Catulin Based Reporter System to Track and Characterize the Population of Invasive Cancer Cells in the Head and Neck Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Results
2.1. Development of a Novel Catulin-Promoter-Based Reporter System to Label and Track the Population of Invasive Cancer Cells of HNSCC
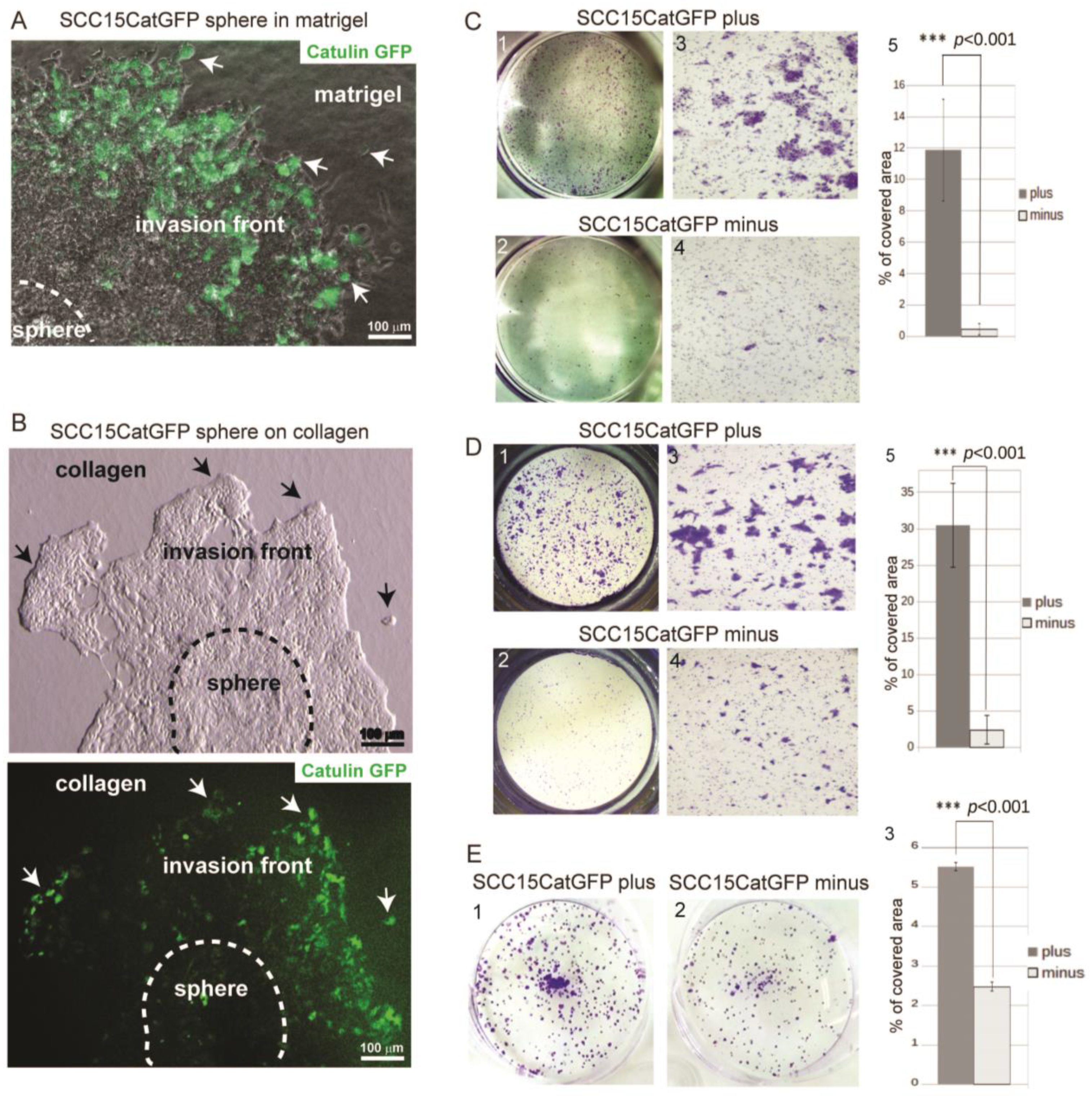
2.2. Catulin-GFP Reporter System Labels Cells with Higher Migratory, Invasive, and Tumorigenic Potential In Vitro
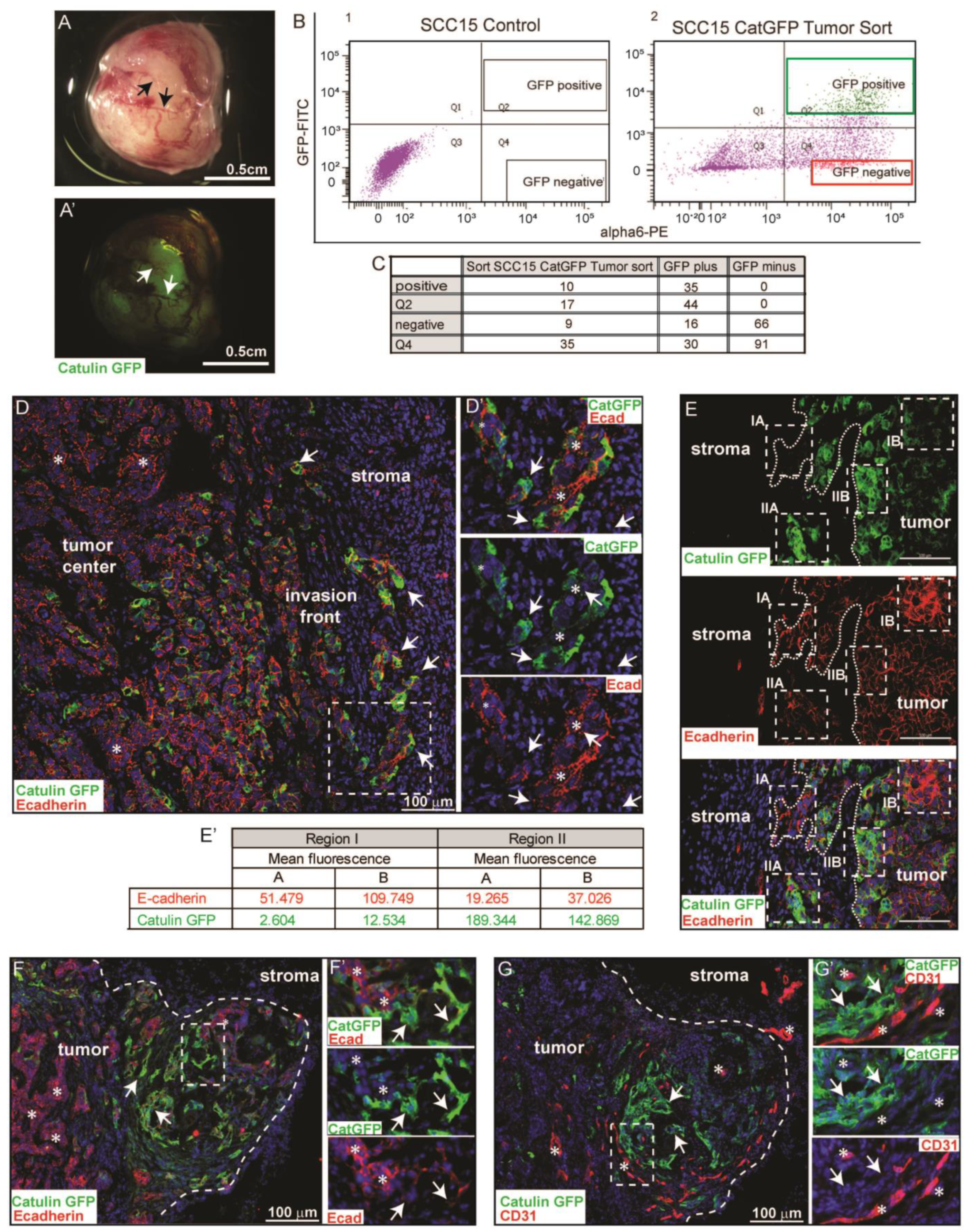
2.3. Catulin-GFP Reporter System Marks a Small Population of Tumor Cells at the Invasive Front in a Xenograft Model of HNSCC That Looses Epithelial Marker E-Cadherin, Indicative of a Partial EMT
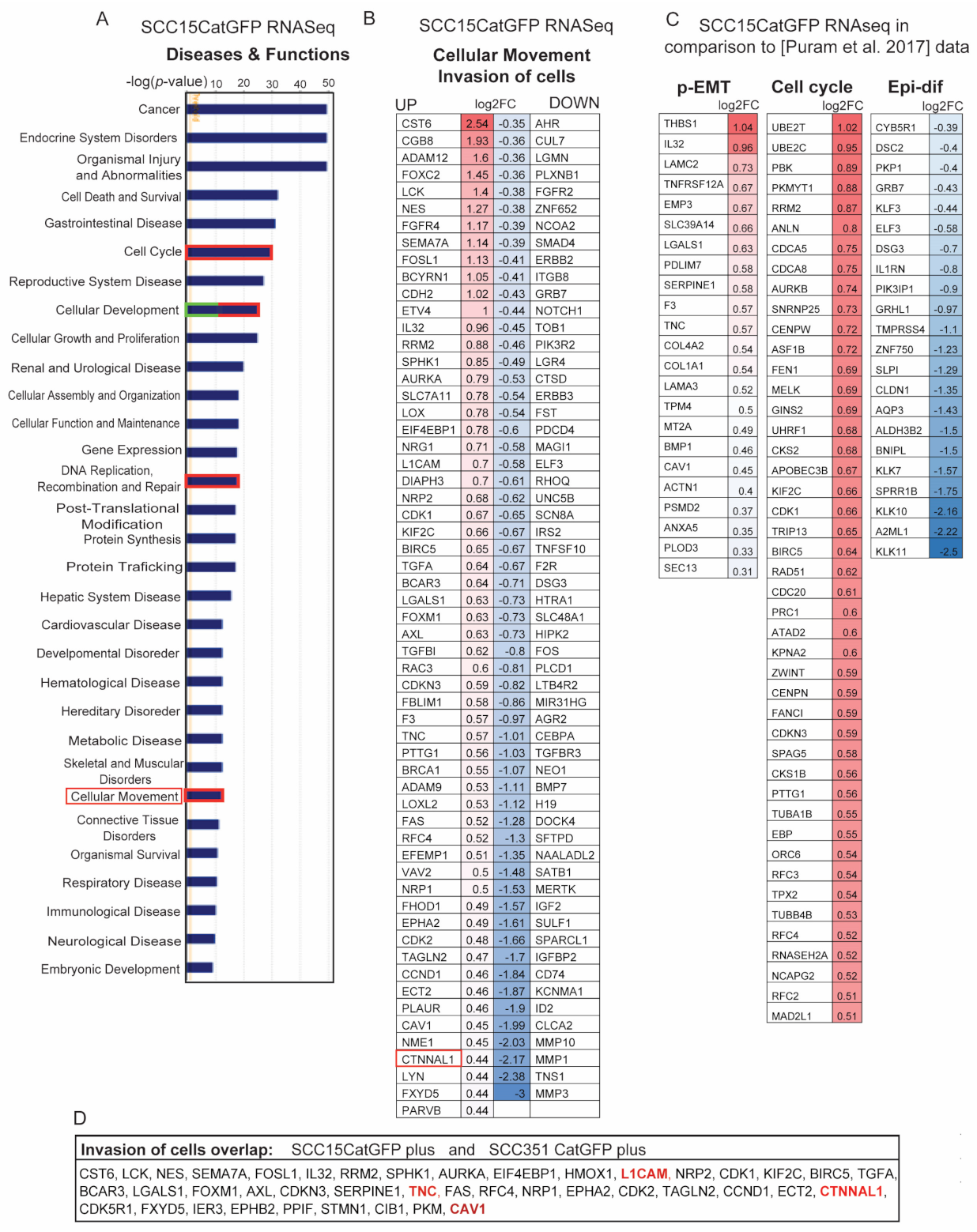
2.4. Identification of Signature Genes of Invasive Catulin-GFP Reporter-Labeled Cancer Cells
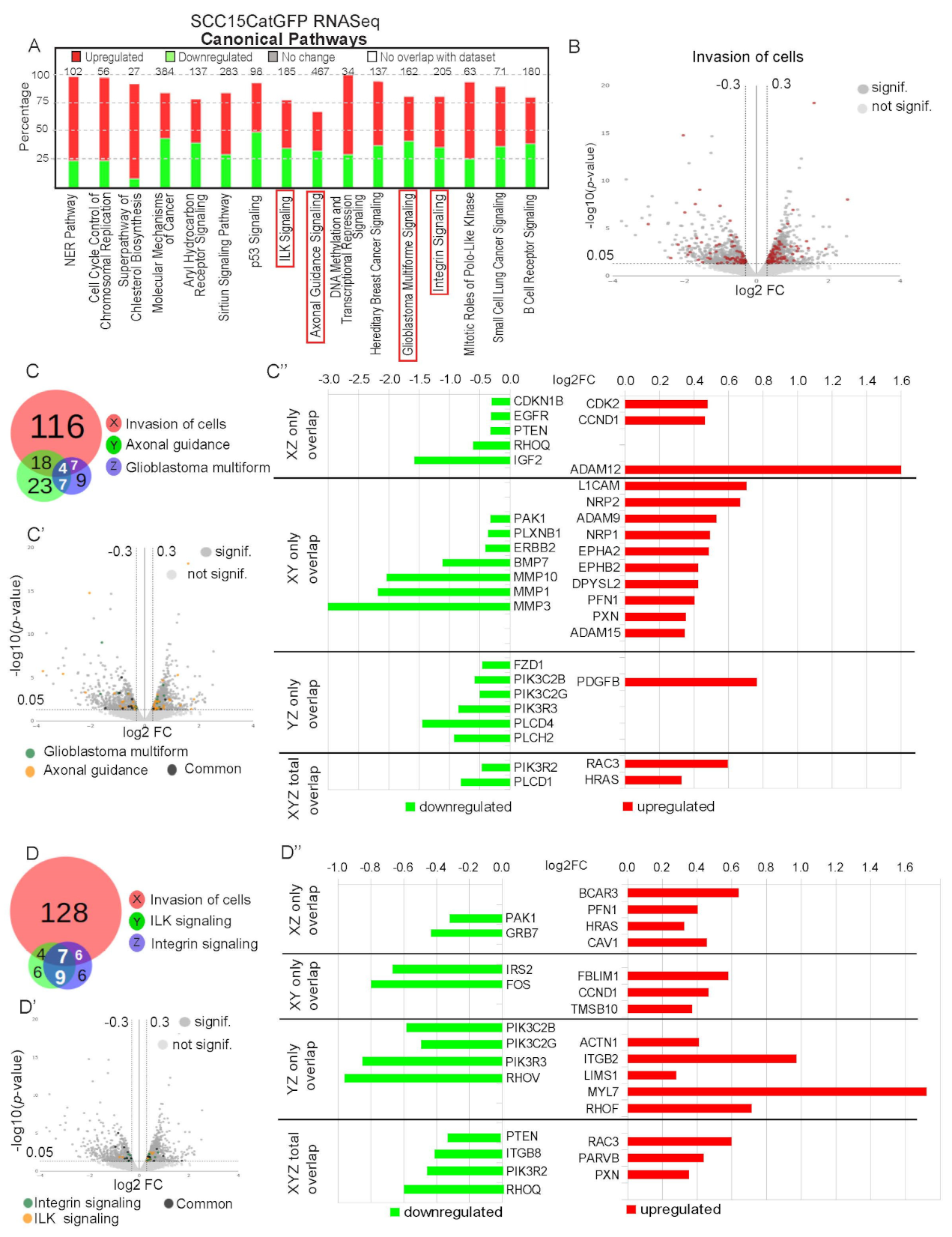
2.5. Genetic Signature of Catulin-GFP Reporter-Labeled Cancer Cells Indicates an Enrichment in Genes Involved in Axonal Guidance, Glioblastoma Multiforme, ILK, and Integrin Signaling Pathways
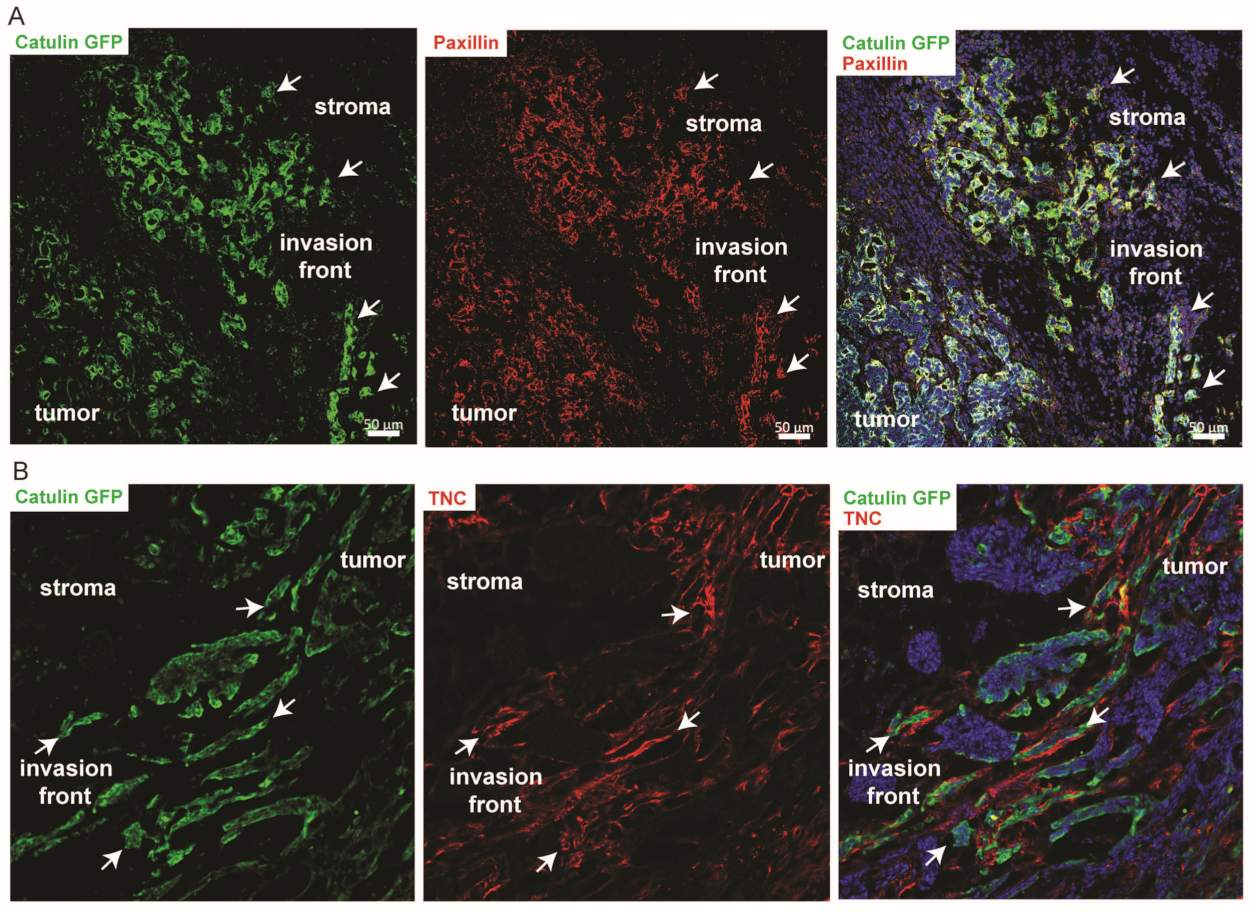
2.6. Known Invasion Markers PXN and Tenascin-C Are Expressed in the Cells Marked by the Catulin Reporter System at the Tumor Invasion Front
2.7. CAV1 Is Expressed at the Invasion Front in Tumors Derived from Cells with the Catulin Reporter System and in the Human Specimens of HNSCC
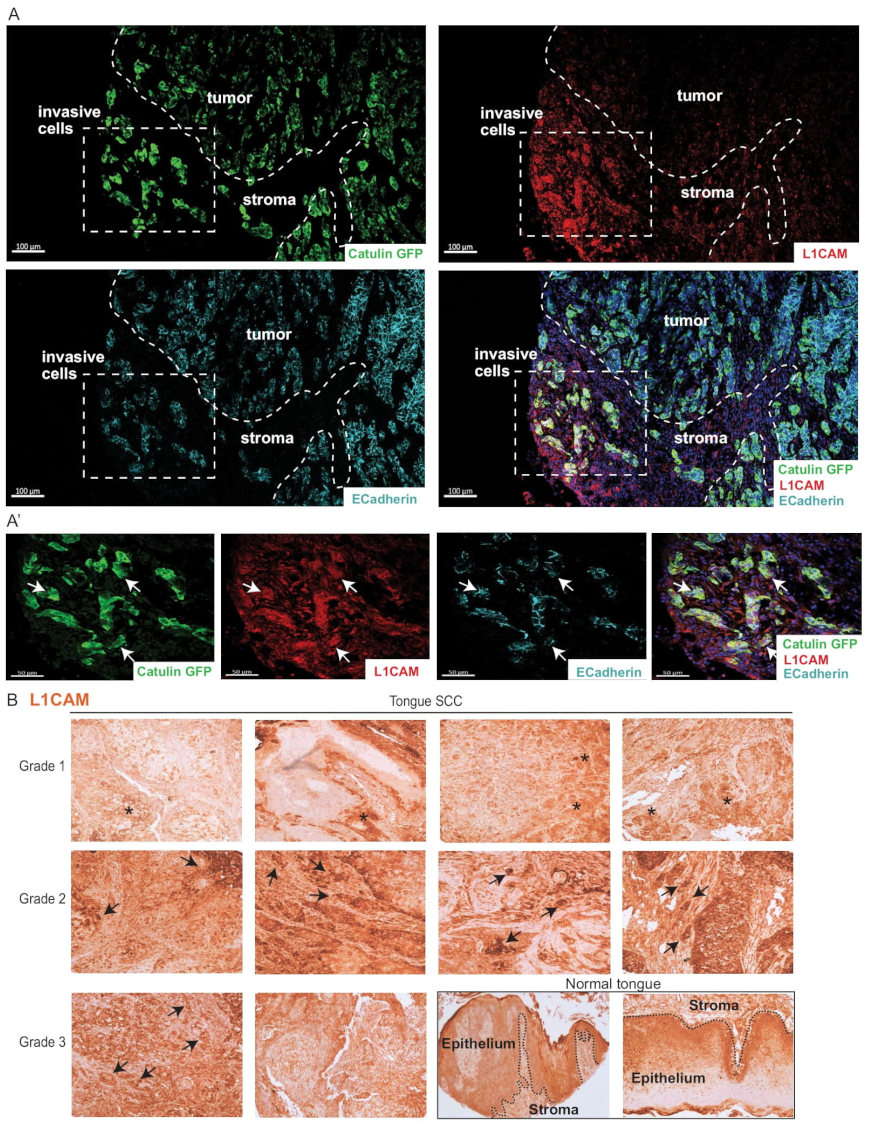
2.8. Invasive Cells Marked by the Catulin Reporter System and Human Specimens of HNSCC Express a Perineural Invasion Marker L1CAM at the Tumor–Stroma Border
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Generation of the Catulin Reporter System
4.2. FACS and Flow Cytometry Fluorescence Analysis
4.3. Protein Isolation and Western Blot Analysis
4.4. Sphere Invasion
4.5. Migration and Invasion Assay
4.6. Colony Formation Assay
4.7. Xenograft Transplants and Experimental Animals
4.8. Dissection Scope Pictures
4.9. Tissue Preparation and Cell Isolation
4.10. Hematoxlin and Eosin (H&E) Staining and Immunofluorescence
4.11. Immunohistochemistry
4.12. RNA Isolation
4.13. RNAseq Analysis
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Ctnnal1 | catenin alpha-like 1 |
| HNSCC | head and neck squamous cell carcinoma |
| TME | tumor microenvironment |
| CAF | cancer-associated fibroblast |
| EMT | epithelial-to-mesenchymal transition |
References
- Wynder, E.L.; Bross, I.J. Aetiological factors in mouth cancer; an approach to its prevention. Br. Med. J. 1957, 1, 1137–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.Y.; Kahyo, H. Effect of cigarette smoking and alcohol consumption in the aetiology of cancer of the oral cavity, pharynx and larynx. Int. J. Epidemiol. 1991, 20, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Zur Hausen, H. Papillomavirus infections—A major cause of human cancers. Biochim. Biophys. Acta 1996, 1288, F55–F78. [Google Scholar] [CrossRef]
- Winn, D.M.; Diehl, S.R.; Horowitz, A.M.; Gutkind, S.; Sandberg, A.L.; Kleinman, D.V. Scientific progress in understanding oral and pharyngeal cancers. J. Am. Dent. Assoc. 1998, 129, 713–718. [Google Scholar] [CrossRef]
- Harras, A.; Edwards, B.K.; Blot, W.J.; Ries, L.A. Cancer Rates and Risk (NIH Publication No. 96-691); National Institutes of Health, Cancer Statistics Branch, Division of Cancer Prevention and Control, Department of Health and Human Services: Washington, DC, USA, 1996. [Google Scholar]
- Bhave, S.L.; Teknos, T.N.; Pan, Q. Molecular parameters of head and neck cancer metastasis. Crit. Rev. Eukaryot. Gene Exp. 2011, 21, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zheng, Q.; Lu, Z.; Wang, L.; Ding, L.; Xia, L.; Zhang, H.; Wang, M.; Chen, Y.; Li, G. Role of the nervous system in cancers: A review. Cell Death Discov. 2021, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhuang, X.; Lin, L.; Liangyu, L.; Wang, Y.; Shi, Y.; Hu, G.; Sun, Y. New horizons in tumor microenvironment biology: Challenges and opportunities. BMC Med. 2015, 13, 45. [Google Scholar] [CrossRef] [Green Version]
- Knops, A.M.; South, A.; Rodeck, U.; Martinez-Outschoorn, U.; Harshyne, L.A.; Johnson, J.; Luginbuhl, A.J.; Curry, J.M. Cancer-Associated Fibroblast Density, Prognostic Characteristics, and Recurrence in Head and Neck Squamous Cell Carcinoma: A Meta-Analysis. Front. Oncol. 2020, 10, 565306. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.-H.; Hong, T.-M.; Cheng, H.-W.; Pan, S.-H.; Liang, Y.-R.; Hong, H.-C.; Chiang, W.-F.; Wong, T.-Y.; Shieh, D.-B.; Shiau, A.-L.; et al. Galectin-1-mediated tumor invasion and metastasis, up-regulated matrix metalloproteinase expression, and reorganized actin cytoskeletons. Mol. Cancer Res. 2009, 7, 311–318. [Google Scholar] [CrossRef] [Green Version]
- Deryugina, E.I.; Quigley, J.P. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006, 25, 9–34. [Google Scholar] [CrossRef]
- Fidler, I.J. The pathogenesis of cancer metastasis: The ’seed and soil’ hypothesis revisited. Nat. Rev. Cancer 2003, 3, 453–458. [Google Scholar] [CrossRef]
- Batsakis, J.G. Nerves and neurotropic carcinomas. Ann. Otol. Rhinol. Laryngol. 1985, 94 Pt 1, 426–427. [Google Scholar]
- Liebig, C.; Ayala, G.; Wilks, J.A.; Berger, D.H.; Albo, D. Perineural invasion in cancer: A review of the literature. Cancer 2009, 115, 3379–3391. [Google Scholar] [CrossRef]
- Saidak, Z.; Lailler, C.; Clatot, F.; Galmiche, A. Perineural invasion in head and neck squamous cell carcinoma: Background, mechanisms, and prognostic implications. Curr. Opin. Otolaryngol. Head Neck Surg. 2020, 28, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Chinn, S.; Spector, M.E.; Bellile, E.L.; McHugh, J.B.; Gernon, T.J.; Bradford, C.R.; Wolf, G.T.; Eisbruch, A.; Chepeha, D.B. Impact of perineural invasion in the pathologically N0 neck in oral cavity squamous cell carcinoma. Otolaryngol. Head Neck Surg. 2013, 149, 893–899. [Google Scholar] [CrossRef] [Green Version]
- Fagan, J.J.; Collins, B.; Barnes, L.; D’Amico, F.; Myers, E.N.; Johnson, J.T. Perineural invasion in squamous cell carcinoma of the head and neck. Arch. Otolaryngol. Head Neck Surg. 1998, 124, 637–640. [Google Scholar] [CrossRef] [Green Version]
- Soo, K.-C.; Carter, R.L.; O’Brien, C.J.; Barr, L.; Bliss, J.M.; Shaw, H.J. Prognostic implications of perineural spread in squamous carcinomas of the head and neck. Laryngoscope 1986, 96, 1145–1148. [Google Scholar] [CrossRef] [PubMed]
- Thierauf, J.L.; Veit, J.A.; Hess, J. Epithelial-to-Mesenchymal Transition in the Pathogenesis and Therapy of Head and Neck Cancer. Cancers 2017, 9, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, M.; Myers, J.N.; Lippman, S.M.; Johnson, F.M.; Williams, M.D.; Rayala, S.; Ohshiro, K.; Rosenthal, D.I.; Weber, R.S.; Gallick, G.E.; et al. Epithelial to mesenchymal transition in head and neck squamous carcinoma: Association of Src activation with E-cadherin down-regulation, vimentin expression, and aggressive tumor features. Cancer 2008, 112, 2088–2100. [Google Scholar] [CrossRef] [PubMed]
- Thierauf, J.; Veit, J.A.; Hess, J. Expression of E-cadherin and vimentin correlates with metastasis formation in head and neck squamous cell carcinoma patients. Radiother. Oncol. 2011, 99, 344–348. [Google Scholar]
- Smith, A.; Teknos, T.N.; Pan, Q. Epithelial to mesenchymal transition in head and neck squamous cell carcinoma. Oral Oncol. 2013, 49, 287–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobielak, A.; Fuchs, E. Links between alpha-catenin, NF-kappaB, and squamous cell carcinoma in skin. Proc. Natl. Acad. Sci. USA 2006, 103, 2322–2327. [Google Scholar] [CrossRef] [Green Version]
- Cao, C.; Chen, Y.; Masood, R.; Sinha, U.K.; Kobielak, A. α-Catulin marks the invasion front of squamous cell carcinoma and is important for tumor cell metastasis. Mol. Cancer Res. MCR 2012, 10, 892–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karpińska, K.; Cao, C.; Yamamoto, V.; Gielata, M.; Kobielak, A. Alpha-Catulin, a New Player in a Rho Dependent Apical Constriction That Contributes to the Mouse Neural Tube Closure. Front. Cell Dev. Biol. 2020, 8, 154. [Google Scholar] [CrossRef] [Green Version]
- Kreiseder, B.; Orel, L.; Bujnow, C.; Buschek, S.; Pflueger, M.; Schuett, W.; Hundsberger, H.; de Martin, R.; Wiesner, C. α-Catulin downregulates E-cadherin and promotes melanoma progression and invasion. Int. J. Cancer. 2013, 132, 521–530. [Google Scholar] [CrossRef]
- Kreiseder, B.; Holper-Schichl, Y.M.; Muellauer, B.; Jacobi, N.; Pretsch, A.; Schmid, J.; De Martin, R.; Hundsberger, H.; Eger, A.; Wiesner, C. Alpha-catulin contributes to drug-resistance of melanoma by activating NF-κB and AP-1. PLoS ONE 2015, 10, e0119402. [Google Scholar]
- Liang, C.H.; Chiu, S.Y.; Hsu, I.L.; Wu, Y.Y.; Tsai, Y.T.; Ke, J.Y.; Pan, S.H.; Hsu, Y.C.; Li, K.C.; Yang, P.C.; et al. α-Catulin drives metastasis by activating ILK and driving an αvβ3 integrin signaling axis. Cancer Res. 2013, 73, 428–438. [Google Scholar] [CrossRef] [Green Version]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e24. [Google Scholar] [CrossRef] [Green Version]
- Campos, A.; Burgos-Ravanal, R.; González, M.F.; Huilcaman, R.; González, L.L.; Quest, A.F.G. Cell Intrinsic and Extrinsic Mechanisms of Caveolin-1-Enhanced Metastasis. Biomolecules 2019, 9, 314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wehinger, S.; Ortiz, R.; Diaz, N.; Diaz, J.; Lobos-Gonzalez, L.; Quest, A. Caveolin-1 in cell migration and metastasis. Curr. Mol. Med. 2014, 14, 255–274. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, L.; Bruschini, R.; Giugliano, G.; Ostuni, A.; Maffini, F.; Massaro, M.A.; Santoro, L.; Navach, V.; Preda, L.; Alterio, D.; et al. Compartmental tongue surgery: Long term oncologic results in the treatment of tongue cancer. Oral Oncol. 2011, 47, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.J.; Ewald, A.J. Illuminating breast cancer invasion: Diverse roles for cell-cell interactions. Curr. Opin. Cell Biol. 2014, 30, 99–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaggioli, C. Collective invasion of carcinoma cells: When the fibroblasts take the lead. Cell Adh. Migr. 2008, 2, 45–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, R.; Wewer, U.M.; Zurakowski, D.; Pories, S.E.; Moses, M.A. ADAM 12 cleaves extracellular matrix proteins and correlates with cancer status and stage. J. Biol. Chem. 2004, 279, 51323–51330. [Google Scholar] [CrossRef] [Green Version]
- Duffy, M.J.; Lynn, D.J.; Lloyd, A.T.; O’Shea, C.M. The ADAMs family of proteins: From basic studies to potential clinical applications. Thromb. Haemost. 2003, 89, 622–631. [Google Scholar]
- Lu, P.; Takai, K.; Weaver, V.M.; Werb, Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 2011, 3, 12. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, P.; Etcheverry, A.; Aubry, M.; Missey, A.; Lachat, C.; Perrard, J.; Hendrick, E.; Delage-Mourroux, R.; Mosser, J.; Borg, C.; et al. EMT is associated with an epigenetic signature of ECM remodeling genes. Cell Death Dis. 2019, 10, 205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jolly, M.K.; Boareto, M.; Huang, B.; Jia, D.; Lu, M.; Ben-Jacob, E.; Onuchic, J.N.; Levine, H. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front. Oncol. 2015, 5, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanken, H.; Gröbe, A.; Cachovan, G.; Smeets, R.; Simon, R.; Sauter, G.; Heiland, M.; Blessmann, M. CCND1 amplification and cyclin D1 immunohistochemical expression in head and neck squamous cell carcinomas. Clin. Oral Investig. 2014, 18, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, V.; Verma, J.; Misra, V.; Srivastav, S.; Hasan, F. Evaluation of Cyclin D1 expression in Head and Neck Squamous Cell Carcinoma. J. Clin. Diagn. Res. 2017, 11, ec01–ec04. [Google Scholar] [CrossRef] [PubMed]
- Soffar, A.; Storch, K.; Aleem, E.; Cordes, N. CDK2 knockdown enhances head and neck cancer cell radiosensitivity. Int. J. Radiat. Biol. 2013, 89, 523–531. [Google Scholar] [CrossRef]
- Qin, X.; Guo, H.; Wang, X.; Zhu, X.; Yan, M.; Wang, X.; Xu, Q.; Shi, J.; Lu, E.; Chen, W.; et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 2019, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.L.; Schrecengost, R.S.; Guerrero, M.S.; Thomas, K.S.; Bouton, A.H. Breast cancer antiestrogen resistance 3 (BCAR3) promotes cell motility by regulating actin cytoskeletal and adhesion remodeling in invasive breast cancer cells. PLoS ONE 2013, 8, e65678. [Google Scholar] [CrossRef] [Green Version]
- Xie, G.-F.; Zhao, L.-D.; Chen, Q.; Tang, D.; Chen, Q.-Y.; Lu, H.-F.; Cai, J.-R.; Chen, Z. High ACTN1 Is Associated with Poor Prognosis, and ACTN1 Silencing Suppresses Cell Proliferation and Metastasis in Oral Squamous Cell Carcinoma. Drug Des. Dev. Ther. 2020, 14, 1717–1727. [Google Scholar] [CrossRef]
- Yamaji, S.; Suzuki, A.; Kanamori, H.; Mishima, W.; Yoshimi, R.; Takasaki, H.; Takabayashi, M.; Fujimaki, K.; Fujisawa, S.; Ohno, S.; et al. Affixin interacts with alpha-actinin and mediates integrin signaling for reorganization of F-actin induced by initial cell-substrate interaction. J. Cell Biol. 2004, 165, 539–551. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.-J.; Zhong, J.; Yu, J.-G.; Zhao, F.; Xiang, Y. The structure and functions of paxillin and its roles in neovascularization. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1768–1773. [Google Scholar] [PubMed]
- Eslami, A.; Miyaguchi, K.; Mogushi, K.; Watanabe, H.; Okada, N.; Shibuya, H.; Mizushima, H.; Miura, M.; Tanaka, H. PARVB overexpression increases cell migration capability and defines high risk for endophytic growth and metastasis in tongue squamous cell carcinoma. Br. J. Cancer 2015, 112, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Stiegler, A.L.; Draheim, K.M.; Li, X.; Chayen, N.E.; Calderwood, D.A.; Boggon, T.J. Structural basis for paxillin binding and focal adhesion targeting of β-parvin. J. Biol. Chem. 2012, 287, 32566–32577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theocharis, S.; Kotta-Loizou, I.; Giaginis, C.; Alexandrou, P.; Danas, E.; Tsourouflis, G.; Tsoukalas, N.; Coutts, R.H.; Tasoulas, J.; Klijanienko, J. Expression and Clinical Significance of Concomitant FAK/SRC and p-Paxillin in Mobile Tongue Squamous Cell Carcinoma. Anticancer Res. 2017, 37, 1313–1319. [Google Scholar] [PubMed]
- Wu, D.-W.; Chuang, C.-Y.; Lin, W.-L.; Sung, W.-W.; Cheng, Y.-W.; Lee, H. Paxillin promotes tumor progression and predicts survival and relapse in oral cavity squamous cell carcinoma by microRNA-218 targeting. Carcinogenesis 2014, 35, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Tampakis, A.; Tampaki, E.C.; Nonni, A.; Tsourouflis, G.; Posabella, A.; Patsouris, E.; Kontzoglou, K.; Von Flüe, M.; Nikiteas, N.; Kouraklis, G. L1CAM expression in colorectal cancer identifies a high-risk group of patients with dismal prognosis already in early-stage disease. Acta Oncol. 2020, 59, 55–59. [Google Scholar] [CrossRef]
- Altevogt, P.; Doberstein, K.; Fogel, M. L1CAM in human cancer. Int. J. Cancer 2016, 138, 1565–1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben, Q.-W.; Wang, J.-C.; Liu, J.; Zhu, Y.; Yuan, F.; Yao, W.-Y.; Yuan, Y.-Z. Positive expression of L1-CAM is associated with perineural invasion and poor outcome in pancreatic ductal adenocarcinoma. Ann. Surg. Oncol. 2010, 17, 2213–2221. [Google Scholar] [CrossRef] [PubMed]
- Na’ara, S.; Amit, M.; Gil, Z. L1CAM induces perineural invasion of pancreas cancer cells by upregulation of metalloproteinase expression. Oncogene 2019, 38, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Castellani, V.; Chedotal, A.; Schachner, M.; Faivre-Sarrailh, C.; Rougon, G. Analysis of the L1-deficient mouse phenotype reveals cross-talk between Sema3A and L1 signaling pathways in axonal guidance. Neuron 2000, 27, 237–249. [Google Scholar] [CrossRef] [Green Version]
- Kawakami, T.; Tokunaga, T.; Hatanaka, H.; Kijima, H.; Yamazaki, H.; Abe, Y.; Osamura, Y.; Inoue, H.; Ueyama, Y.; Nakamura, M. Neuropilin 1 and neuropilin 2 co-expression is significantly correlated with increased vascularity and poor prognosis in nonsmall cell lung carcinoma. Cancer 2002, 95, 2196–2201. [Google Scholar] [CrossRef] [PubMed]
- Niland, S.; Eble, J.A. Neuropilins in the Context of Tumor Vasculature. Int. J. Mol. Sci. 2019, 20, 639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stasikowska-Kanicka, O.; Wągrowska-Danilewicz, M.; Danilewicz, M. Immunohistochemical study on neuropilin 1 (NRP1) immunoexpression in oral squamous cell carcinoma. Folia Histochem. Cytobiol. 2018, 1, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Gao, Z.; Sun, M.; Li, H.; Fan, H.; Chen, D.; Zheng, J. Prognostic significance of VEGF-C, semaphorin 3F, and neuropilin-2 expression in oral squamous cell carcinomas and their relationship with lymphangiogenesis. J. Surg. Oncol. 2015, 111, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, R.C.; Han, K.Y.; Rojas, J.; Ozturk, O.; Kim, D.J.; Jain, S.; Chang, J.-H.; Azar, D.T. Semaphorin 7A promotes angiogenesis in an experimental corneal neovascularization model. Curr. Eye Res. 2011, 36, 989–996. [Google Scholar] [CrossRef] [Green Version]
- Allegra, M.; Zaragkoulias, A.; Vorgia, E.; Ioannou, M.; Litos, G.; Beug, H.; Mavrothalassitis, G. Semaphorin-7a reverses the ERF-induced inhibition of EMT in Ras-dependent mouse mammary epithelial cells. Mol. Biol. Cell. 2012, 23, 3873–3881. [Google Scholar] [CrossRef]
- Saito, T.; Kasamatsu, A.; Ogawara, K.; Miyamoto, I.; Saito, K.; Iyoda, M.; Suzuki, T.; Endo-Sakamoto, Y.; Shiiba, M.; Tanzawa, H.; et al. Semaphorin7A Promotion of Tumoral Growth and Metastasis in Human Oral Cancer by Regulation of G1 Cell Cycle and Matrix Metalloproteases: Possible Contribution to Tumoral Angiogenesis. PLoS ONE 2015, 10, e0137923. [Google Scholar]
- Lazova, R.; Rothberg, B.E.G.; Rimm, D.; Scott, G. The semaphorin 7A receptor Plexin C1 is lost during melanoma metastasis. Am. J. Dermatopathol. 2009, 31, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Scott, G.A.; McClelland, L.A.; Fricke, A.F.; Fender, A. Plexin C1, A Receptor for Semaphorin 7A, Inactivates Cofilin and Is a Potential Tumor Suppressor for Melanoma Progression. J. Investig. Dermatol. 2009, 129, 954–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Su, L.; Müller, S.; Tighiouart, M.; Xu, Z.; Zhang, X.; Shin, H.J.C.; Hunt, J.; Sun, S.-Y.; Shin, D.M.; et al. Restoration of caveolin-1 expression suppresses growth and metastasis of head and neck squamous cell carcinoma. Br. J. Cancer. 2008, 99, 1684–1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, A.; Ray, A.-M.; Ramolu, L.; Macabre, C.; Simon, F.; Noulet, F.; Blandin, A.-F.; Renner, G.; Lehmann, M.; Choulier, L.; et al. Caveolin-1-negative head and neck squamous cell carcinoma primary tumors display increased epithelial to mesenchymal transition and prometastatic properties. Oncotarget 2015, 6, 41884–41901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, A.; Ray, A.-M.; Ramolu, L.; Macabre, C.; Simon, F.; Noulet, F.; Blandin, A.-F.; Renner, G.; Lehmann, M.; Choulier, L.; et al. Involvement of the TGF-β1 pathway in caveolin-1-associated regulation of head and neck tumor cell metastasis. Oncol. Lett. 2020, 19, 1298–1304. [Google Scholar]
- Seki, N.; Nohata, N.; Hanazawa, T.; Kikkawa, N.; Mutallip, M.; Fujimura, L.; Yoshino, H.; Kawakami, K.; Chiyomaru, T.; Enokida, H.; et al. Caveolin-1 mediates tumor cell migration and invasion and its regulation by miR-133a in head and neck squamous cell carcinoma. Int. J. Oncol. 2011, 38, 209–217. [Google Scholar] [CrossRef] [Green Version]
- Masuelli, L.; Budillon, A.; Marzocchella, L.; Mrozek, M.-A.; Vitolo, D.; Di Gennaro, E.; Losito, S.; Sale, P.; Longo, F.; Ionna, F.; et al. Caveolin-1 overexpression is associated with simultaneous abnormal expression of the E-cadherin/α-β catenins complex and multiple ErbB receptors and with lymph nodes metastasis in head and neck squamous cell carcinomas. J. Cell Physiol. 2012, 227, 3344–3353. [Google Scholar] [CrossRef]
- Liebertz, D.J.; Lechner, M.G.; Masood, R.; Sinha, U.K.; Han, J.; Puri, R.K.; Correa, A.J.; Epstein, A.L. Establishment and characterization of a novel head and neck squamous cell carcinoma cell line USC-HN1. Head Neck Oncol. 2010, 2, 5. [Google Scholar] [CrossRef] [Green Version]
- Young, L.; Sung, J.; Stacey, G.; Masters, J.R. Detection of Mycoplasma in cell cultures. Nat. Protoc. 2010, 5, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, B.; Lin, L.-L.; Zhao, S. Evaluation and comparison of computational tools for RNA-seq isoform quantification. BMC Genomics 2017, 18, 583. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Karolchik, D.; Hinrichs, A.S.; Furey, T.S.; Roskin, K.M.; Sugnet, C.W.; Haussler, D.; Kent, W.J. The UCSC Table Browser data retrieval tool. Nucl. Acids Res. 2004, 32 (Suppl. S1), D493–D496. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Hulsen, T.; de Vlieg, J.; Alkema, W. BioVenn—A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom. 2008, 9, 488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karpińska, K.; Gielata, M.; Gwiazdowska, A.; Boryń, Ł.; Kobielak, A. Catulin Based Reporter System to Track and Characterize the Population of Invasive Cancer Cells in the Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 140. https://doi.org/10.3390/ijms23010140
Karpińska K, Gielata M, Gwiazdowska A, Boryń Ł, Kobielak A. Catulin Based Reporter System to Track and Characterize the Population of Invasive Cancer Cells in the Head and Neck Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2022; 23(1):140. https://doi.org/10.3390/ijms23010140
Chicago/Turabian StyleKarpińska, Kamila, Mateusz Gielata, Aleksandra Gwiazdowska, Łukasz Boryń, and Agnieszka Kobielak. 2022. "Catulin Based Reporter System to Track and Characterize the Population of Invasive Cancer Cells in the Head and Neck Squamous Cell Carcinoma" International Journal of Molecular Sciences 23, no. 1: 140. https://doi.org/10.3390/ijms23010140
APA StyleKarpińska, K., Gielata, M., Gwiazdowska, A., Boryń, Ł., & Kobielak, A. (2022). Catulin Based Reporter System to Track and Characterize the Population of Invasive Cancer Cells in the Head and Neck Squamous Cell Carcinoma. International Journal of Molecular Sciences, 23(1), 140. https://doi.org/10.3390/ijms23010140





