alcala, S.; Rodriguez-Arabaolaza, I.; Ramirez, J.C.; et al. Nodal/Activin Signaling Drives Self-Renewal and Tumorigenicity of Pancreatic Cancer Stem Cells and Provides a Target for Combined Drug Therapy. Cell Stem Cell 2011, 9, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, F.; Wang, Y.; Li, T.; Xiu, P.; Zhong, J.; Sun, X.; Li, J. Verteporfin Suppresses Cell Survival, Angiogenesis and Vasculogenic Mimicry of Pancreatic Ductal Adenocarcinoma via Disrupting the YAP-TEAD Complex. Cancer Sci. 2017, 108, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Bloomston, M.; Frankel, W.L.; Petrocca, F.; Volinia, S.; Alder, H.; Hagan, J.P.; Liu, C.-G.; Bhatt, D.; Taccioli, C.; Croce, C.M. MicroRNA Expression Patterns to Differentiate Pancreatic Adenocarcinoma From Normal Pancreas and Chronic Pancreatitis. JAMA 2007, 297, 1901–1908. [Google Scholar] [CrossRef]
- Lee, E.J.; Gusev, Y.; Jiang, J.; Nuovo, G.J.; Lerner, M.R.; Frankel, W.L.; Morgan, D.L.; Postier, R.G.; Brackett, D.J.; Schmittgen, T.D. Expression Profiling Identifies MicroRNA Signature in Pancreatic Cancer. Int. J. Cancer 2007, 120, 1046–1054. [Google Scholar] [CrossRef]
- Schultz, N.A.; Dehlendorff, C.; Jensen, B.V.; Bjerregaard, J.K.; Nielsen, K.R.; Bojesen, S.E.; Calatayud, D.; Nielsen, S.E.; Yilmaz, M.; Holländer, N.H.; et al. MicroRNA Biomarkers in Whole Blood for Detection of Pancreatic Cancer. JAMA 2014, 311, 392–404. [Google Scholar] [CrossRef]
- Reddy, K.B. MicroRNA (MiRNA) in Cancer. Cancer Cell Int. 2015, 15, 38. [Google Scholar] [CrossRef]
- Singh, S.; Chitkara, D.; Kumar, V.; Behrman, S.W.; Mahato, R.I. MiRNA Profiling in Pancreatic Cancer and Restoration of Chemosensitivity. Cancer Lett. 2013, 334, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Ali, S.; Banerjee, S.; Wang, Z.; Logna, F.; Azmi, A.S.; Kong, D.; Ahmad, A.; Li, Y.; Padhye, S.; et al. Curcumin Analogue CDF Inhibits Pancreatic Tumor Growth by Switching on Suppressor MicroRNAs and Attenuating EZH2 Expression. Cancer Res. 2012, 72, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Hong, T.H.; Park, I.Y. MicroRNA Expression Profiling of Diagnostic Needle Aspirates from Surgical Pancreatic Cancer Specimens. Ann. Surg Treat. Res. 2014, 87, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Schultz, N.A.; Werner, J.; Willenbrock, H.; Roslind, A.; Giese, N.; Horn, T.; Wøjdemann, M.; Johansen, J.S. MicroRNA Expression Profiles Associated with Pancreatic Adenocarcinoma and Ampullary Adenocarcinoma. Mod. Pathol. 2012, 25, 1609–1622. [Google Scholar]
- Calatayud, D.; Dehlendorff, C.; Boisen, M.K.; Hasselby, J.P.; Schultz, N.A.; Werner, J.; Immervoll, H.; Molven, A.; Hansen, C.P.; Johansen, J.S. Tissue MicroRNA Profiles as Diagnostic and Prognostic Biomarkers in Patients with Resectable Pancreatic Ductal Adenocarcinoma and Periampullary Cancers. Biomark Res. 2017, 5, 8. [Google Scholar] [CrossRef]
- Papaconstantinou, I.G.; Manta, A.; Gazouli, M.; Lyberopoulou, A.; Lykoudis, P.M.; Polymeneas, G.; Voros, D. Expression of MicroRNAs in Patients with Pancreatic Cancer and Its Prognostic Significance. Pancreas 2013, 42, 67–71. [Google Scholar] [CrossRef]
- Morimura, R.; Komatsu, S.; Ichikawa, D.; Takeshita, H.; Tsujiura, M.; Nagata, H.; Konishi, H.; Shiozaki, A.; Ikoma, H.; Okamoto, K.; et al. Novel Diagnostic Value of Circulating MiR-18a in Plasma of Patients with Pancreatic Cancer. Br. J. Cancer 2011, 105, 1733–1740. [Google Scholar] [CrossRef]
- Olson, P.; Lu, J.; Zhang, H.; Shai, A.; Chun, M.G.; Wang, Y.; Libutti, S.K.; Nakakura, E.K.; Golub, T.R.; Hanahan, D. MicroRNA Dynamics in the Stages of Tumorigenesis Correlate with Hallmark Capabilities of Cancer. Genes Dev. 2009, 23, 2152–2165. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Ou, Y.; Wu, K.; Chen, Y.; Sun, W. MiR-143 Inhibits the Metastasis of Pancreatic Cancer and an Associated Signaling Pathway. Tumour Biol. 2012, 33, 1863–1870. [Google Scholar] [CrossRef] [PubMed]
- Frampton, A.E.; Krell, J.; Jacob, J.; Stebbing, J.; Castellano, L.; Jiao, L.R. Loss of MiR-126 Is Crucial to Pancreatic Cancer Progression. Expert Rev. Anticancer 2012, 12, 881–884. [Google Scholar] [CrossRef]
- Li, Y.; Vandenboom, T.G.; Wang, Z.; Kong, D.; Ali, S.; Philip, P.A.; Sarkar, F.H. MiR-146a Suppresses Invasion of Pancreatic Cancer Cells. Cancer Res. 2010, 70, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Di, Y.; Liang, M.; Yang, F.; Yao, L.; Hao, S.; Li, J.; Jiang, Y.; Jin, C.; Fu, D. The MicroRNA-218 and ROBO-1 Signaling Axis Correlates with the Lymphatic Metastasis of Pancreatic Cancer. Oncol. Rep. 2013, 30, 651–658. [Google Scholar] [CrossRef]
- He, H.; Hao, S.-J.; Yao, L.; Yang, F.; Di, Y.; Li, J.; Jiang, Y.-J.; Jin, C.; Fu, D.-L. MicroRNA-218 Inhibits Cell Invasion and Migration of Pancreatic Cancer via Regulating ROBO1. Cancer Biol. 2014, 15, 1333–1339. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Zhang, Y.; Li, J.; Cao, G.; Yang, W. High Expression of MicroRNA-4295 Contributes to Cell Proliferation and Invasion of Pancreatic Ductal Adenocarcinoma by the down-Regulation of Glypican-5. Biochem. Biophys. Res. Commun. 2018, 497, 73–79. [Google Scholar] [CrossRef]
| S. No. | Detection Technology | Advantages | Drawbacks and Limitations |
|---|---|---|---|
| 1. | Fluorescence-activated cell sorting (FACS) | Highly flexible technique with a large range of stem cell sorting capabilities Very precise Multiparameter isolation | Complicated method Viability of recovered cells is low High cost Time-consuming There is no universal marker for identifying CSCs Require cells in suspension, and in this state, cells clump together, and metabolism may be altered |
| 2. | Magnetic-activated cell sorting (MACS) | Fast and easy method in the isolation of CSCs with the capability of isolating small populations of the cells within the tumor bulk High specificity | Monoparameter separation Involves a cell suspension solution rather than a solid sample There is no universal marker for identifying CSCs |
| 3. | Aldehyde dehydrogenase 1 (ALDH1) activity | Stability than the cell surface markers ALDH1-positive cells displayed increased sphere formation capability, self-renewal properties, tumorigenicity and high expression of stemness genes | Low specificity (It can be used either for the normal or CSC)ALDH1 may not be a proper CSC marker for all tumor types |
| 4. | Spheroid formation assay |
Simple assay There is no need for expensive laboratory facilities |
Heterogeneity and presence of differentiated cells In spheroid formation, there is no quiescent CSCs |
| 5. | Colony formation | Simple and easy | Freshly prepared required To ensure that each colony results from a single cell, proper cell dilution is needed |
| 6. | SP assays | Easier and reliable method Promising method for identifying stem cell and progenitor populations in different tissues and numerous cancers There are no unique cellular markers needed for CSC isolation | Lack of homogeneity in the SP staining protocols Unspecified method for SP population in various tumors Low specificity Lack of purity Toxicity of Hoechst 33342 |
| S. No. | Signaling Pathway | Therapeutic Agents (Function)/Small Molecule Compounds | References |
|---|---|---|---|
| 1. | Notch | Curcumin (diferuloylmethane), genistein (soy isoflavonoid), quercetin (polyphenol and flavonoid), sulforaphane (phytochemical), PF-03084014 (γ-secretase inhibitor), MRK-003 (γ-secretase inhibitor) | [77,78,79,80,81,82,83,84,85,86,87,88] |
| 2. | Wnt, EMT | Salinomycin, azithromycin, tigecycline, and ketamine (anesthetic and antidepressant), vantictumab (monoclonal antibody) | [93,94,95,96,97,98,99,100,101,102,103] |
| 3. | Hedgehog | Curcumin (diferuloylmethane), cyclopamine (phytochemical), crocetinic acid, chloroquine (antimalarial agent), sulforaphane (phytochemical), quercetin (polyphenol and flavonoid), IPI-269609, and GDC-0449 | [111,112,113,114,115,116,117,118,119,120,121,122] |
| 4. | Hippo-signaling | Verteporfin (porphyrin molecule) | [153] |
| 5. | JAK-STAT pathway | AG-490, curcumin (diferuloylmethane), resveratrol (polyphenol), indole-3-carbinol (I3C) and genistein | [137,138,139,140] |
| 6. | PI3K/Akt/mTOR-signaling | Rapamycin, AZD8055, NVP-LDE-225, NVP-LDE-225, NVP-BEZ-235, δ-tocotrienol (vitamin E) | [141,142,143] |
| 7. | MAPK-ERK pathway | Metformin | [147] |
| 8. | CXCR4-signaling | AMD3100 (small-molecule inhibitor), chloroquine (antimalarial agent) | [64] |
| 9. | NODAL/ACTIVIN-signaling | SB431542 | [152] |
| miRNA/s | Sample Type/Site of Action | Regulation | Target (−ve)/(+ve) | Implication | Reference |
|---|---|---|---|---|---|
| miR-146 | Metastatic pancreatic cancer tissues vs. normal control | Up | [157] | ||
| miRNA-205, miRNA-7 | Down | ||||
| miR-26a, miR-200b | PDAC samples vs. normal control | EZH2, EpCAM, pancreatospheres | [158] | ||
| miR-21, miR-27a, miR-146a, miR200a and miR-196a | Pancreatic cancer tissue vs. paraneoplastic normal pancreatic tissues | Up | 51 | [160] | |
| miR-217, miR-20a, and miR-96 | Down | 107 | |||
| miR-198, miR-650, | Pancreatic adenocarcinomas and chronic pancreatitis vs. normal pancreas | Up | 43 | [161] | |
| miR-130b, miR-141, miR-194 and miR-219-1-3p | Down | ||||
| 41 | |||||
| miR-21-5p, -23a-3p, -31-5p, -34c-5p, -93-3p, -135b-3p, -155-5p, -186-5p, -196b-5p, -203, -205-5p, -210, -222-3p, -451, -492, -614, and miR-622 | Pancreatic cancer vs. healthy control | Up | 17 | [162] | |
| miR-122-5p, -130b-3p, -216b, -217, and miR-375 | Down | 5 | |||
| miR-21, miR-155, miR-210, miR-221, and miR-222 | PDAC vs. healthy control | Up | 5 | [163] | |
| miR-31, miR-122, miR-145, and miR-146a | Down | 4 | |||
| miR-18a | Plasma of pancreatic cancer patient vs. healthy control | Up | [164] | ||
| miR-21 | Plasma of pancreatic cancer patient vs. healthy control | Up | 54 | [165] | |
| miR-146a | Down | 37 | |||
| miR-143 | Metastatic pancreatic cancer | Down | GEF1, GEF2, K-RAS, MMP-2, and MMP-9 (−ve) | Metastasis, invasive potential ↑, EMT ↑ | [166] |
| miR-126 | PDAC progressive samples with metastasis | Down | ADAM9 (−ve) | Metastasis, invasive potential ↑, EMT ↑ | [167] |
| miR-146a | Pancreatic cancer vs. normal human pancreatic duct | Down | EGFR, MTA-2, IRAK-1, NFkB (−ve) | Invasive potential ↑ | [168] |
| miR-218 | Metastatic pancreatic cancer; microarray analysis/pancreatic cancer sample | Down | ROBO1 ↑ | Progression and lymphatic metastasis ↑, Invasion and migration potential ↑ | [169,170] |
| miR-4295 | PDAC cells | Up | GPC5 ↓ | Proliferation, invasion and Wnt/β-catenin signaling ↑ | [171] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barman, S.; Fatima, I.; Singh, A.B.; Dhawan, P. Pancreatic Cancer and Therapy: Role and Regulation of Cancer Stem Cells. Int. J. Mol. Sci. 2021, 22, 4765. https://doi.org/10.3390/ijms22094765
Barman S, Fatima I, Singh AB, Dhawan P. Pancreatic Cancer and Therapy: Role and Regulation of Cancer Stem Cells. International Journal of Molecular Sciences. 2021; 22(9):4765. https://doi.org/10.3390/ijms22094765
Chicago/Turabian StyleBarman, Susmita, Iram Fatima, Amar B. Singh, and Punita Dhawan. 2021. "Pancreatic Cancer and Therapy: Role and Regulation of Cancer Stem Cells" International Journal of Molecular Sciences 22, no. 9: 4765. https://doi.org/10.3390/ijms22094765
APA StyleBarman, S., Fatima, I., Singh, A. B., & Dhawan, P. (2021). Pancreatic Cancer and Therapy: Role and Regulation of Cancer Stem Cells. International Journal of Molecular Sciences, 22(9), 4765. https://doi.org/10.3390/ijms22094765







