Characterization of the HCN Interaction Partner TRIP8b/PEX5R in the Intracardiac Nervous System of TRIP8b-Deficient and Wild-Type Mice
Abstract
1. Introduction
2. Results
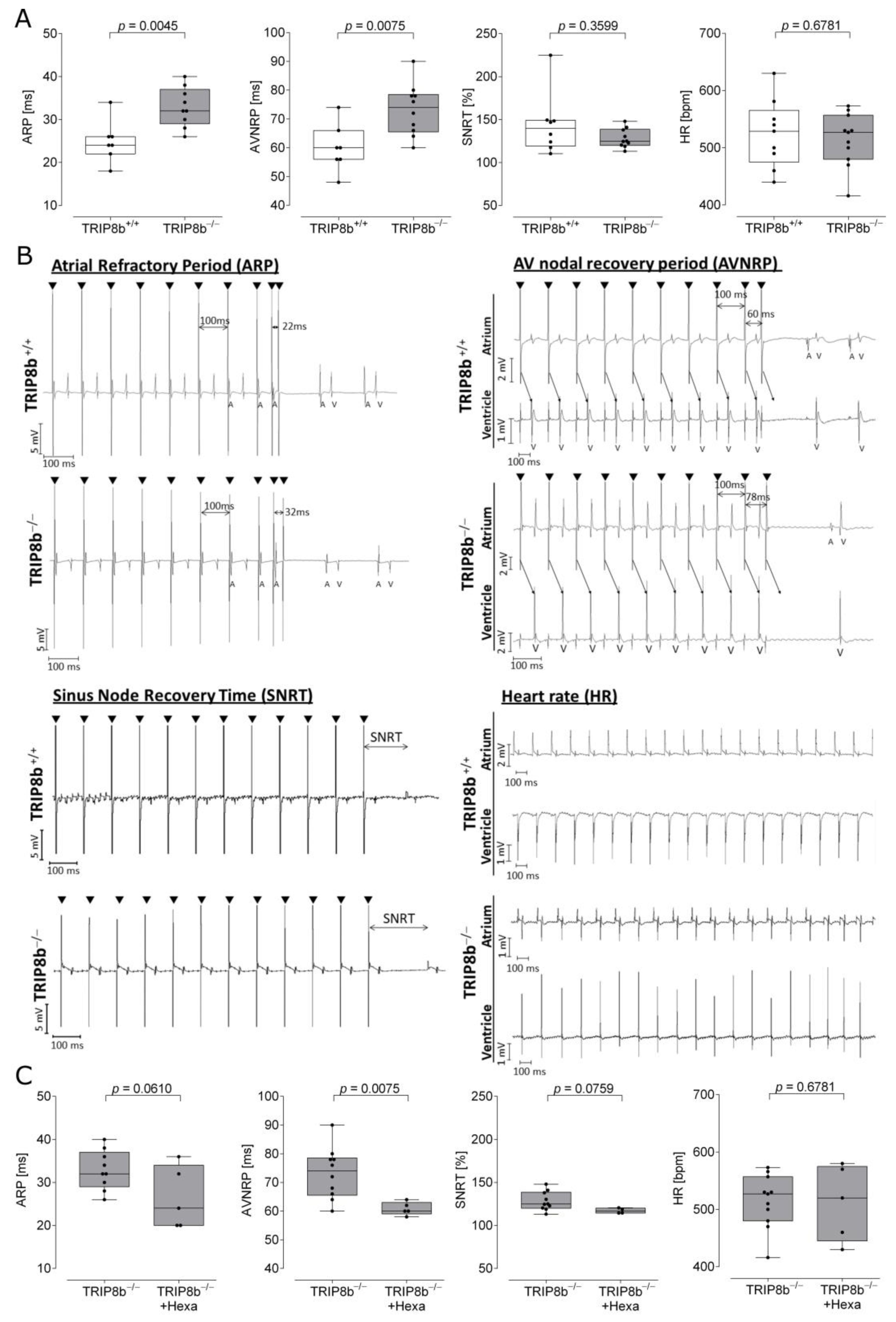
2.1. Electrophysiological Recordings from Wild-Type and TRIP8b-Deficient Mouse Heart Preparations
2.2. Morphological Analyses
2.3. Gene Expression Analysis of Trip8b
2.4. Protein Expression Analysis of TRIP8b
2.5. HCN Channel Expression Analysis in Cardiac Ganglia
3. Discussion
4. Materials and Methods
4.1. TRIP8b-Deficient Mice
4.2. Langendorff Perfusion
4.3. In Vitro Electrophysiological Study
4.4. Histology and Immunohistochemistry
4.5. Staining of Murine Atria and Vibratome Sections
4.6. RNAscope In Situ Hybridization
4.7. Microscopy
4.8. mRNA Quantification
4.9. Western Blot Analysis
4.10. Gene Expression Analyses
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herrmann, S.; Schnorr, S.; Ludwig, A. HCN Channels—Modulators of Cardiac and Neuronal Excitability. Int. J. Mol. Sci. 2015, 16, 1429–1447. [Google Scholar] [CrossRef] [PubMed]
- Baruscotti, M.; Barbuti, A.; Bucchi, A. The cardiac pacemaker current. J. Mol. Cell. Cardiol. 2010, 48, 55–64. [Google Scholar] [CrossRef]
- Zolles, G.; Wenzel, D.; Bildl, W.; Schulte, U.; Hofmann, A.; Müller, C.S.; Thumfart, J.-O.; Vlachos, A.; Deller, T.; Pfeifer, A.; et al. Association with the Auxiliary Subunit PEX5R/Trip8b Controls Responsiveness of HCN Channels to cAMP and Adrenergic Stimulation. Neuron 2009, 62, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.S.; Vaidya, S.P.; Blaiss, C.A.; Liu, Z.; Stoub, T.R.; Brager, H.; Chen, X.; Bender, R.A.; Estep, C.M.; Popov, A.B.; et al. Deletion of the HCN channel auxiliary subunit TRIP8b impairs hippocampal Ih localization and function and promotes antidepressant behavior in mice. J. Neurosci. 2011, 31, 7424–7440. [Google Scholar] [CrossRef]
- Piskorowski, R.; Santoro, B.; Siegelbaum, S.A. TRIP8b Splice Forms Act in Concert to Regulate the Localization and Expression of HCN1 Channels in CA1 Pyramidal Neurons. Neuron 2011, 70, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Santoro, B.; Wainger, B.J.; Siegelbaum, S.A. Regulation of HCN Channel Surface Expression by a Novel C-Terminal Protein-Protein Interaction. J. Neurosci. 2004, 24, 10750–10762. [Google Scholar] [CrossRef]
- Saponaro, A.; Cantini, F.; Porro, A.; Bucchi, A.; DiFrancesco, D.; Maione, V.; Donadoni, C.; Introini, B.; Mesirca, P.; Mangoni, M.E.; et al. A synthetic peptide that prevents cAMP regulation in mammalian hyperpolarization-activated cyclic nucleotide-gated (HCN) channels. eLife 2018, 7, 085201. [Google Scholar] [CrossRef]
- Ardell, J.L.; Armour, J.A. Neurocardiology: Structure-Based Function. Compr. Physiol. 2016, 6, 1635–1653. [Google Scholar] [CrossRef] [PubMed]
- Campos, I.D.; Pinto, V.; Sousa, N.; Pereira, V.H. A brain within the heart: A review on the intracardiac nervous system. J. Mol. Cell. Cardiol. 2018, 119, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rysevaite, K.; Saburkina, I.; Pauziene, N.; Vaitkevicius, R.; Noujaim, S.F.; Jalife, J.; Pauza, D.H. Immunohistochemical characterization of the intrinsic cardiac neural plexus in whole-mount mouse heart preparations. Hear. Rhythm. 2011, 8, 731–738. [Google Scholar] [CrossRef]
- Pauza, D.H.; Skripka, V.; Pauziene, N.; Stropus, R. Morphology, distribution, and variability of the epicardiac neural ganglionated subplexuses in the human heart. Anat. Record 2000, 259, 353–382. [Google Scholar] [CrossRef]
- Armour, J.A. Cardiac neuronal hierarchy in health and disease. Am. J. Physiol. Integr. Comp. Physiol. 2004, 287, R262–R271. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, J.D.; Lawrence, Y.T.; Parsons, R.L. Enhancement of Ih, but not inhibition of IM, is a key mechanism underlying the PACAP-induced increase in excitability of guinea pig intrinsic cardiac neurons. Am. J. Physiol. Integr. Comp. Physiol. 2009, 297, R52–R59. [Google Scholar] [CrossRef] [PubMed]
- Merriam, L.A.; Barstow, K.L.; Parsons, R.L. Pituitary adenylate cyclase-activating polypeptide enhances the hyperpolarization-activated nonselective cationic conductance, Ih, in dissociated guinea pig intracardiac neurons. Regul. Pept. 2004, 123, 123–133. [Google Scholar] [CrossRef]
- Emery, E.C.; Young, G.T.; Berrocoso, E.M.; Chen, L.; McNaughton, P.A. HCN2 Ion Channels Play a Central Role in Inflammatory and Neuropathic Pain. Science 2011, 333, 1462–1466. [Google Scholar] [CrossRef]
- E McGovern, A.; Robusto, J.; Rakoczy, J.; Simmons, D.G.; Phipps, S.; Mazzone, S.B. The effect of hyperpolarization-activated cyclic nucleotide-gated ion channel inhibitors on the vagal control of guinea pig airway smooth muscle tone. Br. J. Pharmacol. 2014, 171, 3633–3650. [Google Scholar] [CrossRef]
- Kullmann, P.H.M.; Sikora, K.M.; Clark, K.L.; Arduini, I.; Springer, M.G.; Horn, J.P. HCN hyperpolarization-activated cation channels strengthen virtual nicotinic EPSPs and thereby elevate synaptic amplification in rat sympathetic neurons. J. Neurophysiol. 2016, 116, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Doan, T.N.; Stephans, K.; Ramirez, A.N.; Glazebrook, P.A.; Andresen, M.C.; Kunze, D.L. Differential Distribution and Function of Hyperpolarization-Activated Channels in Sensory Neurons and Mechanosensitive Fibers. J. Neurosci. 2004, 24, 3335–3343. [Google Scholar] [CrossRef]
- Jungen, C.; Scherschel, K.; Eickholt, C.; Kuklik, P.; Klatt, N.; Bork, N.; Salzbrunn, T.; Alken, F.; Angendohr, S.; Klene, C.; et al. Disruption of cardiac cholinergic neurons enhances susceptibility to ventricular arrhythmias. Nat. Commun. 2017, 8, 14155. [Google Scholar] [CrossRef] [PubMed]
- Pauza, D.H.; Rysevaite, K.; Inokaitis, H.; Jokubauskas, M.; Pauza, A.G.; Brack, K.E.; Pauziene, N. Innervation of sinoatrial nodal cardiomyocytes in mouse. A combined approach using immunofluorescent and electron microscopy. J. Mol. Cell. Cardiol. 2014, 75, 188–197. [Google Scholar] [CrossRef]
- Bankston, J.R.; DeBerg, H.A.; Stoll, S.; Zagotta, W.N. Mechanism for the inhibition of the cAMP dependence of HCN ion channels by the auxiliary subunit TRIP8b. J. Biol. Chem. 2017, 292, 17794–17803. [Google Scholar] [CrossRef]
- Santoro, B.; Piskorowski, R.A.; Pian, P.; Hu, L.; Liu, H.; Siegelbaum, S.A. TRIP8b Splice Variants Form a Family of Auxiliary Subunits that Regulate Gating and Trafficking of HCN Channels in the Brain. Neuron 2009, 62, 802–813. [Google Scholar] [CrossRef]
- Silbernagel, N.; Walecki, M.; Schäfer, M.K.-H.; Kessler, M.; Zobeiri, M.; Rinné, S.; Kiper, A.K.; Komadowski, M.A.; Vowinkel, K.S.; Wemhöner, K.; et al. The VAMP-associated protein VAPB is required for cardiac and neuronal pacemaker channel function. FASEB J. 2018, 32, 6159–6173. [Google Scholar] [CrossRef]
- Lau, D.H.; Mackenzie, L.; Kelly, D.J.; Psaltis, P.J.; Worthington, M.; Rajendram, A.; Kelly, D.R.; Nelson, A.J.; Zhang, Y.; Kuklik, P.; et al. Short-term hypertension is associated with the development of atrial fibrillation substrate: A study in an ovine hypertensive model. Hear. Rhythm. 2010, 7, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Noble, P.J.; Xiao, G.; Abdelrahman, M.; Dobrzynski, H.; Boyett, M.R.; Lei, M.; Noble, D. Role of pacemaking current in cardiac nodes: Insights from a comparative study of sinoatrial node and atrioventricular node. Prog. Biophys. Mol. Biol. 2008, 96, 294–304. [Google Scholar] [CrossRef]
- Hou, Y.; Scherlag, B.J.; Lin, J.; Zhang, Y.; Lu, Z.; Truong, K.; Patterson, E.; Lazzara, R.; Jackman, W.M.; Po, S.S. Ganglionated Plexi Modulate Extrinsic Cardiac Autonomic Nerve Input. J. Am. Coll. Cardiol. 2007, 50, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, P.S.; Challis, R.C.; Fowlkes, C.C.; Hanna, P.; Tompkins, J.D.; Jordan, M.C.; Hiyari, S.; Gabris-Weber, B.A.; Greenbaum, A.; Chan, K.Y.; et al. Identification of peripheral neural circuits that regulate heart rate using optogenetic and viral vector strategies. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Brack, K.E.; Coote, J.H.; Ng, G.A. Vagus nerve stimulation protects against ventricular fibrillation independent of muscarinic receptor activation. Cardiovasc. Res. 2011, 91, 437–446. [Google Scholar] [CrossRef]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.-C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.-T.; Ma, X.-J.; Luo, Y. RNAscope: A Novel in Situ RNA Analysis Platform for Formalin-Fixed, Paraffin-Embedded Tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Tu, H.; Zhang, L.; Tran, T.; Muelleman, R.; Li, Y. Diabetes alters protein expression of hyperpolarization-activated cyclic nucleotide-gated channel subunits in rat nodose ganglion cells. Neuroscience 2010, 165, 39–52. [Google Scholar] [CrossRef]
- Schauerte, P.; Scherlag, B.J.; Pitha, J.; Scherlag, M.A.; Reynolds, D.; Lazzara, R.; Jackman, W.M. Catheter Ablation of Cardiac Autonomic Nerves for Prevention of Vagal Atrial Fibrillation. Circulation 2000, 102, 2774–2780. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Scherlag, B.J.; Patterson, E.; Ikeda, A.; Lockwood, D.; Jackman, W.M. Pathophysiologic basis of autonomic ganglionated plexus ablation in patients with atrial fibrillation. Hear. Rhythm. 2009, 6, S26–S34. [Google Scholar] [CrossRef]
- Nishida, K.; Michael, G.; Dobrev, D.; Nattel, S. Animal models for atrial fibrillation: Clinical insights and scientific opportunities. Europace 2009, 12, 160–172. [Google Scholar] [CrossRef]
- Jungen, C.; Scherschel, K.; Bork, N.I.; Kuklik, P.; Eickholt, C.; Kniep, H.; Klatt, N.; Willems, S.; Nikolaev, V.O.; Meyer, C. Impact of Intracardiac Neurons on Cardiac Electrophysiology and Arrhythmogenesis in an Ex Vivo Langendorff System. J. Vis. Exp. 2018, e57617. [Google Scholar] [CrossRef] [PubMed]
- Schrickel, J.W.; Stöckigt, F.; Krzyzak, W.; Paulin, D.; Li, Z.; Lübkemeier, I.; Fleischmann, B.; Sasse, P.; Linhart, M.; Lewalter, T.; et al. Cardiac conduction disturbances and differential effects on atrial and ventricular electrophysiological properties in desmin deficient mice. J. Interv. Card. Electrophysiol. 2010, 28, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Schrickel, J.W.; Bielik, H.; Yang, A.; Schimpf, R.; Shlevkov, N.; Burkhardt, D.; Meyer, R.; Grohe, C.; Fink, K.; Tiemann, K.; et al. Induction of atrial fibrillation in mice by rapid transesophageal atrial pacing. Basic Res. Cardiol. 2002, 97, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Scherschel, K.; Hedenus, K.; Jungen, C.; Lemoine, M.D.; Rübsamen, N.; Veldkamp, M.W.; Klatt, N.; Lindner, D.; Westermann, D.; Casini, S.; et al. Cardiac glial cells release neurotrophic S100B upon catheter-based treatment of atrial fibrillation. Sci. Transl. Med. 2019, 11, eaav7770. [Google Scholar] [CrossRef]
- Rysevaite, K.; Saburkina, I.; Pauziene, N.; Noujaim, S.F.; Jalife, J.; Pauza, D.H. Morphologic pattern of the intrinsic ganglionated nerve plexus in mouse heart. Hear. Rhythm. 2011, 8, 448–454. [Google Scholar] [CrossRef]
- Dent, J.A.; Polson, A.G.; Klymkowsky, M.W. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development 1989, 105, 61–74. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Merritt, C.R.; Ong, G.T.; Church, S.E.; Barker, K.; Danaher, P.; Geiss, G.; Hoang, M.; Jung, J.; Liang, Y.; McKay-Fleisch, J.; et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 2020, 38, 586–599. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scherschel, K.; Bräuninger, H.; Mölders, A.; Erlenhardt, N.; Amin, E.; Jungen, C.; Pape, U.; Lindner, D.; Chetkovich, D.M.; Klöcker, N.; et al. Characterization of the HCN Interaction Partner TRIP8b/PEX5R in the Intracardiac Nervous System of TRIP8b-Deficient and Wild-Type Mice. Int. J. Mol. Sci. 2021, 22, 4772. https://doi.org/10.3390/ijms22094772
Scherschel K, Bräuninger H, Mölders A, Erlenhardt N, Amin E, Jungen C, Pape U, Lindner D, Chetkovich DM, Klöcker N, et al. Characterization of the HCN Interaction Partner TRIP8b/PEX5R in the Intracardiac Nervous System of TRIP8b-Deficient and Wild-Type Mice. International Journal of Molecular Sciences. 2021; 22(9):4772. https://doi.org/10.3390/ijms22094772
Chicago/Turabian StyleScherschel, Katharina, Hanna Bräuninger, Andrea Mölders, Nadine Erlenhardt, Ehsan Amin, Christiane Jungen, Ulrike Pape, Diana Lindner, Dane M. Chetkovich, Nikolaj Klöcker, and et al. 2021. "Characterization of the HCN Interaction Partner TRIP8b/PEX5R in the Intracardiac Nervous System of TRIP8b-Deficient and Wild-Type Mice" International Journal of Molecular Sciences 22, no. 9: 4772. https://doi.org/10.3390/ijms22094772
APA StyleScherschel, K., Bräuninger, H., Mölders, A., Erlenhardt, N., Amin, E., Jungen, C., Pape, U., Lindner, D., Chetkovich, D. M., Klöcker, N., & Meyer, C. (2021). Characterization of the HCN Interaction Partner TRIP8b/PEX5R in the Intracardiac Nervous System of TRIP8b-Deficient and Wild-Type Mice. International Journal of Molecular Sciences, 22(9), 4772. https://doi.org/10.3390/ijms22094772







