Ribonucleic Acid Export 1 Is a Kinetochore-Associated Protein That Participates in Chromosome Alignment in Mouse Oocytes
Abstract
1. Introduction
2. Results
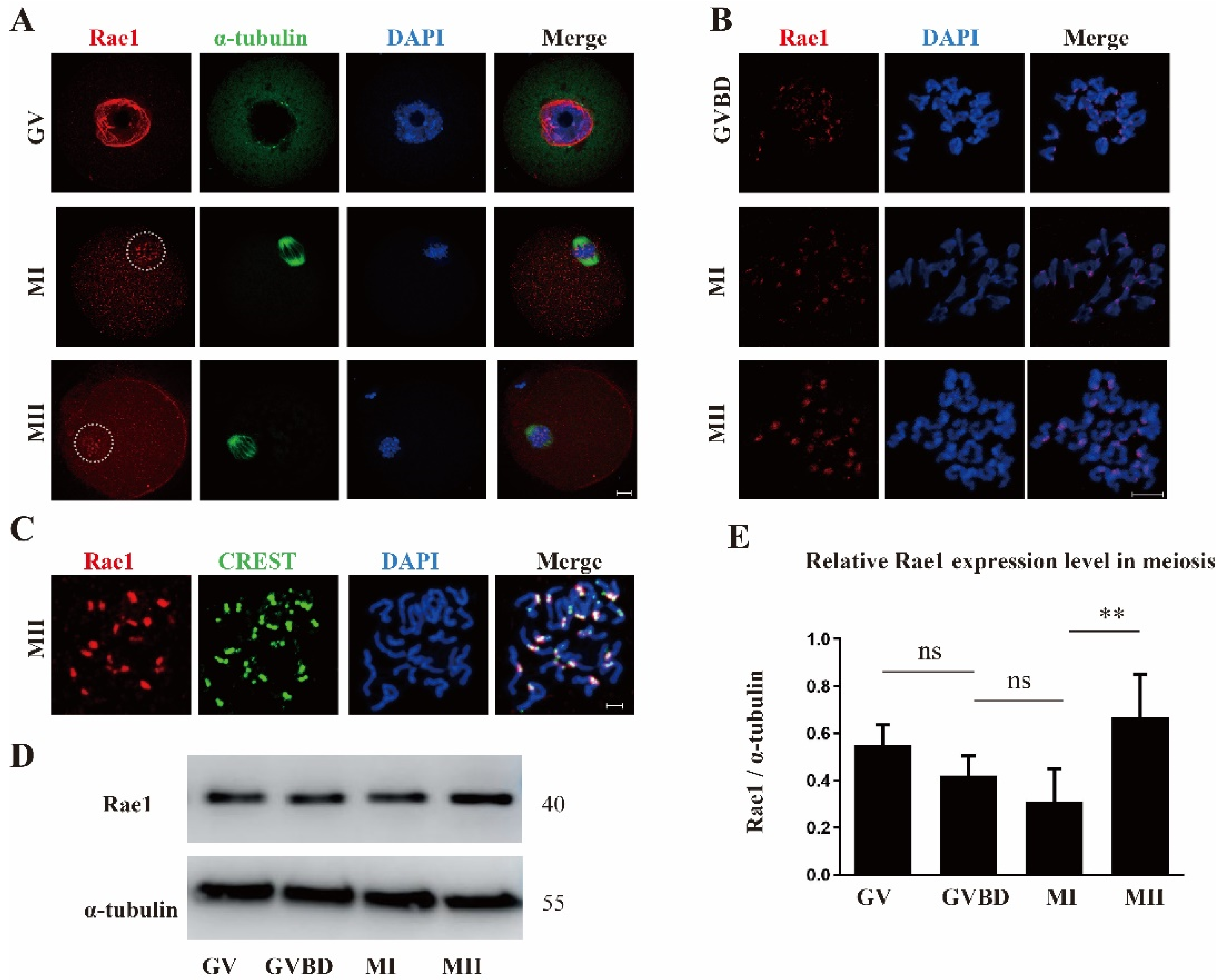
2.1. Subcellular Localization and Expression Pattern of Rae1 during Mouse Oocyte Maturation
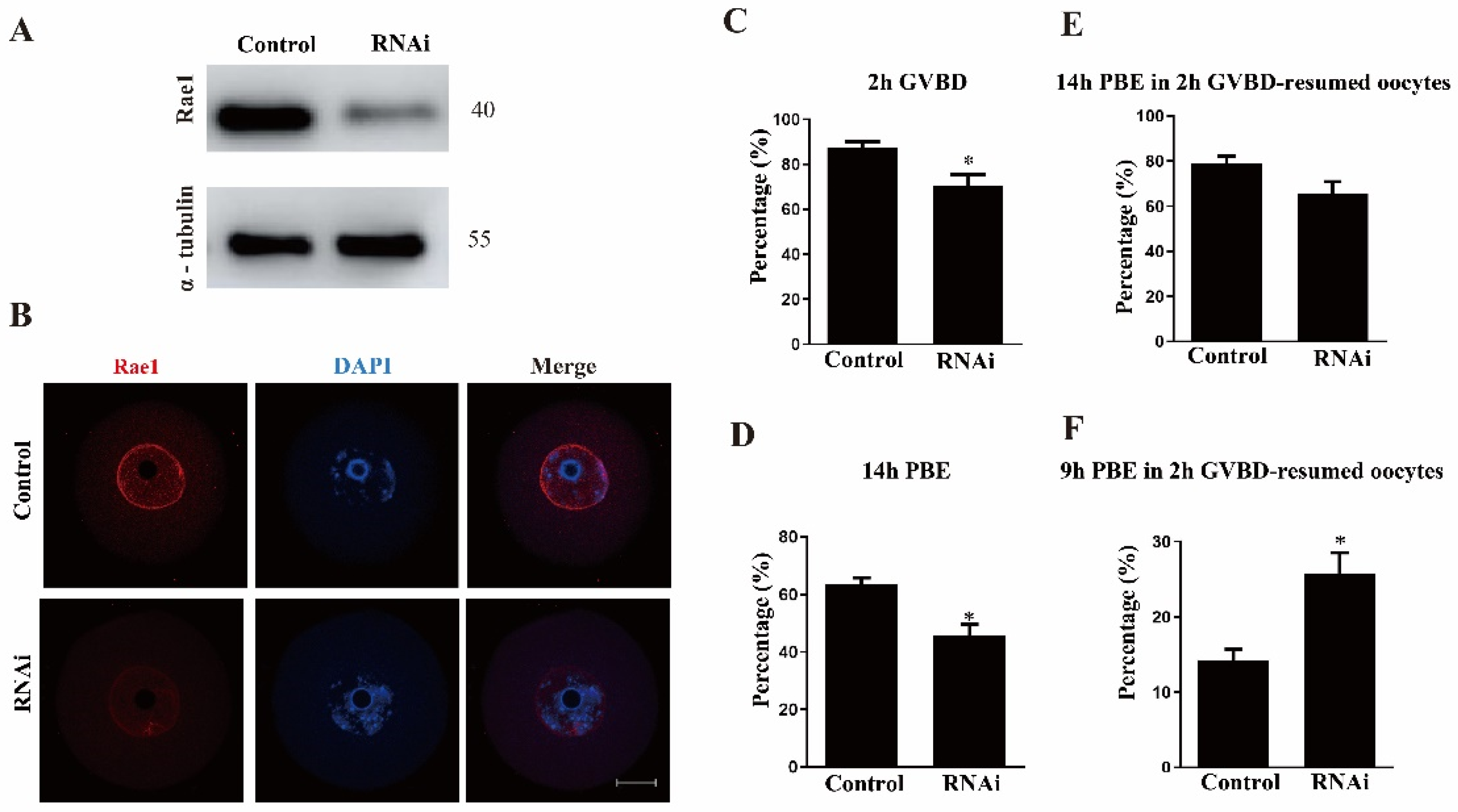
2.2. Rae1 Knockdown Weakens Meiotic Resumption but Precociously Promotes the First Polar Extrusion
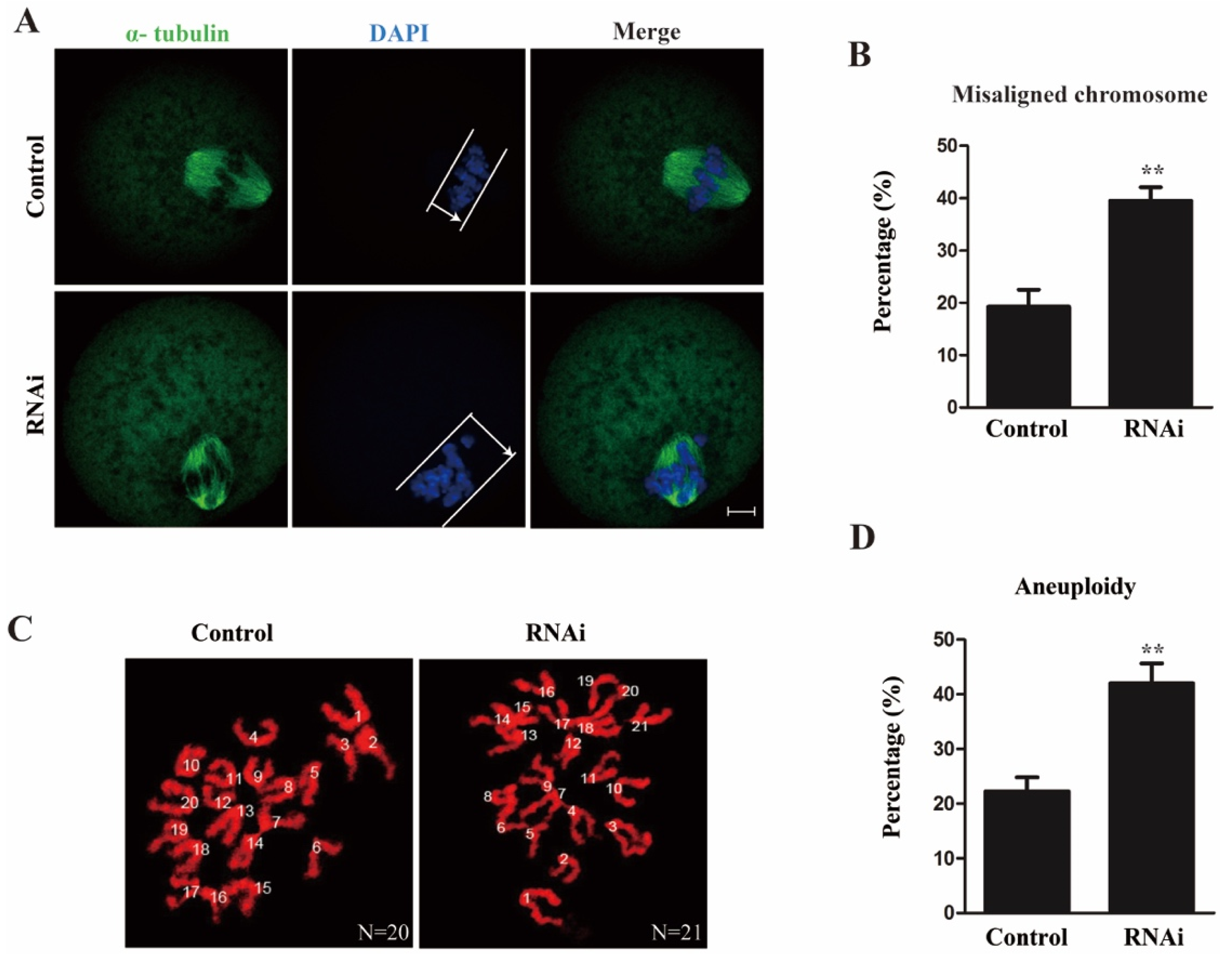
2.3. Rae1 Interference Induces Abnormal Chromosome Alignment and a High Incidence of Aneuploidy
2.4. Rae1 Depletion Compromises K-MT Attachments during Oocyte Meiotic Maturation
2.5. Rae1 Depletion Had No Effects on BubR1 and Mad1 Location
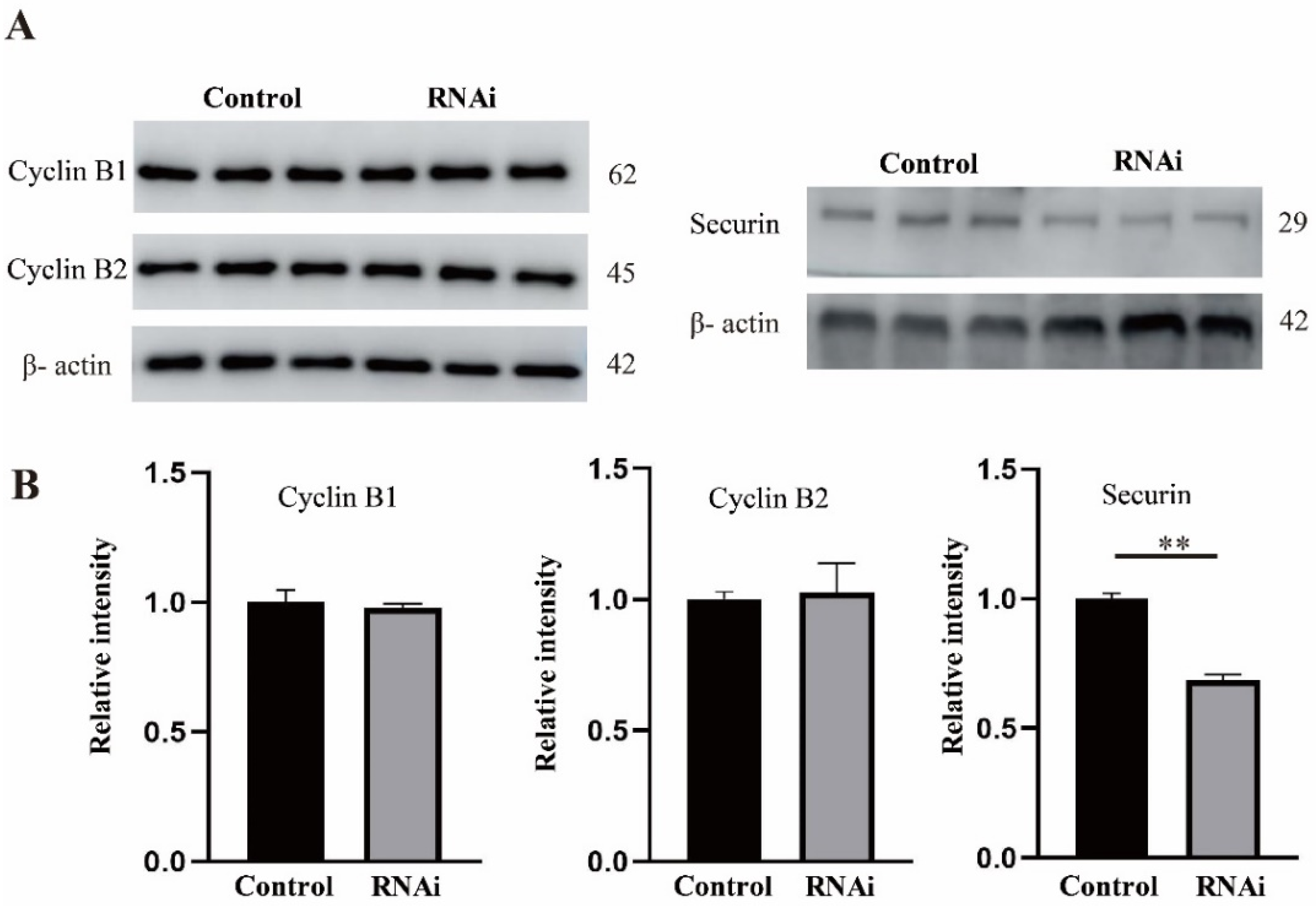
2.6. Rae1 Depletion Decreases the Protein Level of securin but Not Cyclin B
3. Discussion
4. Materials and Methods
4.1. Animal Statement
4.2. Antibodies and Reagents
4.3. Oocyte Retrieval and Culture
4.4. Microinjection of Rae1-Targeted Short Interfering siRNA
4.5. Immunofluorescence and Confocal Microscopy
4.6. Western Blotting
4.7. Oocyte Cold Treatment
4.8. Chromosome Spreading
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Cronshaw, J.M.; Krutchinsky, A.N.; Zhang, W.; Chait, B.T.; Matunis, M.J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002, 158, 915–927. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Bharathi, A.; Ghosh, A.; Whalen, W.; Fitzgerald, E.; Dhar, R. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J. Biol. Chem. 1995, 270, 7411–7419. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.; Watkins, J.L.; Wente, S.R. GLE2. A Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol. Biol. Cell 1996, 7, 1921–1937. [Google Scholar] [CrossRef] [PubMed]
- Whalen, W.A.; Bharathi, A.; Danielewicz, D.; Dhar, R. Advancement through mitosis requires rae1 gene function in fission yeast. Yeast 1997, 13, 1167–1179. [Google Scholar] [CrossRef]
- Blower, M.D.; Nachury, M.; Heald, R.; Weis, K.A. Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 2005, 121, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.W.; Blobel, G.; Coutavas, E. Rae1 interaction with NuMA is required for bipolar spindle formation. Proc. Natl. Acad. Sci. USA 2006, 103, 19783–19787. [Google Scholar] [CrossRef]
- Stockum, A.; Snijders, A.P.; Maertens, G.N. USP11 deubiquitinates RAE1 and plays a key role in bipolar spindle formation. PLoS ONE 2018, 13, e0190513. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.W. Interaction between Rae1 and cohesin subunit SMC1 is required for proper spindle formation. Cell Cycle 2010, 9, 198–200. [Google Scholar] [CrossRef]
- Wong, R.W.; Blobel, G. Cohesin Subunit SMC1 Associates with Mitotic Microtubules at the Spindle Pole. Proc. Natl. Acad. Sci. USA 2018, 105, 15441–15445. [Google Scholar] [CrossRef]
- Reddy, D.M.; Aspatwar, A.; Dholakia, B.B.; Gupta, V.S. Evolutionary analysis of WD40 super family proteins involved in spindle checkpoint and RNA export: Molecular evolution of spindle checkpoint. Bioinformation 2008, 2, 461–468. [Google Scholar] [CrossRef][Green Version]
- Wang, X.; Babu, J.R.; Harden, J.M.; Jablonski, S.A.; Gazi, M.H.; Lingle, W.L.; de Groen, P.C.; Yen, T.J.; van Deursen, J.M. The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLE2p-binding sequence (GLEBS)-containing proteins. J. Biol. Chem. 2001, 276, 26559–26567. [Google Scholar] [CrossRef]
- Babu, J.R.; Jeganathan, K.B.; Baker, D.J.; Wu, X.; Kang-Decker, N.; Van deursen, J.M. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 2003, 160, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Volpi, S.; Bongiorni, S.; Fabbretti, F.; Wakimoto, B.T.; Prantera, G. Drosophila rae1 is required for male meiosis and spermatogenesis. J. Cell Sci. 2013, 126, 3541–3551. [Google Scholar] [CrossRef] [PubMed]
- Verlhac, M.H.; Terret, M.E. Oocyte maturation and development. F1000Res 2016, 5, 309. [Google Scholar] [CrossRef]
- Nagaoka, S.I.; Hassold, T.J.; Hunt, P.A. Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 2012, 13, 493–504. [Google Scholar] [CrossRef]
- Cheeseman, I.M.; Desai, A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008, 9, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A.; Salmon, E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007, 8, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Santaguida, S.; Musacchio, A. The life and miracles of kinetochores. EMBO J. 2009, 28, 2511–2531. [Google Scholar] [CrossRef]
- Chen, F.; Jiao, X.F.; Zhang, J.Y.; Wu, D.; Ding, Z.M.; Wang, Y.S.; Miao, Y.L.; Huo, L.J. Nucleoporin35 is a novel microtubule associated protein functioning in oocyte meiotic spindle architecture. Exp. Cell Res. 2018, 371, 435–443. [Google Scholar] [CrossRef]
- Herbert, M.; Kalleas, D.; Cooney, D.; Lamb, M.; Lister, L. Meiosis and maternal aging: Insights from aneuploid oocytes and trisomy births. Nat. Rev. Mol. Cell Biol. 2015, 7, a017970. [Google Scholar] [CrossRef]
- Godek, K.M.; Kabeche, L.; Compton, D.A. Regulation of kinetochore–microtubule attachments through homeostatic control during mitosis. Nat. Rev. Mol. Cell Biol. 2015, 16, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.F.; Ren, Y.X.; Yuan, P.; Yan, L.Y.; Qiao, J. TRAIP is involved in chromosome alignment and SAC regulation in mouse oocyte meiosis. Sci. Rep. 2016, 6, 29735. [Google Scholar] [CrossRef] [PubMed]
- Horner, K.; Livera, G.; Hinckley, M.; Trinh, K.; Storm, D.; Conti, M. Rodent oocytes express an active adenylyl cyclase required for meiotic arrest. Dev. Biol. 2003, 258, 385–396. [Google Scholar] [CrossRef]
- Liu, J.; Lu, X.; Wang, W.; Zhu, J.; Li, Y.; Luo, L.; Zhang, W. Activity of MPF and expression of its related genes in mouse MI oocytes exposed to cadmium. Food Chem. Toxicol. 2018, 112, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Laurell, E.; Beck, K.; Krupina, K.; Theerthagiri, G.; Bodenmiller, B.; Horvath, P.; Aebersold, R.; Antonin, W.; Kutay, U. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 2011, 144, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Powers, M.A. Nup98-homeodomain fusions interact with endogenous Nup98 during interphase and localize to kinetochores and chromosome arms during mitosis. Mol. Biol. Cell 2010, 2, 1585–1596. [Google Scholar] [CrossRef] [PubMed]
- Jeganathan, K.B.; Malureanu, L.; Van deursen, J.M. The Rae1–Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature 2005, 438, 1036. [Google Scholar] [CrossRef]
- Herbert, M.; Levasseur, M.; Homer, H.; Yallop, K.; Murdoch, A.; McDougall, A. Homologue disjunction in mouse oocytes requires proteolysis of securin and cyclin B1. Nat. Cell Biol. 2003, 5, 1023–1025. [Google Scholar] [CrossRef]
- Nabti, I.; Grimes, R.; Sarna, H.; Marangos, P.; Carroll, J. Maternal age-dependent APC/C-mediated decrease in securin causes premature sister chromatid separation in meiosis II. Nat. Commun. 2017, 8, 15346. [Google Scholar] [CrossRef]
- Lara-Gonzalez, P.; Westhorpe, F.G.; Taylor, S.S. The spindle assembly checkpoint. Curr. Biol. 2012, 22, R966–R980. [Google Scholar] [CrossRef]
- Baker, D.J.; Jeganathan, K.B.; Malureanu, L.; Perez-Terzic, C.; Terzic, A.; Van deursen, J.M. Early aging–associated phenotypes in Bub3/Rae1 haploinsufficient mice. J. Cell Biol. 2006, 172, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Jeganathan, K.B.; Baker, D.J.; Van deursen, J.M. Securin associates with APCCdh1 in prometaphase but its destruction is delayed by Rae1 and Nup98 until the metaphase/anaphase transition. Cell Cycle 2006, 5, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Naylor, R.M.; Jeganathan, K.B.; Cao, X.; Van deursen, J.M. Nuclear pore protein NUP88 activates anaphase-promoting complex to promote aneuploidy. J. Clin. Investig. 2016, 126, 543–559. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.J.; Wu, D.; Jiao, X.F.; Khan, F.A.; Xiong, C.L.; Liu, X.M.; Yang, J.; Yin, T.L.; Huo, L.J. Maternal SENP7 programs meiosis architecture and embryo survival in mouse. Biochim. Biophys. Acta 2017, 1864, 1195–1206. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, F.; Jiao, X.-F.; Meng, F.; Wang, Y.-S.; Ding, Z.-M.; Miao, Y.-L.; Xiong, J.-J.; Huo, L.-J. Ribonucleic Acid Export 1 Is a Kinetochore-Associated Protein That Participates in Chromosome Alignment in Mouse Oocytes. Int. J. Mol. Sci. 2021, 22, 4841. https://doi.org/10.3390/ijms22094841
Chen F, Jiao X-F, Meng F, Wang Y-S, Ding Z-M, Miao Y-L, Xiong J-J, Huo L-J. Ribonucleic Acid Export 1 Is a Kinetochore-Associated Protein That Participates in Chromosome Alignment in Mouse Oocytes. International Journal of Molecular Sciences. 2021; 22(9):4841. https://doi.org/10.3390/ijms22094841
Chicago/Turabian StyleChen, Fan, Xiao-Fei Jiao, Fei Meng, Yong-Sheng Wang, Zhi-Ming Ding, Yi-Liang Miao, Jia-Jun Xiong, and Li-Jun Huo. 2021. "Ribonucleic Acid Export 1 Is a Kinetochore-Associated Protein That Participates in Chromosome Alignment in Mouse Oocytes" International Journal of Molecular Sciences 22, no. 9: 4841. https://doi.org/10.3390/ijms22094841
APA StyleChen, F., Jiao, X.-F., Meng, F., Wang, Y.-S., Ding, Z.-M., Miao, Y.-L., Xiong, J.-J., & Huo, L.-J. (2021). Ribonucleic Acid Export 1 Is a Kinetochore-Associated Protein That Participates in Chromosome Alignment in Mouse Oocytes. International Journal of Molecular Sciences, 22(9), 4841. https://doi.org/10.3390/ijms22094841





