Paternal Exercise Improves the Metabolic Health of Offspring via Epigenetic Modulation of the Germline
Abstract
:1. Introduction
2. Results
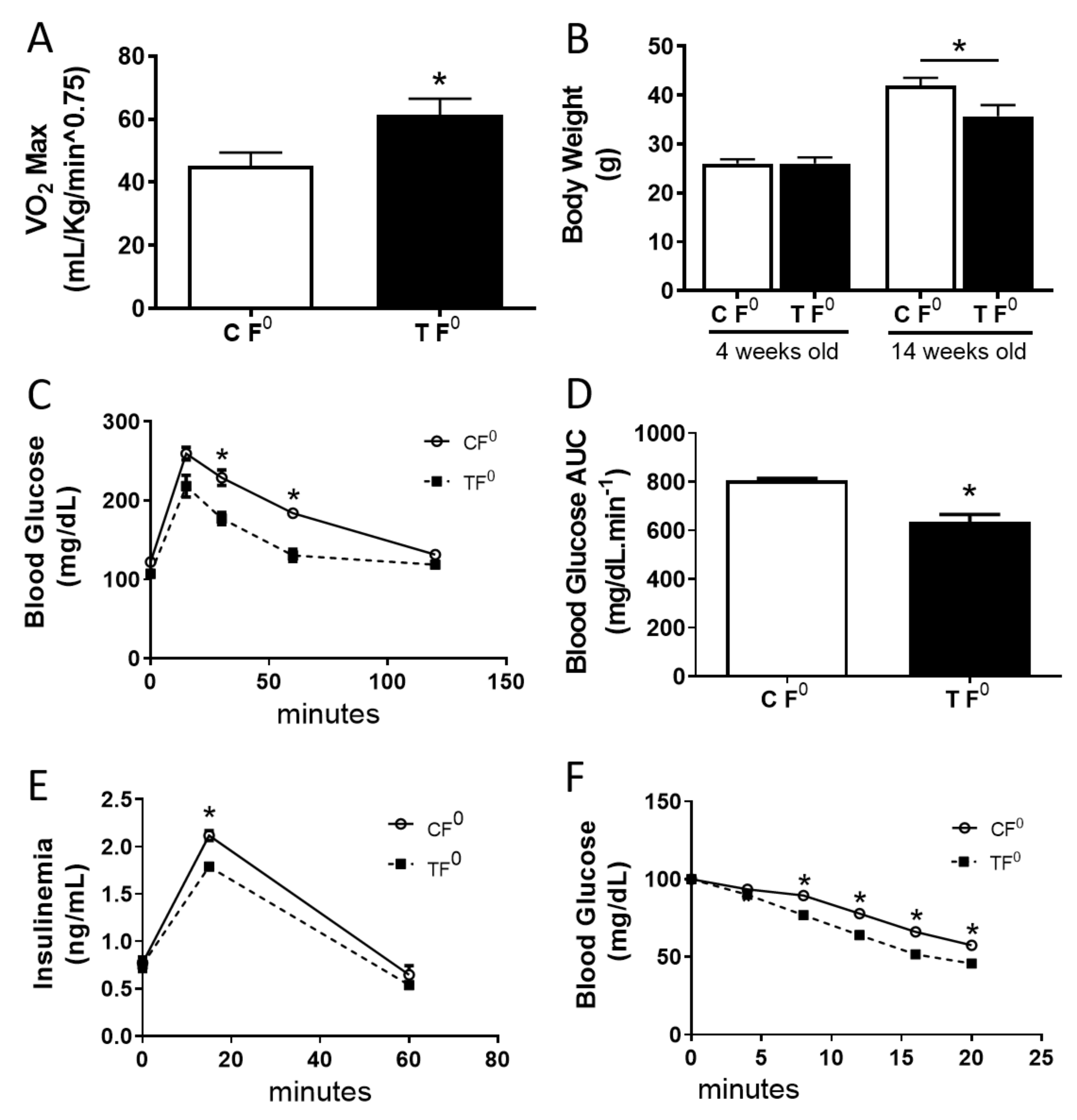
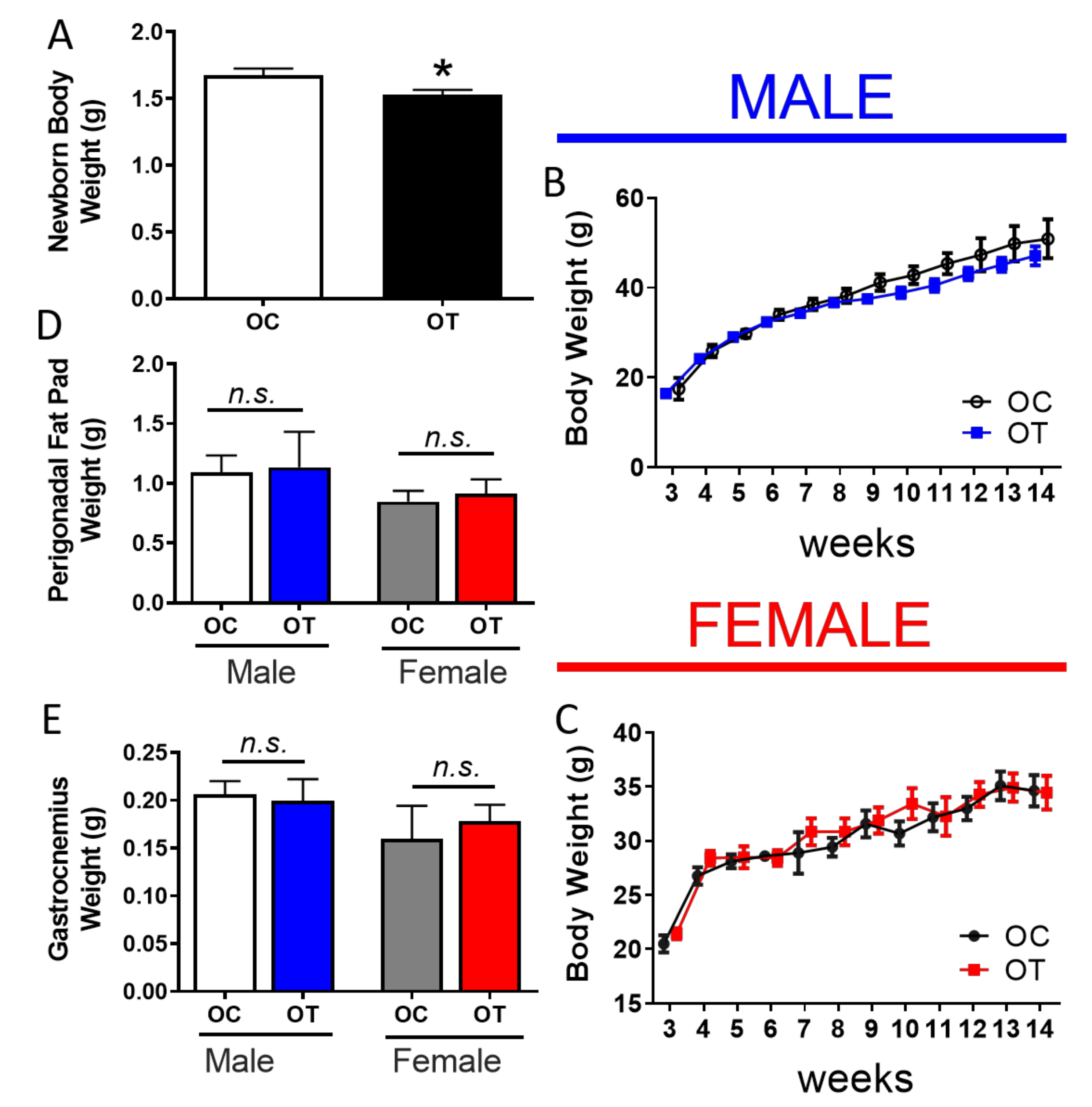
2.1. Paternal Exercise Improves Glucose Homeostasis in the Offspring
2.2. Paternal Exercise Alters DNA Methylation in the Skeletal Muscle of Their Offspring
2.3. Endurance Exercise-Training Alters the DNA Methylation Profile in the Male Progenitor Sperm
3. Discussion
4. Materials and Methods
4.1. Animal Care and Husbandry
4.2. Maximal Oxygen Consumption (VO2Max)
4.3. Mating Scheme
4.4. Intraperitoneal Glucose Tolerance Test
4.5. Intraperitoneal Insulin Tolerance Test
4.6. DNA Methylation Analysis
4.7. Next-Generation Sequencing
4.8. Quantitative Real-Time PCR
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ling, C.; Groop, L. Epigenetics: A molecular link between environmental factors and type 2 diabetes. Diabetes 2009, 58, 2718–2725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornelis, M.C.; Hu, F.B. Gene-environment interactions in the development of type 2 diabetes: Recent progress and continuing challenges. Annu. Rev. Nutr. 2012, 32, 245–259. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Yang, C.-R.; Wei, Y.-P.; Zhao, Z.-A.; Hou, Y.; Schatten, H.; Sun, Q.-Y. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc. Natl. Acad. Sci. USA 2014, 111, 1873–1878. [Google Scholar] [CrossRef] [Green Version]
- Gabory, A.; Jammes, H.; Dandolo, L. The H19 locus: Role of an imprinted non- coding RNA in growth and development. Bioessays 2010, 32, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Rakyan, V.K.; Down, T.A.; Balding, D.J.; Beck, S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011, 12, 529–541. [Google Scholar] [CrossRef] [Green Version]
- Tuomilehto, J.; Lindström, J.; Eriksson, J.G.; Valle, T.T.; Hämäläinen, H.; Ilanne-Parikka, P.; Keinänen-Kiukaanniemi, S.; Laakso, M.; Louheranta, A.; Rastas, M. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N. Engl. J. Med. 2001, 344, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Saaristo, T.; Moilanen, L.; Korpi-Hyövälti, E.; Vanhala, M.; Saltevo, J.; Niskanen, L.; Jokelainen, J.; Peltonen, M.; Oksa, H.; Tuomilehto, J.; et al. ifestyle intervention for prevention of type 2 diabetes in primary health care: One-year follow-up of the Finnish National Diabetes Prevention Program (FIN-D2D). Diabetes Care 2010, 33, 2146–2151. [Google Scholar] [CrossRef] [Green Version]
- Adamo, K.B.; Sigal, R.J.; Kenny, G.; Prud’homme, D.; Tesson, F. Influence of Pro12Ala peroxisome proliferator-activated receptor gamma2 polymorphism on glucose response to exercise training in type 2 diabetes. Diabetologia 2005, 48, 1503–1509. [Google Scholar] [CrossRef] [Green Version]
- Ling, C.; Guerra, S.; Lupi, R.; Rönn, T.; Granhall, C.; Luthman, H.; Masiello, P.; Marchetti, P.; Groop, L.; Prato, S. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia 2008, 51, 615–622. [Google Scholar] [CrossRef] [Green Version]
- Barrès, R.; Yan, J.; Egan, B.; Treebak, J.T.; Rasmussen, M.; Fritz, T.; Caidahl, K.; Krook, A.; O’Gorman, D.J.; Zierath, J.R. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012, 15, 405–411. [Google Scholar] [CrossRef] [Green Version]
- Carone, B.R.; Fauquier, L.; Habib, N.; Shea, J.M.; Hart, C.E.; Li, R.; Bock, C.; Li, C.; Gu, H.; Zamore, P.D.; et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 2010, 143, 1084–1096. [Google Scholar] [CrossRef] [Green Version]
- Glastras, S.J.; Chen, H.; McGrath, R.T.; Zaky, A.A.; Gill, A.J.; Pollock, C.A.; Sadd, S. Effect of GLP-1 Receptor Activation on Offspring Kidney Health in a Rat Model of Maternal Obesity. Sci. Rep. 2016, 6, 23525. [Google Scholar] [CrossRef] [Green Version]
- Soubry, A.; Murphy, S.K.; Wang, F.; Huang, Z.; Vidal, A.C.; Fuemmeler, B.F.; Kurtzberg, J.; Murtha, A.; Jirtle, R.L.; Schildkraut, J.M.; et al. Newborns of obese parents have altered DNA methylation patterns at imprinted genes. Int. J. Obes. 2013, 39, 650–657. [Google Scholar] [CrossRef] [Green Version]
- Tyrrell, J.; Richmond, R.C.; Palmer, T.M.; Feenstra, B.; Rangarajan, J.; Metrustry, S.; Cavadino, A.; Paternoster, L.; Armstrong, L.L.; Silva, N.M.G. Genetic Evidence for Causal Relationships Between Maternal Obesity-Related Traits and Birth Weight. JAMA 2016, 315, 1129–1140. [Google Scholar] [CrossRef] [Green Version]
- Andersson, S.W.; Bengtsson, C.; Hallberg, L.; Lapidus, L.; Niklasson, A.; Wallgren, A.; Hulthén, L. Cancer risk in Swedish women: The relation to size at birth. Br. J. Cancer 2001, 84, 1193–1198. [Google Scholar] [CrossRef] [Green Version]
- Barker, D.J.; Thornburg, K.L. Placental programming of chronic diseases, cancer and lifespan: A review. Placenta 2013, 34, 841–845. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.F.; Ruby, C.Y.; Lin, R.C.Y.; Laybutt, D.R.; Barres, R.; Owens, J.A.; Morris, M.J. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature 2010, 467, 963–966. [Google Scholar] [CrossRef] [PubMed]
- Hales, C.N.; Barker, D.J. Type 2 (non-insulin-dependent) diabetes mellitus: The thrifty phenotype hypothesis. Diabetologia 1992, 35, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.J. The fetal and infant origins of adult disease. BMJ 1990, 301, 1111. [Google Scholar] [CrossRef] [Green Version]
- Han, T.S.; Hart, C.L.; Haig, C.; Logue, J.; Uotin, M.N.; Watt, G.C.; Lean, M.E.J. Contributions of maternal and paternal adiposity and smoking to adult offspring adiposity and cardiovascular risk: The Midspan Family Study. BMJ Open 2015, 5, e007682. [Google Scholar] [CrossRef] [Green Version]
- Finer, S.; Mathews, C.; Lowe, R.; Smart, M.; Hillman, S.; Foo, L.; Sinha, A.; Williams, D.; Rakyan, V.K.; Hitman, G.A. Maternal gestational diabetes is associated with genome-wide DNA methylation variation in placenta and cord blood of exposed offspring. Hum. Mol. Genet. 2015, 24, 3021–3029. [Google Scholar] [CrossRef] [PubMed]
- Young, L.E. Imprinting of genes and the Barker hypothesis. Twin Res. 2001, 4, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Moore, T.; Haig, D. Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 1991, 7, 45–49. [Google Scholar] [CrossRef]
- Wilkins, J.F.; Haig, D. What good is genomic imprinting: The function of parent-specific gene expression. Nat. Rev. Genet. 2003, 4, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, M.Z. Igf2-H19, an Imprinted Tandem Yin-Yanggene and its Emerging Role in Development, Proliferation of Pluripotent Stem Cells, Senescence and Cancerogenesis. J. Stem Cell Res. Ther. 2012, 2, 108. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.J.; Chen, F.; Liu, Q.-G.; Liu, C.-C.; Yao, H.; Yu, B.; Zhang, H.-B.; Yan, H.-X.; Ye, Y.; Chen, T.; et al. Insulin-like growth factor 2 is a key mitogen driving liver repopulation in mice. Cell Death Dis. 2018, 9, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Gui, W.; Tan, B.; Du, Y.; Zhou, J.; Wu, F.; Li, H.; Lin, X. IGF2 deficiency causes mitochondrial defects in skeletal muscle. Clin. Sci. 2021, 135, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Geng, T.; Liu, Y.; Xu, Y.; Jiang, Y.; Zhang, N.; Wang, Z.; Carmichael, G.C.; Taylor, H.S.; Li, D.; Huang, Y. H19 lncRNA Promotes Skeletal Muscle Insulin Sensitivity in Part by Targeting AMPK. Diabetes 2018, 67, 2183–2198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cedernaes, J.; Benedict, C. Comment on Laker. Exercise prevents maternal high-fat diet-induced hypermethylation of the pgc-1α gene and age-dependent metabolic dysfunction in the offspring. Diabetes 2014, 63, 1605–1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McPherson, N.O.; Owens, J.A.; Fullston, T.; Lane, M. Preconception diet or exercise intervention in obese fathers normalizes sperm microRNA profile and metabolic syndrome in female offspring. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E805–E821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guth, L.M.; Ludlow, A.T.; Witkowski, S.; Marshall, M.R.; Lima, L.C.J.; Venezia, A.C.; Xiao, T.; Lee, M.-L.T.; Spangenburg, E.E.; Roth, S.M. Sex-specific effects of exercise ancestry on metabolic, morphological and gene expression phenotypes in multiple generations of mouse offspring. Exp. Physiol. 2013, 98, 1469–1484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murashov, A.K.; Pak, E.S.; Koury, M.; Ajmera, A.; Jeyakumar, M.; Parker, M.; Williams, O.; Ding, J.; Walters, D.; Neufer, P.D. Paternal long-term exercise programs offspring for low energy expenditure and increased risk for obesity in mice. FASEB J. 2016, 30, 775–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guth, L.M.; Roth, S.M. Genetic influence on athletic performance. Curr. Opin. Pediatrics 2013, 25, 653–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontzer, H.; Raichlen, D.A.; Wood, B.M.; Thompson, M.A.; Racette, S.B.; Mabulla, A.Z.P.; Marlowe, F.W. Energy expenditure and activity among Hadza hunter-gatherers. Am. J. Hum. Biol. 2015, 27, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Pontzer, H.; Durazo-Arvizu, R.; Dugas, L.R.; Plange-Rhule, J.; Bovet, P.; Forrester, T.E.; Lambert, E.V.; Cooper, R.S.; Schoeller, D.A.; Luke, A. Constrained Total Energy Expenditure and Metabolic Adaptation to Physical Activity in Adult Humans. Curr. Biol. 2016, 26, 410–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa Júnior, J.M.; Rosa, M.R.; Protzer, A.O.; Paula, F.M.; Ferreira, S.M.; Rezende, L.F.; Vanzela, E.C.; Zoppi, C.C.; Silveira, L.R.; Kettelhut, I.C.; et al. Leucine smentation does not affect protein turnover and impairs the beneficial effects of endurance training on glucose homeostasis in healthy mice. Amino Acids 2015, 47, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Maarbjerg, S.J.; Sylow, L.; Richter, E.A. Current understanding of increased insulin sensitivity after exercise—Emerging candidates. Acta Physiol. 2011, 202, 323–335. [Google Scholar] [CrossRef]
- Egan, B.; Zierath, J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013, 17, 162–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruas, J.L.; White, J.P.; Rao, R.R.; Kleiner, S.; Brannan, K.T.; Harrison, B.C.; Greene, N.P.; Wu, J.; Estall, J.L.; Irving, B.A.; et al. A PGC-1α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 2012, 151, 1319–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Dijk, J.W.; Venema, M.; Mechelen, W.; Stehouwer, C.D.A.; Hartgens, F.; Loon, L.J.C. Effect of moderate-intensity exercise versus activities of daily living on 24-h blood glucose homeostasis in male patients with type 2 diabetes. Diabetes Care 2013, 36, 3448–3453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, N.R.; Marques, S.O.; Luciano, T.F.; Pauli, J.R.; Mpura, L.P.; Caperuto, E.; Pieri, B.L.; Engelmann, J.; Scaini, G.; Streck, E.L.; et al. Retracted: Treadmill Training Increases SIRT-1 and PGC-1α Protein Levels and AMPK Phosphorylation in Quadriceps of Middle-Aged Rats in an Intensity-Dependent Manner. Mediat. Inflamm. 2017, 2017, 8287646. [Google Scholar]
- Fernandez-Twinn, D.S.; Constância, M.; Ozanne, S.E. Intergenerational epigenetic inheritance in models of developmental programming of adult disease. Semin. Cell Dev. Biol. 2015, 43, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Gabory, A.; Attig, L.; Junien, C. Developmental programming and epigenetics. Am. J. Clin. Nutr. 2011, 94 (Suppl. 6), 1943S–1952S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernstein, D.L.; Kameswaran, V.; Le Lay, J.E.; Sheaffer, K.L.; Kaestner, K.H. The BisPCR(2) method for targeted bisulfite sequencing. Epigenetics Chromatin 2015, 8, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huypens, P.; Sass, S.; Wu, M.; Dychoff, D.; Tschöp, M.; Theis, F.; Marschall, S.; Angelis, M.H.; Beckers, J. Epigenetic germline inheritance of diet-induced obesity and insulin resistance. Nat. Genet. 2016, 48, 497–499. [Google Scholar] [CrossRef] [PubMed]
- Reik, W.; Walter, J. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2001, 2, 21–32. [Google Scholar] [CrossRef]
- Mu, Q.; Wang, L.; Yu, F.; Gao, H.; Lei, T.; Li, P.; Liu, P.; Zheng, X.; Hu, X.; Chen, Y.; et al. Imp2 regulates GBM progression by activating IGF2/PI3K/Akt pathway. Cancer Biol. Ther. 2015, 16, 623–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamamura, K.; Zhang, P.; Yokota, H. IGF2-driven PI3 kinase and TGFbeta signaling pathways in chondrogenesis. Cell Biol. Int. 2008, 32, 1238–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, M.; Milligan, L.; Delalbre, A.; Forné, T. Extensive tissue-specific variation of allelic methylation in the Igf2 gene during mouse fetal development: Relation to expression and imprinting. Mech. Dev. 2001, 101, 133–141. [Google Scholar] [CrossRef]
- Connallon, T.; Knowles, L.L. Intergenomic conflict revealed by patterns of sex- biased gene expression. Trends Genet. 2005, 21, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Tycko, B.; Morison, I.M. Physiological functions of imprinted genes. J. Cell. Physiol. 2002, 192, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.A. Seminal plasma and male factor signalling in the female reproductive tract. Cell Tissue Res. 2005, 322, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, R.M. Environmental/lifestyle effects on spermatogenesis. Philos. Trans. R. Soc. Lond B Biol. Sci. 2010, 365, 1697–1712. [Google Scholar] [CrossRef]
- Stanford, K.I.; Rasmussen, M.; Baer, L.A.; Lehnig, A.C.; Rowlnad, L.A.; Whit, J.D.; So, K.; Sousa-Coelho, A.L.; Hisrman, M.F.; Patti, M.E.; et al. Paternal Exercise Improves Glucose Metabolism in Adult Offspring. Diabetes 2018, 67, 2530–2540. [Google Scholar] [CrossRef] [Green Version]
- Safarinejad, M.R.; Azma, K.; Kolahi, A.A. The effects of intensive, long-term treadmill running on reproductive hormones, hypothalamus-pituitary-testis axis, and semen quality: A randomized controlled study. J. Endocrinol. 2009, 200, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, A.O.; Wilde, N.; Gibson, M.; Parks, A.; Carrell, D.T.; Meikle, A.W. Male obesity and alteration in sperm parameters. Fertil. Steril. 2008, 90, 2222–2225. [Google Scholar] [CrossRef] [PubMed]
- Di Luigi, L.; Romanelli, F.; Sgrò, P.; Lenzi, A. Andrological aspects of physical exercise and sport medicine. Endocrine 2012, 42, 278–284. [Google Scholar] [CrossRef]
- Nallella, K.P.; Allamaneni, S.S.R.; Pasqualotti, F.; Sharma, R.K.; Thomas, A.J., Jr.; Agarwal, A. Relationship of interleukin-6 with semen characteristics and oxidative stress in patients with varicocele. Urology 2004, 64, 1010–1013. [Google Scholar] [CrossRef]
- Pedersen, B.K. Muscle as a secretory organ. Compr. Physiol. 2013, 3, 1337–1362. [Google Scholar]
- Ding, G.L.; Wang, F.-F.; Shu, J.; Tian, S.; Jiang, Y.; Zhang, D.; Wang, N.; Luo, Q.; Zhang, Y.; Jin, F.; et al. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes 2012, 61, 1133–1142. [Google Scholar] [CrossRef] [Green Version]
- Rezende, E.L.; Garland, T., Jr.; Chappell, M.A.; Malisch, J.L.; Gomes, F.R. Maximum aerobic performance in lines of Mus selected for high wheel-running activity: Effects of selection, oxygen availability and the mini-muscle phenotype. J. Exp. Biol. 2006, 209 Pt 1, 115–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthews, J.N.; Altman, D.G.; Campbell, M.J.; Royston, P. Analysis of serial measurements in medical research. BMJ 1990, 300, 230–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.Y.; Cokus, S.J.; Pellegrini, M. BS Seeker: Precise mapping for bisulfite sequencing. BMC Bioinform. 2010, 11, 203. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa-Júnior, J.M.; Ferreira, S.M.; Kurauti, M.A.; Bernstein, D.L.; Ruano, E.G.; Kameswaran, V.; Schug, J.; Freitas-Dias, R.; Zoppi, C.C.; Boschero, A.C.; et al. Paternal Exercise Improves the Metabolic Health of Offspring via Epigenetic Modulation of the Germline. Int. J. Mol. Sci. 2022, 23, 1. https://doi.org/10.3390/ijms23010001
Costa-Júnior JM, Ferreira SM, Kurauti MA, Bernstein DL, Ruano EG, Kameswaran V, Schug J, Freitas-Dias R, Zoppi CC, Boschero AC, et al. Paternal Exercise Improves the Metabolic Health of Offspring via Epigenetic Modulation of the Germline. International Journal of Molecular Sciences. 2022; 23(1):1. https://doi.org/10.3390/ijms23010001
Chicago/Turabian StyleCosta-Júnior, José Maria, Sandra Mara Ferreira, Mirian Ayumi Kurauti, Diana L. Bernstein, Elena G. Ruano, Vasumathi Kameswaran, Jonathan Schug, Ricardo Freitas-Dias, Claudio C. Zoppi, Antonio C. Boschero, and et al. 2022. "Paternal Exercise Improves the Metabolic Health of Offspring via Epigenetic Modulation of the Germline" International Journal of Molecular Sciences 23, no. 1: 1. https://doi.org/10.3390/ijms23010001





