Abstract
MicroRNAs (miRNAs) are regulators of the post-transcription stage of gene activity documented to play central roles in flower and fruit development in model plant species. However, little is known about their roles and differences in domesticated and wild Capsicum species. In this study, we used high-throughput sequencing to analyze the miRNA content at three developmental stages (flower, small fruit, and middle fruit) from two cultivated (C. baccatum and C. annuum) and two wild (C. chacoense and C. eximium) pepper species. This analysis revealed 22 known and 27 novel miRNAs differentially expressed across species and tissues. A number of stage- and species-specific miRNAs were identified, and Gene Ontology terms were assigned to 138 genes targeted by the miRNAs. Most Gene Ontology terms were for the categories “genetic information processing”, “signaling and cellular processes”, “amino acid metabolism”, and “carbohydrate metabolism”. Enriched KEGG analysis revealed the pathways amino acids, sugar and nucleotide metabolism, starch and sucrose metabolism, and fructose-mannose metabolism among the principal ones regulated by miRNAs during pepper fruit ripening. We predicted miRNA–target gene interactions regulating flowering time and fruit development, including miR156/157 with SPL genes, miR159 with GaMYB proteins, miR160 with ARF genes, miR172 with AP2-like transcription factors, and miR408 with CLAVATA1 gene across the different Capsicum species. In addition, novel miRNAs play an important role in regulating interactions potentially controlling plant pathogen defense and fruit quality via fructokinase, alpha-L-arabinofuranosidase, and aromatic and neutral amino acid transporter. Overall, the small RNA-sequencing results from this study represent valuable information that provides a solid foundation for uncovering the miRNA-mediated mechanisms of flower and fruit development between domesticated and wild Capsicum species.
1. Introduction
MicroRNAs (miRNAs) are a specific class of 20- to 24-nt endogenous small non-protein coding RNAs involved in post-transcriptional and translational gene expression regulation in plants and animals [1,2]. Mature miRNA, generated from longer pri-RNA via nuclease cleavage processes [3], negatively regulate gene expression by recognition and complementary binding to the open reading frame or untranslated regions (UTRs) of target genes. The expression of multiple genes can be regulated by a single miRNA, and multiple miRNAs can control a single gene expression [4]. In plants, the complementarity between miRNA and their targets is very high, which results in RNA-induced silencing complexes by degrading the target mRNA or inhibiting mRNA translation [5]. Therefore, miRNA-mediated gene silencing plays an important role in several essential plant biological processes, including developmental control [6,7], hormone secretion [8], cell differentiation and proliferation [6], as well as environmental adaptation and response to conditions such as salinity, drought, and low temperature [9,10,11,12]. Similarly, some miRNAs mediate plant–microbe associations, which suggests their participation in processes such as symbiosis events and plant–pest interactions [13,14,15]. Nevertheless, the regulation of miRNAs is known to be spatiotemporally specific, so understanding their regulatory roles in plants is difficult [16].
The rapid advances in bioinformatics and next-generation sequencing technologies have led to the continuously increasing identification of novel miRNAs in plants. Different databases provide relevant information on miRNAs; one is miRbase (v22.1, http://www.mirbase.org, accessed on 1 August 2020), which contains about 38,600 entries representing hairpin precursor miRNAs that express 48,860 mature miRNA products in 271 species [17]: 10,414 belong to 82 different plant species, including the model Arabidopsis thaliana, and crops such as Oryza sativa, Glycine max, and Medicago truncatula. However, the miRNA annotation of other important plants such as those in the Solanaceae family, with more than 3,000 species [18], remains limited. Currently, miRbase contains miRNA information for only a few members of this family—Nicotiana tabacum (164), Solanum lycopersicum (147), and Solanum tuberosum (343). Pepper (Capsicum spp.), which also belongs to the Solanaceae family, exhibits wide diversity, with more than 200 species that vary by color, size, shape, and chemical composition. It is one of the most economically important crops cultivated worldwide because of its economic importance and also its medicinal and nutrimental value. In addition to their dietary and culinary importance, capsaicinoid compounds (capsaicin and dihydrocapsaicin) in pepper have a beneficial effect for humans, including antioxidant, anticarcinogenic, antimutagenic, antiaging, and antibacterial properties [19,20,21].
The release of the pepper (Capsicum annuum) genome sequence [19,20] has offered an opportunity to better understand different molecular mechanisms at a transcriptional level. To date, only a few studies have focused on pepper miRNA profiles in C. annuum [22,23]; however, considering the importance of pepper, we need to understand the function and expression of miRNAs across different Capsicum species. Further information on their expression across the diverse developmental stages may be useful to elucidate the mechanisms involved in gene regulation and can provide insights into the biological processes underlying the environmental adaptation of Capsicum species. In this study, we used high-throughput sequencing to identify pepper miRNAs in two cultivated Capsicum species, C. baccatum and C. annuum, and two wild Capsicum species, C. chacoense and C. eximium, at three different developmental stages (flower, small fruit, and middle fruit). Simultaneously, we investigated the dynamic regulation and studied the evolutionary changes of the identified miRNA genes. The identification of putatively conserved and novel miRNAs in inter/intra-species levels across different members of the Capsicum genus provides valuable insights into the evolution of the microRNAome with respect to domestication and selection events related to fruit development.
2. Results
2.1. Analysis of Small RNAs in Pepper
For comprehensive analysis of the Capsicum miRNAome and to identify putative miRNAs associated with fruit development, we sequenced 12 small-RNA libraries derived from flower, small fruit (6 days post anthesis) and medium fruit (25 days post anthesis) developmental stages of four Capsicum species (Figure 1) by using the Illumina HiSeq 2500 instrument. A total of 162,841,574 raw reads were obtained from the 12 libraries. After removing adaptors, low-quality reads, poly-A sequences, and reads of <18 and >30 nt in length, the number of clean reads ranged from ~3 million in C. annuum small fruit, to ~8 million in C. baccatum small fruit (Table S1). The small RNA clean reads were further classified into different categories by BLAST searches against Rfam and Repbase databases, and noncoding RNAs including rRNA, tRNA, snoRNA, snRNA sequences were removed. The remaining sequences were then mapped to the pepper genome to determine whether they could be candidate miRNAs and were selected based on strict criteria for annotation of plant miRNA. Furthermore, parameters such as length distribution and 5′-end of candidate miRNAs were considered by using the clean reads from all species and tissues (Figure 2). Most of these small RNAs had a 5´-end terminal U or A, which indicates canonical small RNAs [24]. Likewise, the length distribution ranged from 20 to 24 nt, the typical length of canonical miRNAs. Although the size distribution of all small RNAs was diverse and varied across species and tissues, the dominant length was 24 nt followed by 23, 22, and 21 nt in C. baccatum and C. eximium (Figure 2a,d). However, this length pattern was not observed in C. chacoense and C. annuum species, whose predominant lengths were 23 nt followed by 24 nt in C. annuum; 21 nt was the predominant length in C. chacoense at middle fruit stage.
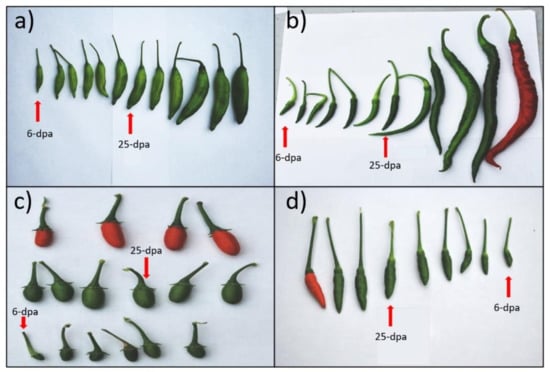
Figure 1.
Morphological features of pepper fruit development stages of four Capsicum species. Harvested fruit at different developmental stages from days after pollination of (a) C. baccatum, (b) C. annuum, (c) C. chacoense, and (d) C. eximium. Red arrows indicate small fruit (6-dpa) and middle fruit (25-dpa) stages that were collected for small RNA sequencing.
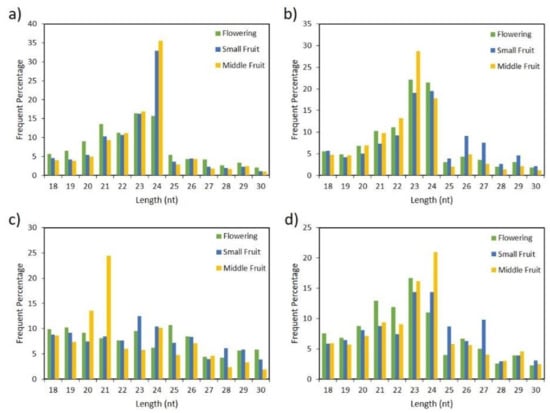
Figure 2.
Length distribution of small RNAs in (a) C. baccatum, (b) C. annuum, (c) C. chacoense, and (d) C. eximium at flowering, small, and medium fruit stages.
2.2. Identification of Known and Novel miRNAs in Pepper
To identify conserved miRNAs in each library, unique sRNA reads were BLASTN searched against currently known mature miRNAs in miRBase (v22.1) allowing one or two mismatches between sequences. Consequently, 22 known miRNAs from 21 conserved families were identified (Table 1). The only miRNA family that showed variants was miR166, with two members (miR166a and miR166b). To predict novel miRNAs in Capsicum species, sRNA reads that were unmatched to the miRbase were aligned with the genome sequences of C. annuum by using miRDeep2. Ultimately, 27 novel miRNA candidates with stable hairpin structures were identified and designated miR01 to miR27. In addition, precursor sequences from the pepper genome database were obtained (Table S2). Notably, candidate precursors for all the 27 predicted novel miRNAs were suggested. Furthermore, the hairpin structures of these novel small RNAs were predicted by using the RNAfold web server. All the novel miRNAs precursors in pepper possessed typical stem-loop structures and had negative folding free energies ranging from -62.6 to -15.9 according to RNAfold. The predicted secondary structures of six randomly selected novel miRNAs candidates are in Figure 3. In addition, we determined whether these novel miRNA candidates contained miRNA* sequences in the 5p or 3p terminus, which resulted in the identification of the miRNA* sequence in most of the candidate miRNAs, with exception of 6 (Table S2). The first cleavage position is critical to determine the mature miRNA sequence and resulting target specificity [25,26]. Therefore, base composition of miRNAs plays another important role because it may affect secondary structures and biological properties. Nucleotide bias analysis revealed that 21- to 22-nt miRNAs more frequently contained G and U at the first position; however, 23- and 24-nt miRNAs had a strong preference for G across tissues and species (Figure S1). Although the nucleotide bias at each position had wide fluctuation, they followed similar patterns through all species and tissues. This was especially observed at the 10th nucleotide, where the predominant base was G followed by C and U, and at the 11th nucleotide, where the four bases were present with dominancy of base C (Figure S2). Previous studies suggested that the first nucleotide is important for miRNA sorting [26] and that the 10th and 11th nucleotides are responsible for guiding the miRNA to cleave the target mRNA [27]. Altogether, these results suggest that novel miRNAs might be involved in regulating similar physiological and biological processes across Capsicum species.

Table 1.
Expression level based on transcripts per million (TPM) of known miRNAs identified in Capsicum species at flowering, small fruit, and middle fruit stages.
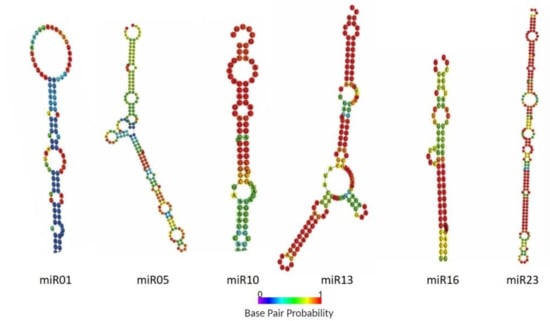
Figure 3.
Predicted secondary hairpin structures of six novel miRNAs identified in Capsicum species. Base pair probability is represented by color: red, high-probability, and purple, low-probability.
2.3. Expression Pattern of Known and Novel miRNAs in Pepper
We analyzed the sequence frequencies of known and novel miRNAs from the 12 libraries to estimate the pattern expression of the miRNAs found at a specific developmental stage or tissue and to infer their possible roles in the different Capsicum species. Seven of 22 known miRNAs were expressed at different densities in all stages and species (Table 1 and Figure 4): 9, 11 and 12 known miRNAs were expressed at flower, small fruit, and medium fruit, respectively, among all Capsicum species. From the miRNA expression at flower stage, C. baccatum, C. annuum, and C. chacoense were clustered together; however, miR157, 166b, 172 and miR319 showed higher expression in C. annuum. Furthermore, the expression of 13 miRNAs (miR156, 159, 160, 162, 166a, 167, 168, 171, 390, 394, 396, 403, and 6478) at small and middle fruit stages was higher in C. baccatum than the other species. Similarly, miR164, 165 and 398 showed high expression in C. chacoense and C. eximium at middle fruit stage, whereas miR408 was highly expressed in C. eximium at small fruit stage. For novel miRNAs, 8 of 27 were expressed in all tissues and species at different levels (Table 2, Figure 5). Additionally, 15, 11, and 10 were shared between Capsicum species at flower, small fruit, and middle fruit stage, respectively (Figure 5b,c,d). Similar to known miRNAs, some novel miRNAs also showed species-specific expression; for instance, 14 miRNAs including miR01, 02, 03, 04, 05, 06, 08, 09, 10, 11, 12, 16, 18, and 27 were upregulated in C. baccatum at small and middle fruit stages. Likewise, at the same stages, miR014 showed high expression in C. annuum, whereas miR07 and 20 were highly expressed in C. chacoense. Moreover, miR17, 22, 23, and 24 showed high abundance at flower stage in C. eximium, but miR25 was highly expressed at the same stage in C. chacoense. Finally, miR13, 15, 19, and 21 showed low expression patterns across all species and stages.
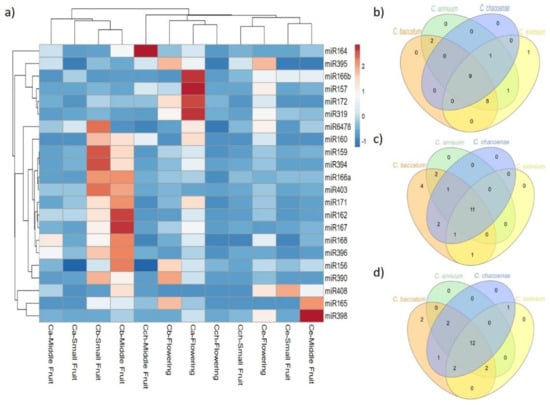
Figure 4.
Differential expression analysis of known miRNAs. (a) Heat map of known miRNA expression profiles in C. baccatum (Cb), C. annuum (Ca), C. chacoense (Cch), and C. eximium (Ce) at different stages analyzed. The expression levels are represented by the color: red, high-expressed; and blue, low-expressed. Venn diagram analysis of shared known miRNAs at (b) flowering, (c) small, and (d) medium fruit stages across Capsicum species.

Table 2.
Expression level based on transcripts per million (TPM) of novel miRNAs identified in Capsicum species at flowering, small fruit, and middle fruit stages.
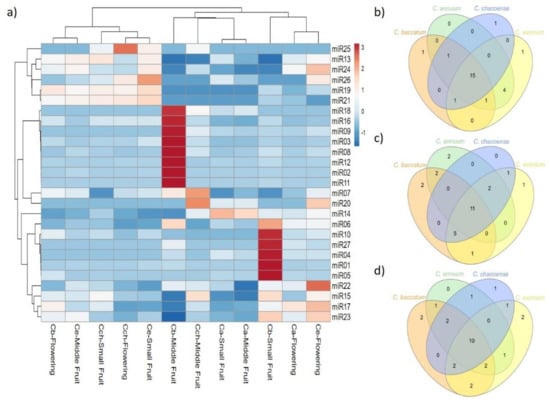
Figure 5.
Differential expression analysis of novel miRNAs. (a) Heat map of novel miRNA expression profiles in C. baccatum (Cb), C. annuum (Ca), C. chacoense (Cch), and C. eximium (Ce) at different stages analyzed. The expression levels are represented by the color: red, high-expressed; and blue, low-expressed. Venn diagram analysis of shared novel miRNAs at (b) flowering, (c) small, and (d) medium fruit stages across Capsicum species.
2.4. Prediction of Putative Targets for Known and Novel miRNAs in Pepper
To better understand the function of the identified miRNAs and assuming that plant miRNAs have a nearly perfect match to their target mRNAs, putative target genes were predicted by using the psRNA target program. Additionally, we performed ‘microRNA:mRNA’ seed sequence similarity analysis between known and novel miRNAs and their target mRNAs because the seed sequence is essential for binding to take place (Mullany et al. 2016) (Table S3). We identified 78 and 60 potential candidate targets for known and novel miRNAs, respectively (Table S4). The number of putative target genes for a single miRNA ranged from 1 to 6 for known miRNAs and from 1 to 4 for novel miRNAs. The set of predicted target proteins was functionally characterized by using the BLASTKOALA sequence similarity tool. BLASTKOALA annotated 54 (39.1%) of 138 proteins identified. A group of 21 genes was thought to be involved in genetic information processing, but other genes were predicted to participate in processes such as signaling and cellular processes (11), metabolism (7), environmental information processing (3), amino acid metabolism (2), energy metabolism (2), xenobiotics biodegradation and metabolism (2), carbohydrate metabolism (2), lipid metabolism (1), metabolism of terpenoids and polyketides (1), metabolism of cofactors and vitamins (1), and glycan biosynthesis and metabolism (1) (Figure S3).
Moreover, to gain insights into a global overview of the regulatory functions of the miRNAs across Capsicum species, we analyzed the GO terms for 116 of the 138 identified targets. GO analysis suggested the putative participation of miRNA targets in multiple biological processes, molecular functions, and cellular component (Figure S4). The major biological processes predicted for these GO-defined target genes were regulation of transcription, DNA-replication (20), oxidation-reduction process (10), protein phosphorylation (10), auxin-activated signaling pathway (5), and hormone-mediated signaling pathway (4). The molecular functions were mostly classified as DNA binding (25), ATP binding (20), protein serine/threonine kinase activity (13), DNA-binding transcription factor (TF) activity (10), and zinc ion binding (8). For cellular components, most genes were related to nucleus (35), membrane (35), plasma membrane (17), cytoplasm (8), and cytosol (8). Furthermore, enriched KEGG pathway analysis of target genes of differentially expressed miRNAs revealed that miRNAs regulate genes involved in amino sugar and nucleotide sugar metabolism, starch and sucrose metabolism, and fructose-mannose metabolism pathways (Figure S5).
2.5. Expression Profile of miRNA Target Genes in C. annuum
To investigate the expression profile of miRNA predicted target genes across different tissues, including leaf, steam, and placenta tissue at 6, 16, and 25 dpa, we used publicly available RNA-seq data for C. annuum cv. CM33428. A heat map was used to visualize tissue-specific expression patterns of target genes (Figure 6). Overall, 112 of 138 predicted genes were identified, and most genes exhibited unique expression profiles. Genes showing exclusively high expression in leaf tissue encoded proteins for ABC transporter (CA06g14420), cellulose synthase (CA10g10190), FGGY carbohydrate kinase (CA07g00430), or class III HD-Zip protein 3 (CA02g10530) or were involved in the PHO system (CA02g17380). Placenta tissue at 6-dpa showed high expression of genes involved in regulation of transcription such as SPL (squamosa promoter binding protein) domain class TF (CA03g12170), TCP TF (CA08g04030), and NAC (NAM, ATAF, and CUC) domain TF (CA06g18770), thus indicating the participation of these genes in fruit development. The same tissue at 16-dpa showed high expression of genes associated with plant hormone signaling: auxin response factor (CA04g09370) and serine/threonine-protein kinase (CA04g16720). Likewise, genes exhibiting the same profile encoded proteins such as class III HD-Zip protein (CA12g13110), protein SUPPRESSOR OF GENE SILENCING 3 (CA03g17450), and fructokinase activity (CA02g24240). Meanwhile, genes upregulated in placenta tissue at 25 dpa were related to carbohydrate metabolism (CA01g27530, CA03g19360, and CA03g29340), aromatic and neutral amino acid transporter (CA04g05160), leucine rich repeat receptor protein kinase CLAVATA1 (CA02g24570), and TFs with key roles in ripening such as AP2 (APETALA2) and NAC TFs (CA11g14070, CA12g13470). Genes showing high expression during pepper fruit development and ripening at 16 and 25-dpa were the SPL TF (CA02g15200) and the response regulator ARR12-like protein (CA07g02260).
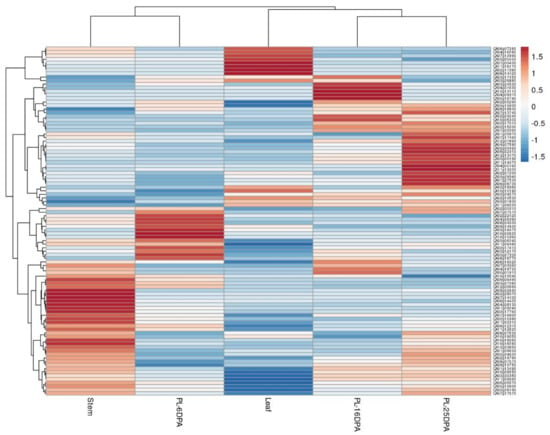
Figure 6.
Expression patterns of known and novel miRNA target genes in leaf, stem, and placenta (PL) tissue at 6, 16, and 25 days post-anthesis of C. annuum var CM344. The expression levels are represented by color: red, upregulated; and blue, downregulated.
2.6. Validation of miRNA and Target Gene Expression
To validate the expression profiles of miRNAs in Capsicum species at flower, small fruit (6-dpa), and middle fruit (25-dpa) stages, we investigated 6 known (miR159, 162, 166a, 319, 396, and 6478) and six novel miRNAs (miR01, 05, 10, 13, 16, and 23) by using stem-loop qRT-PCR based on their sequencing frequencies (Figure 7). In general, expression of most of these miRNAs agreed with sequencing data, with slight variation. The expression profiles of miR162, 166a, 319, 396, 6478, 01, 05, and 16 were consistent with the results from sRNA-sequencing. However, the remaining four miRNAs tested showed inconsistent results between stem-loop qRT-PCR and sRNA sequencing in all or specific Capsicum species. For instance, miR159 showed a discrepancy in C. annuum and C. eximium species, miR10 showed differences in only C. eximium, and miR23 showed a different expression trend in C. annuum. To validate the expression profiles of protein-coding genes targeted by miRNAs, we used qRT-PCR for all Capsicum species at different stages. The expression profiles of auxin-induced protein, class III HD-Zip protein, TCP TF, and serine carboxypeptidase protein, regulated by miR162, 166a, 319, and 016, respectively, showed opposite expression profiles from their target genes, as expected for miRNA targets (Figure 8). As expected, the expression of the remaining predicted targets was not opposite to their miRNA profiles in at least one species, which suggests that the activity of these genes could be determined by translational repression and/or by multiple miRNAs.
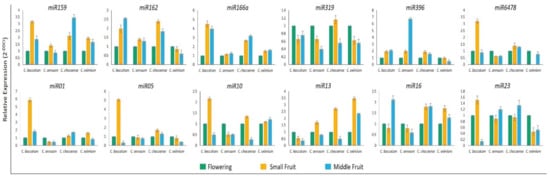
Figure 7.
Validation of six known and six novel miRNAs by stem-loop RT-qPCR in Capsicum species at flowering, small fruit, and middle fruit stages. The small nuclear RNA (snRNA) U6 was a housekeeping gene. Data are mean ± SD from three biological replicates.
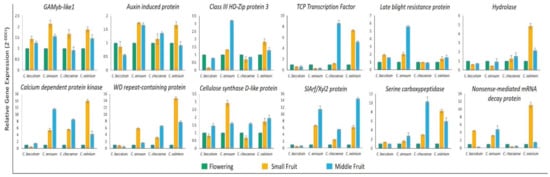
Figure 8.
Expression of target genes of known and novel miRNAs in Capsicum species at flowering, small fruit, and middle fruit stages. B-tub gene was a housekeeping gene. Data are mean ± SD from three biological replicates.
3. Discussion
Small RNAs, including miRNAs, are key regulators of biological processes such as biotic and abiotic stress tolerance, plant growth and development, metabolic pathways, and morphogenesis [28,29]. In plants, miRNAs regulate gene expression mainly by targeting mRNAs for cleavage and/or by translation inhibition of the target mRNAs during or after transcription [30]. Thus, miRNAs usually negatively regulate the accumulation of mRNAs and show an inverse correlated expression pattern in the same plant cells [31]. Such changes in the expression profile of plant genes play an important role in establishing specific phenotypes between plant species [32]. However, the molecular mechanisms underlying these changes are largely unknown.
Although miRNAs have been identified in pepper fruits by using small RNA-seq [22,23], we lack a systemic comparison study of miRNA expression in pepper between domesticated and wild species, despite the worldwide importance of these fruits. In this study, we investigated the contribution of gene expression regulation by miRNAs in cultivated (C. baccatum and C. annuum) and wild (C. chacoense and C. eximium) Capsicum species at flower and two fruit development stages (6- and 25-dpa fruits, namely small and medium fruit stages). Our study identified 22 known and 27 novel miRNAs differentially expressed across Capsicum species that may be closely involved in biological processes controlling flower and fruit ripening, including fruit development, morphogenesis, pigmentation, and quality in wild and cultivated pepper species.
3.1. Role of miRNAs and Their Regulators in Flowering across Capsicum Species
Flowering is an important biological process for plants that represents the phase transition from vegetative growth to reproductive growth, ensuring success in plant reproduction [33,34]. Several miRNAs have been reported to be a main regulator of the floral phase transition in many plants, including Arabidopsis [35], tomato [36], and apple [37]. For instance, miR156/157 and 172 are the two main key members of the aging pathway regulating SPL and AP2-like TFs [34]. These miRNAs play antagonistic but related roles in Arabidopsis flower induction, whereas a high level of miR156 extends the juvenile phase and delays flowering, and miR172 accumulation leads to early flowering [38,39]. As shown in Figure 4a, miR172 was highly accumulated in flowers in domesticated species, especially C. annuum, so cultivated species may exhibit early flowering as compared with wild species, which could be beneficial for pepper productivity. Similar to miR172, miR319 regulating TCP TFs showed high expression in C. annuum. Although TCP TFs have been more associated with leaf development and petal growth [40], recently Li et al. [41] described the function of these TFs as key regulators of flower development. Likewise, Wang et al. [42] reported that overexpression of TCP8 significantly delayed flowering in Arabidopsis under long- and short-day conditions; additionally, high expression of TCP4 promoted pistil abortion in Prunus mume [43]. Our results suggest that overexpression of miR319 in C. annuum negatively regulates TCP TF expression, which results in early flowering in this cultivated species and needs further confirmation.
Besides miR156/157, 172, and 319, many other miRNA families involved in the control of plant flowering time include miR159, 165/166, 167, 169, 171, 319, 390, and 399 [44]. miR159 regulates GAMYB TFs involved in the gibberellic acid (GA) pathway in different land plants as well as meristem formation and seed development [45,46]. Our results revealed a higher expression of miR159 than other miRNAs across species and stages; however, specifically at flowering stage, all species with the exception of C. chacoense showed similar expression. These results may suggest that GA content is an important player in flower initiation and development across Capsicum species. Furthermore, miR166b and 390 showed high expression in C. annuum and C. baccatum, respectively, and regulated LRR receptor-like serine/threonine-protein kinase (LRR-RLK). Previous studies of Arabidopsis reported the role of these proteins in floral organ abscission [47], in the main abscisic acid-mediated (ABA) signaling pathway and in early ABA perception [48]. Thus, miR166b and 390 may participate as negative regulators of flower organ development through the ABA pathway in cultivated species.
Along with the known miRNAs mentioned above, novel miRNAs were upregulated during flower stage, mainly in wild species. For instance, miR17, 22, 23, and 24 showed high expression in C. eximium, whereas miR25 was highly expressed in C. chacoense. These miRNAs regulate genes coding for disease resistance proteins, suppressor of gene silencing 3-like (SGS3) proteins, and nonsense-mediated mRNA decay proteins, involved in plant pathogen defense and destruction of aberrant mRNAs [49,50]. The novel miRNAs we identified may be linked to plant pathogen protection against virus infection during flowering in wild Capsicum species that represent valuable germplasm resources for crop improvement.
3.2. Role of miRNAs and Their Regulators in Pepper Fruit Development
Capsicum species are highly diverse, and fruit attributes are one of the principal phenotypic differences among accessions [51]. Domestication events and continuous selection have increased the variability of fruit features such as shape, size, color, and aroma. Domesticated fruit-bearing crop species have largely increased their fruit size compared with those normally found in progenitor wild species [52,53]. As shown in Figure 1, domesticated species used in this study had large fruits, C. annuum having the largest. However, wild species had smaller fruits, with a round and elongated fruit shape for C. chacoense and C. eximium, respectively. miRNAs orchestrate different fruit development process such as fruit set, formation, shape, size, ripening, and quality in multiple plants including tomato [36,54,55], rice [56], cassava [57], Lycium barbarum [58], sweet potato [59], orange [60], blueberry [61], and diverse cucurbit species [62].
The hierarchical clustering in Figure 4 and Figure 5 for known and novel miRNAs, respectively, revealed that domesticated species were grouped, sharing similar miRNA expression patterns at fruit stages; however, some miRNAs showed high expression in C. baccatum versus other Capsicum species (Figure 5c,d). In addition to their role in plant vegetative versus reproductive phase change, miR156 and 172 have also been associated with fruit size in different species depending on the fruit type [63,64]. For instance, overexpression of miR156a reduced the fruit size and yield in tomato [65], whereas overexpression of miR172 in apple led to reduced fruit size and weight [66]. In our study, miR172 was uniquely expressed in C. baccatum at small fruit stage, whereas miR156 was highly abundant in C. baccatum at both fruit stages. Likewise, miR159 displayed different levels of upregulation in C. baccatum at both fruit stages. Overexpression of miR159 has been associated with abnormal ovule development that affects the initial fruit set, precocious fruit initiation, and seedless fruits in tomato [67]; in contrast, in strawberry, miR159 repression inhibited receptacle ripening and color formation [68]. The increased expression of miR156 and 159 in C. baccatum may interfere somehow in the fruit set and size of this species, which remains to be addressed.
Furthermore, Nimmakayala et al. [69] reported an Ankyrin protein with acyltransferase activity associated with fruit weight in C. annuum. Similarly, Qin et al. [20] reported an acyltransferase involved in the selective sweep signals in the cultivated C. annuum L. (Zunla-1) versus its wild progenitor Chiltepin (C. annuum var. glabriusculum). Our results showed that miR12 with high expression in C. baccatum at middle fruit stage was related to regulation of an acyltransferase protein, which may be involved in the differences in fruit size between cultivated and wild pepper species. As well, we identified a CLAVATA1 gene regulated by miR408. It has been reported in tomato that a mutation in the CLAVATA3 gene involved in the CLAVATA-WUSCHEL pathway promoted stem-proliferation, increasing the meristem size, and resulting in the development of extra organs in flowers and larger fruits [70,71]. These results suggest that CLAVATA genes are implicated in pepper fruit size and are regulated by novel miRNAs. Moreover, Qin et al. [20] also reported an ABC transporter and Pleiotropic drug resistance protein 1 (PDR1) genes likely associated with the morphological and physiological differences between wild and domesticated pepper species. Likewise, it has been reported that the ABC transporter is a candidate gene at the fw3.2 locus associated with fruit weight in tomato [72]. In pepper, ABC transporter has also been associated with fruit weight [69] and capsaicinoid content [73]. In our study, we identified two novel miRNAs, miR20 and miR21, regulating an ABC transporter and PDR4 proteins, suggesting their importance for differences in pepper fruit development between wild and domesticated species.
In addition to their involvement in fruit size, miRNAs are also important for their involvement in the auxin signaling pathway regulating auxin responsive factors (ARFs) [74]. Auxin is a key hormone implicated in most plant organ developmental processes [75] and has been reported to affect fruit shape and development [32]. miR160 regulates ARFs in different plants such as Arabidopsis [76], cotton [77], and tomato [78]; however, in our study, miR162 was also found to regulate an auxin-induced protein. miR160 was downregulated across Capsicum species and fruit stages, whereas miR162 showed high expression at middle fruit stage in C. baccatum. Previous studies have reported that upregulation of miR160 targeting ARF10 affects fruit shape in tomato [79], whereas downregulation alters the abscission of petal and anther, having an effect on tomato fruit set [78]. Together, miR160 and 162 may play an important role in the auxin signaling pathway in Capsicum species and thus are involved in regulating fruit shape and development.
Quality and flavor are two important attributes of pepper fruits. Both features are strongly related to sugar and acid composition [23]. KEGG pathway analysis revealed that some of the miRNAs regulate major candidate genes involved in amino sugar and nucleotide sugar metabolism, starch and sucrose metabolism, and fructose-mannose metabolism pathways. Carbohydrate metabolism-related genes such as cellulose synthase, FGGY carbohydrate kinase, SlArf/Xyl2 and fructokinase proteins were regulated by miR10, 11, 13, and 15, respectively, which suggests that these novel miRNAs may be involved in modulating pepper fruit quality. Particularly, miR13 and 15 were downregulated in all species, whereas miR10 and 11 showed high expression at both fruit stages in C. baccatum. Likewise, novel miR06, which targets a methylenetetrahydrofolate reductase and is involved in folate metabolism, was upregulated in C. baccatum at both fruit stages, so novel miRNAs may also be involved in the concentration of the B vitamin group across Capsicum species. Furthermore, the novel miRNAs miR23 and 24 regulated a gene encoding 1-aminocyclopropane-1-carboxylate oxidase enzyme (ACO), which is directly involved in ethylene biosynthesis [80]. ACO is the rate-limiting step in ethylene production during certain dedicated processes such as fruit ripening in tomato [81,82]. ACO is regulated by the known miR396 in citrus, which inhibits ethylene biosynthesis [83]. Although pepper fruit ripening is widely classified as non-climacteric, patterns of ethylene production and respiratory rates vary across pepper species due to their wide diversity [84,85]. For instance, hot pepper (C. frutescens) fruit ripening is regulated by ethylene and ABA biosynthesis [86]. In addition, some varieties such as ‘Camelot’, ‘King Arthur’, and ‘Tabasco’ exhibit characteristics intermediate between climacteric and non-climacteric fruit ripening [87]. Another miRNA related to fruit ripening in tomato and Lycium barbarum involving the ethylene biosynthesis is miR164, regulating NAC TFs [58]. In tomato, SlNAC4 RNAi-knockout plants showed delayed fruit ripening and a reduction of 30% of total carotenoid content, which suggests that SlNAC4 is a positive regulator of ripening and carotenoid accumulation [88]. In our study, miR164 showed high accumulation at middle fruit stage in C. chacoense, which suggests its role as a negative regulator of fruit ripening in this species. Moreover, fruit ripening and softness are influenced by alpha-N-acetylglucosaminidase and hydrolase (hydrolyzing O-glycosyl compounds) [89,90], which are regulated by miRNAs such as the identified miR168 and 6478.
Narrow genetic diversity which typifies cultivars of domesticated crops has increased the susceptibility of domesticated plants to major diseases [91]. Wild relatives of cultivated crops are recognized as a rich source of genes for disease resistance and stress tolerance. Our results showed different known and novel miRNAs regulating disease resistant genes. For instance, miRNAs such as miR165 and 398 were upregulated in the wild pepper relative C. eximium; these miRNAs were found to target aspartic proteinases CDR1 (constitutive disease resistance 1). Along with their roles in plant development, aspartic proteinases have been associated with immune defense in pepper plants [92]. Additionally, we identified miR394 and miR403 that regulate an F-box protein. F-box genes are well known to be involved in hormone signaling and in response to abiotic stress in pepper [93]. Lim et al. [94] reported a novel F-box protein, CaDIF1 (C. annuum Drought-Induced F-box Protein 1) in pepper. CaDIF1-silenced pepper plants exhibited a drought-sensitive phenotype, whereas CaDIF1-overexpressing plants exhibited ABA-sensitive and drought-tolerant phenotypes. MiR394 and miR403 were upregulated at both fruit stages in C. baccatum, which may suggest a lower accumulation of F-box proteins in this domesticated pepper species and therefore a high susceptibility of C. baccatum to drought stress. In addition, we identified miR403 which regulates an Argonaute (AGO2) protein. AGO proteins bind to small-interfering (si)RNAs and micro (mi)RNAs to target RNA silencing against viruses [95], suggesting that miR403 is directly involved in regulation of pepper defense response to virus infection. Furthermore, miR390 and miR22 associated with a leucine-rich repeat receptor kinase (LRR receptor-like protein) and miR396 associated with a late blight resistance protein were identified. Qin et al. [20] reported 10 disease resistance genes such as LRR and late blight resistance proteins showing a strong selective sweep signals in the cultivated peppers, indicating that these genes seemed to have been affected by selection during domestication. Our results support the involvement of miR390, miR396, and miR22 in pepper domestication. Overall, our results suggest that miRNAs identified, and their target genes, likely serve crucial regulatory roles in pepper related to vegetative to reproductive phase changes, fruit development and quality, and disease resistance across domesticated and wild pepper species. Further research to characterize the microRNAome in Capsicum will be required to confirm these inferences.
4. Materials and Methods
4.1. Plant Materials
C. baccatum cv. Lemon drop, C. annuum cv. Cayenne, C. chacoense cv. PI 669106, and C. eximium cv. PI 645681 were grown in the greenhouse at West Virginia State University with the following conditions: 16 h of light at 26 °C and 8 h of darkness at 20 °C, relative humidity of 75 ± 2%, and plants were watered daily using manual irrigation. Flowers were uniformly sampled for unopened and fully matured buds from all cultivars, while fruit samples were collected at 6 and 25 days post-anthesis (dpa) from all four cultivars. All samples were harvested in three biological replications and frozen immediately in liquid nitrogen and stored at −80 °C for RNA isolation.
4.2. Construction and Sequencing of Small RNA Libraries
Total RNA was isolated by using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA purity was achieved by using the RNA clean and concentrator kit (Zymo Research, Irvine, CA, USA) along with on-column DNaseI digestion to remove genomic DNA. Total RNA quality and quantity were measured by using the Agilent 2100 Bioanalyzer and Qubit 4 Fluorometer (Invitrogen, Carlsbad, CA, USA), respectively. Total RNA from three biological replicates was pooled for each sample before small RNA-Seq library preparation. At least 1.5 µg high-quality total RNA was used to construct each small RNA library and for sequencing by the Beijing Genomics Institute (Hong Kong) with the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA). The small RNA-seq dataset was deposited in the NCBI SRA database under the accession number PRJNA718887.
4.3. Bioinformatics Analysis of miRNAs
After sequencing, the quality of raw reads was ascertained by checking the adapter, GC distribution, average base content, and quality score of the distribution by using the fastqc program. The Cutadapt toolkit was used for removal of poor-quality reads, sequences with a poly-A tail, low-quality reads with ambiguous bases (“N”), reads with <18 or >30 bases, and adapter sequences [96]. Furthermore, the clean reads were BLAST-searched against the Rfam database (v14.1) [97] and Repbase [98] to identify and exclude known noncoding RNAs including rRNAs, scRNA, snoRNAs, snRNAs, and tRNAs. The remaining clean reads were searched against the miRBase database (v22.1, http://www.mirbase.org/, accessed on 1 August 2020) to identify known putative miRNAs [17]. The final miRNAs dataset underwent sequence length distribution and nucleotide preference analysis at each position. Meanwhile, the remaining unannotated sRNA sequences that were not mapped to any pre-miRNAs in miRbase were analyzed by using miRDeep2 [99] to predict potential novel miRNAs using the C. annuum cv. CM334 reference genome [19]. The prediction of novel miRNAs was based on previously reported criteria including (a) no more than four mismatches between the small RNA and the target (G-U bases count as 0.5 mismatches), (b) no more than two adjacent mismatches in the miRNA/target duplex, (c) no adjacent mismatches in positions 2 to 12 of the miRNA/target duplex (5′ of miRNAs), (d) no mismatches in positions 10 to 11 of the miRNA/target duplex, (e) no more than 2.5 mismatches in positions 1 to 12 of the miRNA/target duplex (5′ of miRNAs), and (f) a minimum free energy (MFE) of the miRNA/target > 75% [100,101]. Additionally, the putative precursor sequence was folded for each candidate novel miRNA by using RNAfold from the Vienna RNA software package [102]. Similarly, as for known miRNAs, for all the predicted novel miRNAs, properties including miRNA count, length, and nucleotide bias at each position were determined.
4.4. Analysis of Differentially Expressed miRNAs
We used the R package DEGseq [103] to identify differentially expressed known and novel miRNAs across species and stages. The fold changes of miRNA expression were normalized by transcripts per million according to the formula: Normalized expression (NE) = Actual miRNA reads count/Total count of clean reads × 1,000,000 (Zhou et al., 2010).
4.5. Prediction of miRNA Targets and Enrichment Analyses
The putative target genes of identified miRNAs were predicted by using psRNATarget [104] with a pepper transcriptome dataset [20]. Genes with a final sequence score ≤ 5 were considered potential candidate miRNA targets. The target genes were functionally annotated by using the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/, accessed on 1 September 2020) pathway database with the BlastKOALA sequence similarity tool [105]. Pathways with false discovery rate (FDR) ≤ 0.5 were considered significantly enriched. Gene Ontology (GO) analysis of these genes involved using Blast2GO with a cutoff E-value of 10−5 (http://www.blast2go.com, accessed on 4 May 2021) [106]. The Reads Per Kilobase of transcript expression values from leaf, stem, and placenta tissues (6, 16, 25 dpa) for the identified target genes were retrieved from published RNA-seq data [19] and used to generate a heatmap with the ClustVis web tool (https://biit.cs.ut.ee/clustvis/, accessed on 1 September 2020).
4.6. Quantitative RT-qPCR and Stem-Loop RT-qPCR
To validate our results from the bioinformatics-based analysis, we used stem-loop RT-qPCR for miRNAs and RT-qPCR for target genes in flower, small fruit (6-dpa) and medium fruit (25-dpa) tissues with three biological replications across the Capsicum species analyzed. The RT-qPCR primers were designed from miRNA sequences as described [107] (Table S5). Total RNA used for small RNA sequencing was used for reverse transcription using Superscript IV reverse transcriptase. RT-qPCR analysis was achieved with the Universal SYBR Green Master Mix (Thermo Fisher Scientific, USA) on the StepOne Plus Real-Time PCR System (Applied Biosystems, USA) with the following conditions: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 56 °C for 30s, and 72 °C for 15 s, and melting curve analysis from 65 to 95 °C. To normalize gene expression, small nuclear RNA (snRNA) U6 and β-tubulin were internal controls for miRNA and target genes, respectively. All RT-qPCR reactions involved three technical replicates, and the relative gene expression of miRNAs and targets was estimated by the 2-ΔΔCt method [108]. The oligonucleotide primers corresponding to the predicted target genes are in Table S6.
5. Conclusions
Considering the global importance for pepper worldwide, we used high-throughput sequencing and identified 22 known miRNAs and 27 novel miRNAs differentially expressed in flower and pepper fruits at different developmental stages across 4 Capsicum species. Analysis of differential expression patterns combined with target prediction suggested key roles for these miRNAs in controlling flower time and pepper fruit development in cultivated and wild species. The results expand the study of miRNAs in plants by providing a better understanding of their essential roles in miRNA-based regulation processes in pepper. Similarly, our results provide insight into the biology, evolution, and domestication process of Capsicum species, accelerating the agricultural applications of the miRNAs, the genes of their biogenesis pathway and providing targets for future investigation in pepper and other plants.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22094866/s1.
Author Contributions
Conceptualization, U.K.R. and P.N. (Padma Nimmakayala); Methodology, C.L.-O., M.B., V.L.A., and P.N. (Purushothaman Natarajan); Software, C.L.-O. and P.N. (Purushothaman Natarajan); Validation, C.L.-O. and M.B.; Formal analysis, C.L.-O.; Investigation, C.L.-O.; Data curation, C.L.-O.; Visualization, C.L.-O.; Writing—original draft preparation, C.L.-O. and Y.P.-G.; Writing—review and editing, P.N. (Padma Nimmakayala), J.S., and U.K.R.; Supervision, P.N. (Padma Nimmakayala) and U.K.R.; Project administration, P.N. (Padma Nimmakayala) and U.K.R.; Funding acquisition, P.N. (Padma Nimmakayala) and U.K.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institute of Food and Agriculture USDA-NIFA (grants no. 2017-38821-26434 and 2019-38821-29064) and Department of Defense award (agreement no. W911NF-16-1-0423) for the next-generation sequencing instrument.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The small RNA-seq dataset was deposited in the NCBI SRA database under the accession number PRJNA718887 https://www.ncbi.nlm.nih.gov/sra/PRJNA718887, accessed on 4 May 2021.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Song, Q.-X.; Liu, Y.-F.; Hu, X.-Y.; Zhang, W.-K.; Ma, B.; Chen, S.-Y.; Zhang, J.-S. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 2011, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Murchison, E.P.; Hannon, G.J. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 2004, 16, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; van Haarst, J.C.; Maliepaard, C.; van de Geest, H.; Bovy, A.G.; Lammers, M.; Angenent, G.C.; de Maagd, R.A. Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J. Exp. Bot. 2013, 64, 1863–1878. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Reinhart, B.J.; Jones-Rhoades, M.W.; Tang, G.; Zamore, P.D.; Barton, M.K.; Bartel, D.P. MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 2004, 23, 3356–3364. [Google Scholar] [CrossRef]
- Bartel, B.; Bartel, D.P. MicroRNAs: At the root of plant development? Plant Physiol. 2003, 132, 709–717. [Google Scholar] [CrossRef]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.-K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Sunkar, R. MicroRNAs with macro-effects on plant stress responses. Semin. Cell Dev. Biol. 2010, 21, 805–811. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, F.; Cao, H.; Peng, H.; Ni, Z.; Sun, Q.; Yao, Y. TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 2012, 7, e48445. [Google Scholar] [CrossRef]
- Xu, M.Y.; Zhang, L.; Li, W.W.; Hu, X.L.; Wang, M.-B.; Fan, Y.L.; Zhang, C.Y.; Wang, L. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 89–101. [Google Scholar] [CrossRef]
- Subramanian, S.; Fu, Y.; Sunkar, R.; Barbazuk, W.B.; Zhu, J.-K.; Yu, O. Novel and nodulation-regulated microRNAs in soybean roots. BMC Genom. 2008, 9, 160. [Google Scholar] [CrossRef]
- Pandey, S.P.; Shahi, P.; Gase, K.; Baldwin, I.T. Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuata. Proc. Natl. Acad. Sci. USA 2008, 105, 4559–4564. [Google Scholar] [CrossRef]
- Hewezi, T.; Howe, P.; Maier, T.R.; Baum, T.J. Arabidopsis small RNAs and their targets during cyst nematode parasitism. Mol. Plant Microbe Interact. 2008, 21, 1622–1634. [Google Scholar] [CrossRef]
- Liu, H.; Jin, T.; Liao, R.; Wan, L.; Xu, B.; Zhou, S.; Guan, J. miRFANs: An integrated database for Arabidopsis thaliana microRNA function annotations. BMC Plant Biol. 2012, 12, 1–8. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Knapp, S. Tobacco to tomatoes: A phylogenetic perspective on fruit diversity in the Solanaceae. J. Exp. Bot. 2002, 53, 2001–2022. [Google Scholar] [CrossRef]
- Kim, S.; Park, M.; Yeom, S.-I.; Kim, Y.-M.; Lee, J.M.; Lee, H.-A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.-T. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef]
- Perla, V.; Nadimi, M.; Reddy, R.; Hankins, G.R.; Nimmakayala, P.; Harris, R.T.; Valluri, J.; Sirbu, C.; Reddy, U.K. Effect of ghost pepper on cell proliferation, apoptosis, senescence and global proteomic profile in human renal adenocarcinoma cells. PLoS ONE 2018, 13, e0206183. [Google Scholar] [CrossRef]
- Hwang, D.-G.; Park, J.H.; Lim, J.Y.; Kim, D.; Choi, Y.; Kim, S.; Reeves, G.; Yeom, S.-I.; Lee, J.-S.; Park, M. The hot pepper (Capsicum annuum) microRNA transcriptome reveals novel and conserved targets: A foundation for understanding microRNA functional roles in hot pepper. PLoS ONE 2013, 8, e64238. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Ou, L.; Kang, L.; Liu, Y.; Lv, J.; Wei, G.; Yang, B.; Yang, S.; Chen, W. Identification and characterization of novel microRNAs for fruit development and quality in hot pepper (Capsicum annuum L.). Gene 2017, 608, 66–72. [Google Scholar] [CrossRef]
- Czech, B.; Hannon, G.J. Small RNA sorting: Matchmaking for Argonautes. Nat. Rev. Genet. 2011, 12, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Bologna, N.G.; Voinnet, O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014, 65, 473–503. [Google Scholar] [CrossRef]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef]
- Martinez, J.; Tuschl, T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004, 18, 975–980. [Google Scholar] [CrossRef]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. 2009, 25, 21–44. [Google Scholar] [CrossRef]
- Samad, A.F.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.; Zainal, Z.; Ismail, I. MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef]
- Djami-Tchatchou, A.T.; Sanan-Mishra, N.; Ntushelo, K.; Dubery, I.A. Functional roles of microRNAs in agronomically important plants—potential as targets for crop improvement and protection. Front. Plant Sci. 2017, 8, 378. [Google Scholar] [CrossRef]
- D’Ario, M.; Griffiths-Jones, S.; Kim, M. Small RNAs: Big impact on plant development. Trends Plant Sci. 2017, 22, 1056–1068. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-Q.; Allan, A.C.; Yin, X.-R. Small RNAs With a Big Impact on Horticultural Traits. Crit. Rev. Plant Sci. 2020, 39, 30–43. [Google Scholar] [CrossRef]
- Hong, Y.; Jackson, S. Floral induction and flower formation—the role and potential applications of mi RNA s. Plant Biotechnol. J. 2015, 13, 282–292. [Google Scholar] [CrossRef]
- Waheed, S.; Zeng, L. The Critical Role of miRNAs in Regulation of Flowering Time and Flower Development. Genes 2020, 11, 319. [Google Scholar] [CrossRef]
- Todesco, M.; Rubio-Somoza, I.; Paz-Ares, J.; Weigel, D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001031. [Google Scholar] [CrossRef]
- Karlova, R.; Rosin, F.M.; Busscher-Lange, J.; Parapunova, V.; Do, P.T.; Fernie, A.R.; Fraser, P.D.; Baxter, C.; Angenent, G.C.; de Maagd, R.A. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 2011, 23, 923–941. [Google Scholar] [CrossRef]
- Porto, D.D.; Bruneau, M.; Perini, P.; Anzanello, R.; Renou, J.-P.; Santos, H.P.d.; Fialho, F.B.; Revers, L.F. Transcription profiling of the chilling requirement for bud break in apples: A putative role for FLC-like genes. J. Exp. Bot. 2015, 66, 2659–2672. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.-W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.-Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef]
- Nag, A.; King, S.; Jack, T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 22534–22539. [Google Scholar] [CrossRef]
- Li, D.; Zhang, H.; Mou, M.; Chen, Y.; Xiang, S.; Chen, L.; Yu, D. Arabidopsis class II TCP transcription factors integrate with the FT–FD module to control flowering. Plant Physiol. 2019, 181, 97–111. [Google Scholar] [CrossRef]
- Wang, X.; Xu, X.; Mo, X.; Zhong, L.; Zhang, J.; Mo, B.; Kuai, B. Overexpression of TCP8 delays Arabidopsis flowering through a FLOWERING LOCUS C-dependent pathway. BMC Plant Biol. 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Wang, W.; Shi, T.; Ni, X.; Xu, Y.; Qu, S.; Gao, Z. The role of miR319a and its target gene TCP4 in the regulation of pistil development in Prunus mume. Genome 2018, 61, 43–48. [Google Scholar] [CrossRef]
- Ma, J.; Zhao, P.; Liu, S.; Yang, Q.; Guo, H. The Control of Developmental Phase Transitions by microRNAs and Their Targets in Seed Plants. Int. J. Mol. Sci. 2020, 21, 1971. [Google Scholar] [CrossRef]
- Yanhui, C.; Xiaoyuan, Y.; Kun, H.; Meihua, L.; Jigang, L.; Zhaofeng, G.; Zhiqiang, L.; Yunfei, Z.; Xiaoxiao, W.; Xiaoming, Q. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef]
- Millar, A.A.; Lohe, A.; Wong, G. Biology and Function of miR159 in Plants. Plants 2019, 8, 255. [Google Scholar] [CrossRef]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef]
- Osakabe, Y.; Maruyama, K.; Seki, M.; Satou, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 2005, 17, 1105–1119. [Google Scholar] [CrossRef]
- Rayson, S.; Arciga-Reyes, L.; Wootton, L.; Zabala, M.D.T.; Truman, W.; Graham, N.; Grant, M.; Davies, B. A role for nonsense-mediated mRNA decay in plants: Pathogen responses are induced in Arabidopsis thaliana NMD mutants. PLoS ONE 2012, 7, e31917. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Zhou, X. SGS3 cooperates with RDR6 in triggering geminivirus-induced gene silencing and in suppressing geminivirus infection in Nicotiana benthamiana. Viruses 2017, 9, 247. [Google Scholar] [CrossRef]
- Tripodi, P.; Greco, B. Large scale phenotyping provides insight into the diversity of vegetative and reproductive organs in a wide collection of wild and domesticated peppers (Capsicum spp.). Plants 2018, 7, 103. [Google Scholar] [CrossRef] [PubMed]
- Paran, I.; Van Der Knaap, E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J. Exp. Bot. 2007, 58, 3841–3852. [Google Scholar] [CrossRef] [PubMed]
- Paran, I.; Fallik, E. Breeding for fruit quality in pepper (Capsicum spp.). Breed. Fruit Qual. 2011, 307–322. [Google Scholar] [CrossRef]
- FAN, S.-s.; LI, Q.-n.; GUO, G.-j.; GAO, J.-c.; WANG, X.-x.; GUO, Y.-m.; Snyder, J.C.; DU, Y.-c. Identification of microRNAs in two species of tomato, Solanum lycopersicum and Solanum habrochaites, by deep sequencing. J. Integr. Agric. 2015, 14, 42–49. [Google Scholar] [CrossRef]
- Chen, W.; Kong, J.; Lai, T.; Manning, K.; Wu, C.; Wang, Y.; Qin, C.; Li, B.; Yu, Z.; Zhang, X. Tuning LeSPL-CNR expression by SlymiR157 affects tomato fruit ripening. Sci. Rep. 2015, 5, 1–6. [Google Scholar] [CrossRef]
- Swetha, C.; Basu, D.; Pachamuthu, K.; Tirumalai, V.; Nair, A.; Prasad, M.; Shivaprasad, P.V. Major domestication-related phenotypes in indica rice are due to loss of miRNA-mediated laccase silencing. Plant Cell 2018, 30, 2649–2662. [Google Scholar] [CrossRef]
- Chen, X.; Xia, J.; Xia, Z.; Zhang, H.; Zeng, C.; Lu, C.; Zhang, W.; Wang, W. Potential functions of microRNAs in starch metabolism and development revealed by miRNA transcriptome profiling of cassava cultivars and their wild progenitor. BMC Plant Biol. 2015, 15, 1–11. [Google Scholar] [CrossRef]
- Zeng, S.; Liu, Y.; Pan, L.; Hayward, A.; Wang, Y. Identification and characterization of miRNAs in ripening fruit of Lycium barbarum L. using high-throughput sequencing. Front. Plant Sci. 2015, 6, 778. [Google Scholar] [CrossRef]
- Saminathan, T.; Alvarado, A.; Lopez, C.; Shinde, S.; Gajanayake, B.; Abburi, V.L.; Vajja, V.G.; Jagadeeswaran, G.; Reddy, K.R.; Nimmakayala, P. Elevated carbon dioxide and drought modulate physiology and storage-root development in sweet potato by regulating microRNAs. Funct. Integr. Genom. 2019, 19, 171–190. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, Y.; Zhu, A.; Wu, X.; Ye, J.; Yu, K.; Guo, W.; Deng, X. Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genom. 2010, 11, 1–17. [Google Scholar] [CrossRef]
- Hou, Y.; Zhai, L.; Li, X.; Xue, Y.; Wang, J.; Yang, P.; Cao, C.; Li, H.; Cui, Y.; Bian, S. Comparative analysis of fruit ripening-related miRNAs and their targets in blueberry using small RNA and degradome sequencing. Int. J. Mol. Sci. 2017, 18, 2767. [Google Scholar] [CrossRef]
- Manohar, S.; Jagadeeswaran, G.; Nimmakayala, P.; Tomason, Y.; Almeida, A.; Sunkar, R.; Levi, A.; Reddy, U.K. Dynamic regulation of novel and conserved miRNAs across various tissues of diverse cucurbit species. Plant Mol. Biol. Report. 2013, 31, 335–343. [Google Scholar] [CrossRef]
- Chung, M.Y.; Vrebalov, J.; Alba, R.; Lee, J.; McQuinn, R.; Chung, J.D.; Klein, P.; Giovannoni, J. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 2010, 64, 936–947. [Google Scholar] [CrossRef]
- Yao, J.-L.; Tomes, S.; Xu, J.; Gleave, A.P. How microRNA172 affects fruit growth in different species is dependent on fruit type. Plant Signal. Behav. 2016, 11, 417–427. [Google Scholar] [CrossRef]
- Zhang, X.; Zou, Z.; Zhang, J.; Zhang, Y.; Han, Q.; Hu, T.; Xu, X.; Liu, H.; Li, H.; Ye, Z. Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett. 2011, 585, 435–439. [Google Scholar] [CrossRef]
- Yao, J.L.; Xu, J.; Cornille, A.; Tomes, S.; Karunairetnam, S.; Luo, Z.; Bassett, H.; Whitworth, C.; Rees-George, J.; Ranatunga, C. A micro RNA allele that emerged prior to apple domestication may underlie fruit size evolution. Plant J. 2015, 84, 417–427. [Google Scholar] [CrossRef]
- da Silva, E.M.; Silva, G.F.F.e.; Bidoia, D.B.; da Silva Azevedo, M.; de Jesus, F.A.; Pino, L.E.; Peres, L.E.P.; Carrera, E.; López-Díaz, I.; Nogueira, F.T.S. micro RNA 159-targeted Sl GAMYB transcription factors are required for fruit set in tomato. Plant J. 2017, 92, 95–109. [Google Scholar] [CrossRef]
- Vallarino, J.G.; Osorio, S.; Bombarely, A.; Casañal, A.; Cruz-Rus, E.; Sánchez-Sevilla, J.F.; Amaya, I.; Giavalisco, P.; Fernie, A.R.; Botella, M.A. Central role of Fa GAMYB in the transition of the strawberry receptacle from development to ripening. New Phytol. 2015, 208, 482–496. [Google Scholar] [CrossRef]
- Nimmakayala, P.; Abburi, V.L.; Saminathan, T.; Alaparthi, S.B.; Almeida, A.; Davenport, B.; Nadimi, M.; Davidson, J.; Tonapi, K.; Yadav, L. Genome-wide diversity and association mapping for capsaicinoids and fruit weight in Capsicum annuum L. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Durbak, A.R.; Tax, F.E. CLAVATA signaling pathway receptors of Arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems. Genetics 2011, 189, 177–194. [Google Scholar] [CrossRef]
- Muños, S.; Ranc, N.; Botton, E.; Bérard, A.; Rolland, S.; Duffé, P.; Carretero, Y.; Le Paslier, M.-C.; Delalande, C.; Bouzayen, M. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 2011, 156, 2244–2254. [Google Scholar] [CrossRef]
- Zhang, N.; Brewer, M.T.; van der Knaap, E. Fine mapping of fw3. 2 controlling fruit weight in tomato. Theor. Appl. Genet. 2012, 125, 273–284. [Google Scholar] [CrossRef]
- Lopez-Ortiz, C.; Dutta, S.K.; Natarajan, P.; Peña-Garcia, Y.; Abburi, V.; Saminathan, T.; Nimmakayala, P.; Reddy, U.K. Genome-wide identification and gene expression pattern of ABC transporter gene family in Capsicum spp. PLoS ONE 2019, 14, e0215901. [Google Scholar] [CrossRef]
- Chen, C.; Zeng, Z.; Liu, Z.; Xia, R. Small RNAs, emerging regulators critical for the development of horticultural traits. Hortic. Res. 2018, 5, 1–14. [Google Scholar] [CrossRef]
- De Jong, M.; Wolters-Arts, M.; Feron, R.; Mariani, C.; Vriezen, W.H. The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 2009, 57, 160–170. [Google Scholar] [CrossRef]
- Yang, T.; Wang, Y.; Teotia, S.; Wang, Z.; Shi, C.; Sun, H.; Gu, Y.; Zhang, Z.; Tang, G. The interaction between miR160 and miR165/166 in the control of leaf development and drought tolerance in Arabidopsis. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Liu, J.; Pei, W.; Li, X.; Wang, N.; Ma, J.; Zang, X.; Zhang, J.; Yu, S.; Wu, M. Analysis of the MIR160 gene family and the role of MIR160a_A05 in regulating fiber length in cotton. Planta 2019, 250, 2147–2158. [Google Scholar] [CrossRef]
- Damodharan, S.; Zhao, D.; Arazi, T. A common mi RNA 160-based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J. 2016, 86, 458–471. [Google Scholar] [CrossRef]
- Hendelman, A.; Buxdorf, K.; Stav, R.; Kravchik, M.; Arazi, T. Inhibition of lamina outgrowth following Solanum lycopersicum AUXIN RESPONSE FACTOR 10 (SlARF10) derepression. Plant Mol. Biol. 2012, 78, 561–576. [Google Scholar] [CrossRef]
- Houben, M.; Van de Poel, B. 1-Aminocyclopropane-1-carboxylic acid oxidase (ACO): The enzyme that makes the plant hormone ethylene. Front. Plant Sci. 2019, 10, 695. [Google Scholar] [CrossRef]
- Grierson, D. 10 Ethylene Biosynthesis. In Fruit Ripening: Physiology, Signalling and Genomics; CABI: London, UK, 2014; p. 178. [Google Scholar]
- Van de Poel, B.; Vandenzavel, N.; Smet, C.; Nicolay, T.; Bulens, I.; Mellidou, I.; Vandoninck, S.; Hertog, M.L.; Derua, R.; Spaepen, S. Tissue specific analysis reveals a differential organization and regulation of both ethylene biosynthesis and E8 during climacteric ripening of tomato. BMC Plant Biol. 2014, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Wang, M.; Zhang, H.-Y.; Liu, J.-H. The miR396b of Poncirus trifoliata functions in cold tolerance by regulating ACC oxidase gene expression and modulating ethylene–polyamine homeostasis. Plant Cell Physiol. 2016, 57, 1865–1878. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Chung, E.-J.; Joung, Y.-H.; Choi, D. Non-climacteric fruit ripening in pepper: Increased transcription of EIL-like genes normally regulated by ethylene. Funct. Integr. Genom. 2010, 10, 135–146. [Google Scholar] [CrossRef]
- Osorio, S.; Alba, R.; Nikoloski, Z.; Kochevenko, A.; Fernie, A.R.; Giovannoni, J.J. Integrative comparative analyses of transcript and metabolite profiles from pepper and tomato ripening and development stages uncovers species-specific patterns of network regulatory behavior. Plant Physiol. 2012, 159, 1713–1729. [Google Scholar] [CrossRef]
- Hou, B.-Z.; Li, C.-L.; Han, Y.-Y.; Shen, Y.-Y. Characterization of the hot pepper (Capsicum frutescens) fruit ripening regulated by ethylene and ABA. BMC Plant Biol. 2018, 18, 1–12. [Google Scholar] [CrossRef]
- Villavicencio, L.E.; Blankenship, S.M.; Sanders, D.C.; Swallow, W.H. Ethylene and carbon dioxide concentrations in attached fruits of pepper cultivars during ripening. Sci. Hortic. 2001, 91, 17–24. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, G.; Zhou, S.; Tu, Y.; Wang, Y.; Dong, T.; Hu, Z. A new tomato NAC (N AM/A TAF1/2/C UC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 2014, 55, 119–135. [Google Scholar] [CrossRef]
- Jagadeesh, B.H.; Prabha, T.N.; Srinivasan, K. Activities of glycosidases during fruit development and ripening of tomato (Lycopersicum esculantum L.): Implication in fruit ripening. Plant Sci. 2004, 166, 1451–1459. [Google Scholar] [CrossRef]
- Ghosh, S.; Meli, V.S.; Kumar, A.; Thakur, A.; Chakraborty, N.; Chakraborty, S.; Datta, A. The N-glycan processing enzymes α-mannosidase and β-D-N-acetylhexosaminidase are involved in ripening-associated softening in the non-climacteric fruits of capsicum. J. Exp. Bot. 2011, 62, 571–582. [Google Scholar] [CrossRef]
- Mammadov, J.; Buyyarapu, R.; Guttikonda, S.K.; Parliament, K.; Abdurakhmonov, I.Y.; Kumpatla, S.P. Wild relatives of maize, rice, cotton, and soybean: Treasure troves for tolerance to biotic and abiotic stresses. Front. Plant Sci. 2018, 9, 886. [Google Scholar] [CrossRef]
- Bae, C.; Kim, S.-m.; Lee, D.J.; Choi, D. Multiple classes of immune-related proteases associated with the cell death response in pepper plants. PLoS ONE 2013, 8, e63533. [Google Scholar] [CrossRef]
- Chen, R.; Guo, W.; Yin, Y.; Gong, Z.-H. A novel F-box protein CaF-box is involved in responses to plant hormones and abiotic stress in pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2014, 15, 2413–2430. [Google Scholar] [CrossRef]
- Lim, J.; Lim, C.W.; Lee, S.C. Functional Analysis of Pepper F-box Protein CaDIF1 and Its Interacting Partner CaDIS1: Modulation of ABA Signaling and Drought Stress Response. Front. Plant Sci. 2019, 10, 1365. [Google Scholar] [CrossRef]
- Harvey, J.J.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstädt, S.; Carr, J.P.; Baulcombe, D.C. An antiviral defense role of AGO2 in plants. PLoS ONE 2011, 6, e14639. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018, 46, D335–D342. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Meyers, B.C.; Axtell, M.J.; Bartel, B.; Bartel, D.P.; Baulcombe, D.; Bowman, J.L.; Cao, X.; Carrington, J.C.; Chen, X.; Green, P.J. Criteria for annotation of plant MicroRNAs. Plant Cell 2008, 20, 3186–3190. [Google Scholar] [CrossRef] [PubMed]
- Hofacker, I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003, 31, 3429–3431. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Varkonyi-Gasic, E.; Hellens, R.P. Quantitative stem-loop RT-PCR for detection of microRNAs. In RNAi and Plant Gene Function Analysis; Springer: Berlin/Heidelberg, Germany, 2011; pp. 145–157. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).