Calmodulin and Its Interactive Proteins Participate in Regulating the Explosive Growth of Alexandrium pacificum (Dinoflagellate)
Abstract
:1. Introduction
2. Results
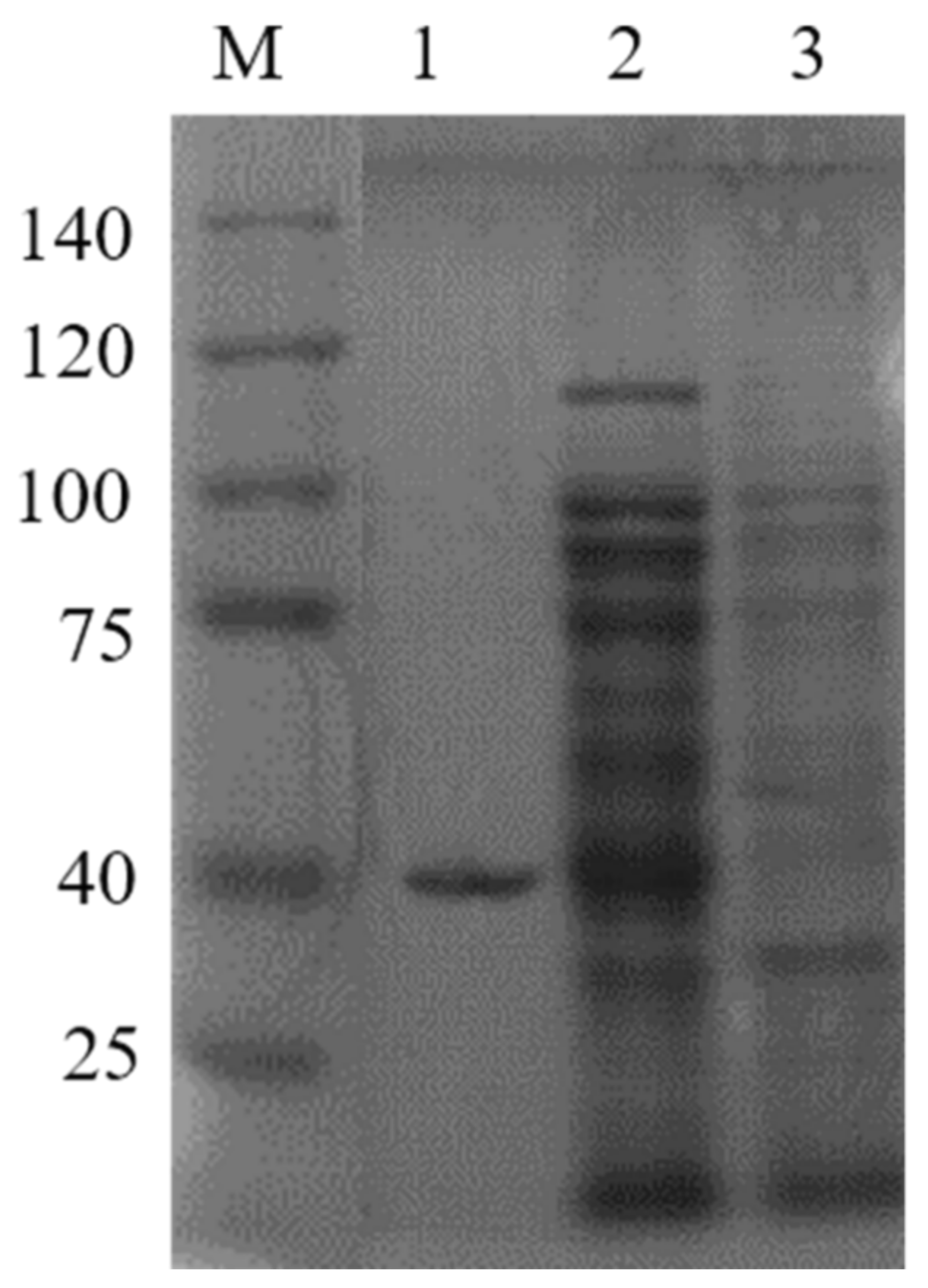
2.1. Heterologous Expression in E. coli and GST-CaM Fusion Protein Purification
2.2. Determination of GST-CaM Specificity
2.3. 2-D and Far Western Blot Analysis of Differential Protein Expression
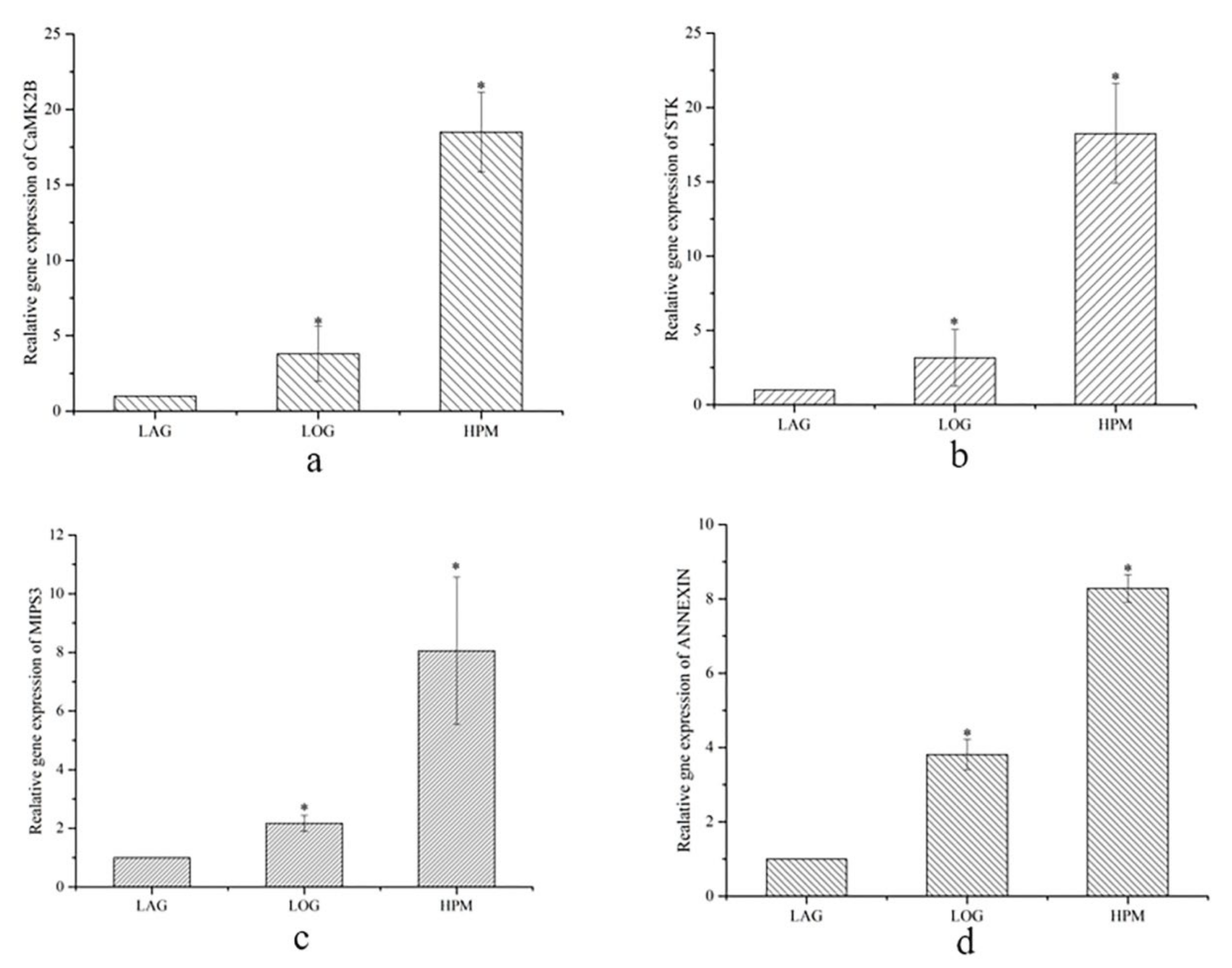
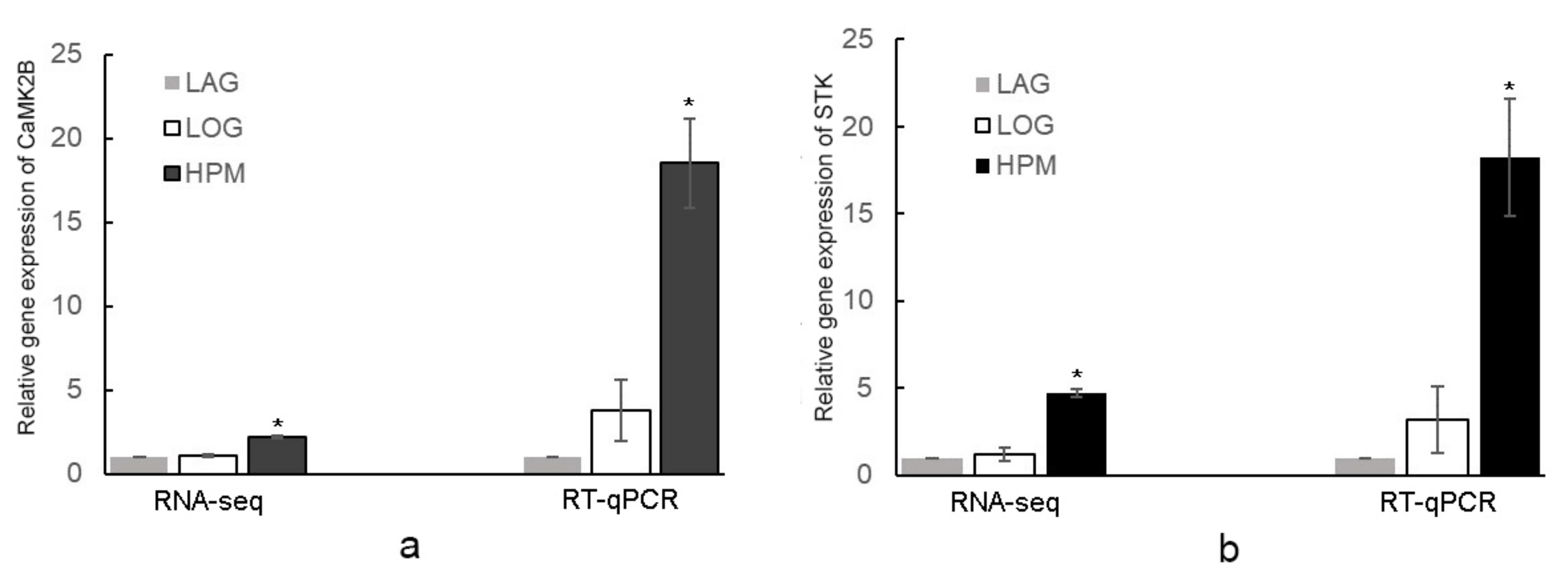
2.4. Gene Expression Analysis of Differentially Expressed Proteins
3. Discussion
3.1. Ca2+ Transport Channel
3.2. CaM and Its Interacting Protein in Cell Cycle Regulation
3.3. Summary of CaM Interacting Proteins
4. Materials and Methods
4.1. Sample Preparation and Collection
4.2. RNA Extraction and First-Strand cDNA Synthesis
4.3. The cDNA Isolation and Sequencing
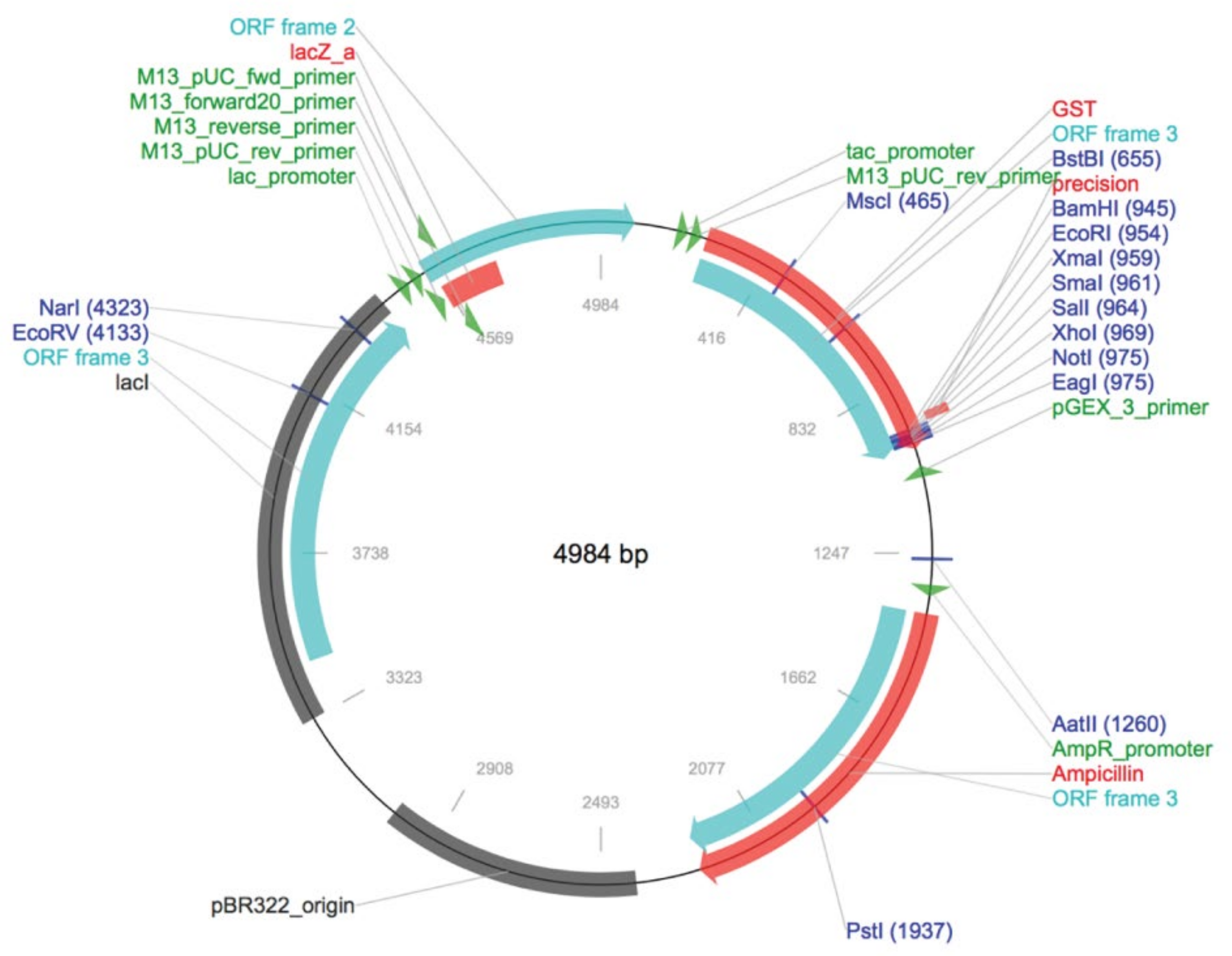
4.4. Construction of Recombinant Plasmids
4.5. Prokaryotic Expression and Purification of GST-CaM Fusion Protein
4.6. Determination of GST-CaM Specificity
4.7. Protein Extraction
4.8. The 2-D Analysis
4.9. Far Western Blot
4.10. MALDI-TOF MS Analysis
4.11. Quantitative PCR to Determine the Gene Expression of Differentially Expressed Protein
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hallegraeff, G.M. A review of harmful algal blooms and their apparent global increase. Phycologia 1993, 32, 79–99. [Google Scholar] [CrossRef] [Green Version]
- Geraci, J.R.; Anderson, D.M.; Timperi, R.J.; Aubin, D.J.S.; Early, G.A.; Prescott, J.H.; Mayo, C.A. Humpback Whales (Megaptera novaeangliae) Fatally Poisoned by Dinoflagellate Toxin. Can. J. Fish. Aquat. Sci. 1989, 46, 1895–1898. [Google Scholar] [CrossRef]
- Scholin, C.A.; Gulland, F.; Doucette, G.J.; Benson, S.; Busman, M.; Chavez, F.P.; Cordaro, J.; Delong, R.; De, V.A.; Harvey, J.; et al. Mortality of sea lions along the central California coast linked to a toxic diatom bloom. Nature 2000, 403, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Alpermann, T.J.; Cembella, A.D.; Collos, Y.; Masseret, E.; Montresor, M. The globally distributed genus Alexandrium: Multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 2012, 14, 10–35. [Google Scholar] [CrossRef] [Green Version]
- Condie, S.A.; Oliver, E.C.J.; Hallegraeff, G.M. Environmental drivers of unprecedented Alexandrium catenella dinoflagellate blooms off eastern Tasmania, 2012–2018. Harmful Algae 2019, 87, 101628. [Google Scholar] [CrossRef]
- Wells, M.L.; Trainer, V.L.; Smayda, T.J.; Karlson, B.S.; Trick, C.G.; Kudela, R.M.; Ishikawa, A.; Bernard, S.; Wulff, A.; Anderson, D.M.; et al. Harmful algal blooms and climate change: Learning from the past and present to forecast the future. Harmful Algae 2015, 49, 68–93. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Gao, H.; Tong, M.; Anderson, D.M. Cell cycle regulation of the mixotrophic dinoflagellate Dinophysis acuminata: Growth, photosynthetic efficiency and toxin production. Harmful Algae 2019, 89, 101672. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, H.K.; Zhang, S.F.; Zhou, Y.; He, Y.B.; Amin, S.A.; Chen, J.W.; Yan, K.Q.; Lin, L.; Liu, S.Q.; et al. Metaproteomics reveals the molecular mechanism underlying bloom maintenance of a marine dinoflagellate under low ambient CO2 and inorganic nutrients. Sci. Total Environ. 2021, 768, 144515. [Google Scholar] [CrossRef]
- Howarth, R.W. Nutrient limitation of net primary production in marine ecosystems. Ann. Rev. Ecol. 1988, 19, 89–110. [Google Scholar] [CrossRef]
- Heisler, J.; Glibert, P.M.; Burkholder, J.M.; Anderson, D.M.; Cochlan, W.; Dennison, W.C.; Dortch, Q.; Gobler, C.J.; Heil, C.A.; Humphries, E.; et al. Eutrophication and Harmful Algal Blooms: A Scientific Consensus. Harmful Algae 2008, 8, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Shi, X.; Guo, C.; Lin, S. Phosphorus Deficiency Inhibits Cell Division but Not Growth in the Dinoflagellate Amphidinium carterae. Front. Microbiol. 2016, 7, 826. [Google Scholar] [CrossRef] [Green Version]
- Leong, S.C.Y.; Mai, M.; Taguchi, S. Carbon and nitrogen acquisition by the toxic dinoflagellate Alexandrium tamarense in response to different nitrogen sources and supply modes. Harmful Algae 2010, 9, 48–58. [Google Scholar] [CrossRef]
- Allen, M.D.; Kropat, J.; Tottey, S.; Del Campo, J.A.; Merchant, S.S. Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 2007, 143, 263–277. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.; Xu, P.; Hu, H.; Hong, H.; Zheng, T. Effects of Fe and Mn on growth and cell size of Alexandrium tamarense under different culture conditions. Acta Sci. Circumstantiae 2000, 20, 537–541. [Google Scholar] [CrossRef]
- Wang, Z.W.; Ren, J.L.; Zhang, R.F.; Xu, D.F.; Wu, Y. Physical and biological controls of dissolved manganese on the northern slope of the South China Sea. Deep. Sea Res. 2019, 167, 25–33. [Google Scholar] [CrossRef]
- Kahl, C.R.; Means, A.R. Regulation of Cell Cycle Progression by Calcium/Calmodulin-Dependent Pathways. Endocr. Rev. 2003, 24, 719–736. [Google Scholar] [CrossRef] [Green Version]
- Berchtold, M.W.; Villalobo, A. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim. Biophys. Acta 2014, 1843, 398–435. [Google Scholar] [CrossRef] [PubMed]
- Poovaiah, B.W.; Xia, M.; Liu, Z.; Wang, W.; Yang, T.; Sathyanarayanan, P.V.; Franceschi, V.R. Developmental regulation of the gene for chimeric calcium/calmodulin-dependent protein kinase in anthers. Planta 1999, 209, 161–171. [Google Scholar] [CrossRef]
- Rasmussen, C.D.; Means, R.L.; Lu, K.P.; May, G.S.; Means, A.R. Characterization and expression of the unique calmodulin gene of Aspergillus nidulans. J. Biol. Chem. 1990, 265, 13767–13775. [Google Scholar] [CrossRef]
- Lu, K.P.; Rasmussent, C.D.; May, G.S.; Means, A.R. Cooperative regulation of cell proliferation by calcium and calmodulin in Aspergillus nidulans. Mol. Endocrinol. 1992, 6, 365–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abassi, S.; Wang, H.; Ki, J.S. Characterization and Ca2+-induced expression of calmodulin (CaM) in marine dinoflagellates. Eur. J. Protistol. 2021, 77, 125765. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.; Sui, Z.; Bao, Z.; Zhou, W.; Wang, C. Isolation and characterization of calmodulin gene of Alexandrium catenella (Dinoflagellate) and its performance in cell growth and heat stress. J. Ocean Univ. 2013, 13, 290–296. [Google Scholar] [CrossRef]
- Taulés, M.; Rius, E.; Talaya, D.; Lópezgirona, A.; Bachs, O.; Agell, N. Calmodulin is essential for cyclin-dependent kinase 4 (Cdk4) activity and nuclear accumulation of cyclin D1-Cdk4 during G1. J. Biol. Chem. 1998, 273, 33279–33286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, T.; Yamamoto, M. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 1987, 84, 3580–3584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moser, M.J.; Flory, M.R.; Davis, T.N. Calmodulin localizes to the spindle pole body of Schizosaccharomyces pombe and performs an essential function in chromosome segregation. J. Cell Sci. 1997, 110 Pt 15, 1805–1812. [Google Scholar] [CrossRef]
- Ohya, Y.; Anraku, Y. A galactose-dependent cmd1 mutant of Saccharomyces cerevisiae: Involvement of calmodulin in nuclear division. Curr. Genet. 1989, 15, 113–120. [Google Scholar] [CrossRef]
- Davis, T.N. A temperature-sensitive calmodulin mutant loses viability during mitosis. J. Cell Biol. 1992, 118, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Morse, D.; Daoust, P.; Benribague, S. A Transcriptome-based Perspective of Cell Cycle Regulation in Dinoflagellates. Protist 2016, 167, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Bhaud, Y.; Guillebault, D.; Lennon, J.; Defacque, H.; Soyer-Gobillard, M.O.; Moreau, H. Morphology and behaviour of dinoflagellate chromosomes during the cell cycle and mitosis. J. Cell Sci. 2000, 113, 1231–1239. [Google Scholar] [CrossRef]
- Michele, B.; Leighfield, T.A.; Soyer-Gobillard, M.O.; Dolah, F.M.V. Permanent Expression of a Cyclin B Homologue in the Cell Cycle of the Dinoflagellate Karenia brevis. J. Eukaryot. Microbiol. 2003, 50, 123–131. [Google Scholar] [CrossRef]
- Dolah, F.M.V.; Leighfield, T.A.; Sandel, H.D.; Hsu, C.K. Cell division in the dinoflagellate Gambierdiscus toxicus is phased to the diurnal cycle and accompanied by activation of the cell cycle regulatory protein, CDC2 kinase. J. Phycol. 1995, 31, 395–400. [Google Scholar] [CrossRef]
- Nurse, P. Ordering S phase and M phase in the cell cycle. Cell 1994, 79, 547–550. [Google Scholar] [CrossRef]
- Lajeunesse, T.C.; Lambert, G.; Andersen, R.A.; Coffroth, M.A.; Galbraith, D.W. Symbiodinium (Pyrrhophyta) genome sizes (DNA content) ARE smallest among dinoflagellates. J. Phycol. 2005, 41, 880–886. [Google Scholar] [CrossRef]
- Bähler, M.; Rhoads, A. Calmodulin signaling via the IQ motif. FEBS Lett. 2002, 513, 107–113. [Google Scholar] [CrossRef]
- Wang, D.Z.; Zhang, H.; Zhang, Y.; Zhang, S.F. Marine dinoflagellate proteomics: Current status and future perspectives. J. Proteom. 2014, 105, 121–132. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, Z.; Qi, J.; Sui, Z.; Shang, E.; Zhang, S.; Niaz, Z. Comparative Transcriptome Profiling Reveals Insights Into the Mechanisms Related to Explosive Growth of Alexandrium pacificum. Front. Mar. Sci. 2021, 8, e751851. [Google Scholar] [CrossRef]
- Tannu, N.S.; Hemby, S.E. Two-dimensional fluorescence difference gel electrophoresis for comparative proteomics profiling. Nat. Protoc. 2006, 1, 1732–1742. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.J. Calcium signalling and cell proliferation. BioEssays 1995, 17, 491–500. [Google Scholar] [CrossRef]
- Bootman, M.D.; Collins, T.J.; Peppiatt, C.M.; Prothero, L.S.; Mackenzie, L.; Smet, P.D.; Travers, M.; Tovey, S.C.; Seo, J.T.; Berridge, M.J.; et al. Calcium signalling- an overview. Semin. Cell Dev. Biol. 2001, 12, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Hickie, R.A.; Wei, J.W.; Blyth, L.M.; Wong, D.Y.; Klaassen, D.J. Cations and calmodulin in normal and neoplastic cell growth regulation. Can. J. Biochem. Cell Biol. 1983, 61, 934–941. [Google Scholar] [CrossRef]
- Santella, L. The Role of Calcium in the Cell Cycle: Facts and Hypotheses. Biochem. Biophys. Res. Commun. 1998, 244, 317–324. [Google Scholar] [CrossRef]
- Kawsar, H.I.; Ohtani, K.; Okumura, K.; Hayashi, H.; Shimizu, T. Organization and transcriptional regulation of myo-inositol operon in Clostridium perfringens. FEMS Microbiol. Lett. 2004, 235, 289–295. [Google Scholar] [CrossRef]
- Hegeman, C.E.; Good, L.L.; Grabau, E.A. Expression of D-myo-inositol-3-phosphate synthase in soybean. Implications for phytic acid biosynthesis. Plant Physiol. 2001, 125, 1941–1948. [Google Scholar] [CrossRef] [Green Version]
- Donahue, J.L.; Alford, S.R.; Torabinejad, J.; Kerwin, R.E.; Nourbakhsh, A.; Ray, W.K.; Hernick, M.; Huang, X.Y.; Lyons, B.M.; Hein, P.P.; et al. The Arabidopsis thaliana Myo-inositol 1-phosphate synthase1 gene is required for Myo-inositol synthesis and suppression of cell death. Plant Cell 2010, 22, 888–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunes, A.C.; Vianna, G.R.; Cuneo, F.; Amayafarfán, J.; Capdeville, G.; Rech, E.L.; Aragão, F.J. RNAi-mediated silencing of the myo-inositol-1-phosphate synthase gene (GmMIPS1) in transgenic soybean inhibited seed development and reduced phytate content. Planta 2006, 224, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Carroll, A.D.; Moyen, C.; Kestteren, P.V.; Tooke, F.; Battey, N.H.; Brownlee, C. Ca2+, annexins, and GTP modulate exocytosis from maize root cap protoplasts. Plant Cell 1998, 10, 1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laohavisit, A.; Mortimer, J.C.; Demidchik, V.; Coxon, K.M.; Stancombe, M.A.; Macpherson, N.; Brownlee, C.; Hofmann, A.; Webb, A.A.; Miedema, H.; et al. Zea mays annexins modulate cytosolic free Ca2+ and generate a Ca2+-permeable conductance. Plant Cell 2009, 21, 479–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortimer, J.C.; Laohavisit, A.; Macpherson, N.; Webb, A.; Brownlee, C.; Battey, N.H.; Davies, J.M. Annexins: Multifunctional components of growth and adaptation. J. Exp. Bot. 2008, 59, 533–544. [Google Scholar] [CrossRef] [Green Version]
- Monastyrskaya, K.; Babiychuk, E.B.; Draeger, A. The annexins: Spatial and temporal coordination of signaling events during cellular stress. Cell. Mol. Life Sci. 2009, 66, 2623–2642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, P.J.; Bowen, H.C.; Demidchik, V.; Nichols, C.; Davies, J.M. Genes for calcium-permeable channels in the plasma membrane of plant root cells. Biochim. Biophys. Acta 2002, 1564, 299–309. [Google Scholar] [CrossRef] [Green Version]
- Roux, S.J. Annexins of Plant Cells. Plant Physiol. 1995, 109, 1133–1139. [Google Scholar] [CrossRef]
- Clark, G.B.; Sessions, A.; Eastburn, D.J.; Roux, S.J. Differential expression of members of the annexin multigene family in Arabidopsis. Plant Physiol. 2001, 126, 1072–1084. [Google Scholar] [CrossRef] [Green Version]
- Hartwell, L.H.; Kastan, M.B. Cell Cycle Control and Cancer. Science 1994, 266, 1821–1828. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, C. DNA replication and cell cycle in plants: Learning from geminiviruses. EMBO J. 2000, 19, 792–799. [Google Scholar] [CrossRef] [Green Version]
- Takuwa, N.; Zhou, W.; Takuwa, Y. Calcium, calmodulin and cell cycle progression. Cell. Signal. 1995, 7, 93–104. [Google Scholar] [CrossRef]
- Fujisawa, H. Regulation of the activities of multifunctional Ca2+/calmodulin-dependent protein kinases. J. Biochem. 2001, 129, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Planas-Silva, M.D.; Means, A.R. Expression of a constitutive form of calcium/calmodulin dependent protein kinase II leads to arrest of the cell cycle in G2. EMBO J. 1992, 11, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Minami, H.; Inoue, S.; Hidaka, H. The Effect of KN-62, Ca2+/Calmodulin Dependent Protein Kinase II Inhibitor on Cell Cycle. Biochem. Biophys. Res. Commun. 1994, 199, 241–248. [Google Scholar] [CrossRef]
- Hudmon, A.; Schulman, H. Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem. J. 2002, 364, 593–611. [Google Scholar] [CrossRef]
- Patel, R.; Holt, M.; Philipova, R.; Moss, S.; Schulman, H.; Hidaka, H.; Whitaker, M. Calcium/calmodulin-dependent phosphorylation and activation of human Cdc25-C at the G2/M phase transition in HeLa cells. J. Biol. Chem. 1999, 274, 7958–7968. [Google Scholar] [CrossRef] [Green Version]
- Dai, G.; Qian, Y.; Chen, J.; Meng, F.L.; Pan, F.Y.; Shen, W.G.; Zhang, S.Z.; Xue, B.; Li, C.J. Calmodulin activation of polo-like kinase 1 is required during mitotic entry. Biochem. Cell Biol. 2013, 91, 287–294. [Google Scholar] [CrossRef]
- Watillon, B.; Kettmann, R.; Boxus, P.; Burny, A. A Calcium/Calmodulin-Binding Serine/Threonine Protein Kinase Homologous to the Mammalian Type II Calcium/Calmodulin-Dependent Protein Kinase Is Expressed in Plant Cells. Plant Physiol. 1993, 101, 1381–1384. [Google Scholar] [CrossRef] [Green Version]
- Patil, S.; Takezawa, D.; Poovaiah, B.W. Chimeric plant calcium/calmodulin-dependent protein kinase gene with a neural visinin-like calcium-binding domain. Proc. Natl. Acad. Sci. USA 1995, 92, 4897–4901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Liu, B.F.; Liang, S.P.; Jones, R.L.; Lu, Y.T. Molecular and biochemical characterization of a calcium/calmodulin-binding protein kinase from rice. Biochem. J. 2002, 368, 145–157. [Google Scholar] [CrossRef]
- Wang, L.; Liang, S.P.; Lu, Y.T. Characterization, physical location and expression of the genes encoding calcium/calmodulin-dependent protein kinases in maize (Zea mays L.). Planta 2001, 213, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, C.K.; Göhlke, U.; Sobott, F.; Good, V.M.; Ali, M.M.U.; Prodromou, C.; Robinson, C.V.; Saibil, H.R.; Pearl, L.H. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006, 23, 697–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polier, S.; Samant, R.S.; Clarke, P.A.; Workman, P.; Prodromou, C.; Pearl, L.H. ATP-competitive inhibitors block protein kinase recruitment to the Hsp90-Cdc37 system. Nat. Chem. Biol. 2013, 9, 307–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S. Genomic understanding of dinoflagellates. Res. Microbiol. 2011, 162, 551–569. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Sui, Z.; Zhang, S.; Du, Q.; Ren, Y.; Liu, Y.; Kong, F.; Zhong, J.; Ma, Q. Identification of microRNAs in the Toxigenic Dinoflagellate Alexandrium catenella by High-Throughput Illumina Sequencing and Bioinformatic Analysis. PLoS ONE 2015, 10, e0138709. [Google Scholar] [CrossRef]
- Galleron, C. Synchronization of the marine dinoflagellate Amphydinium carteri in dense cultures. J. Phycol. 1976, 12, 69–73. [Google Scholar] [CrossRef]
- Machida, K.; Mayer, B.J. Detection of protein-protein interactions by far-western blotting. Methods Mol. Biol. 2009, 536, 313–329. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lupke, M.; Frahm, J.; Lantow, M.; Maercker, C.; Remondini, D.; Bersani, F.; Simkó, M. Gene expression analysis of ELF-MF exposed human monocytes indicating the involvement of the alternative activation pathway. Biochim. Biophys. Acta 2006, 1763, 402–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]

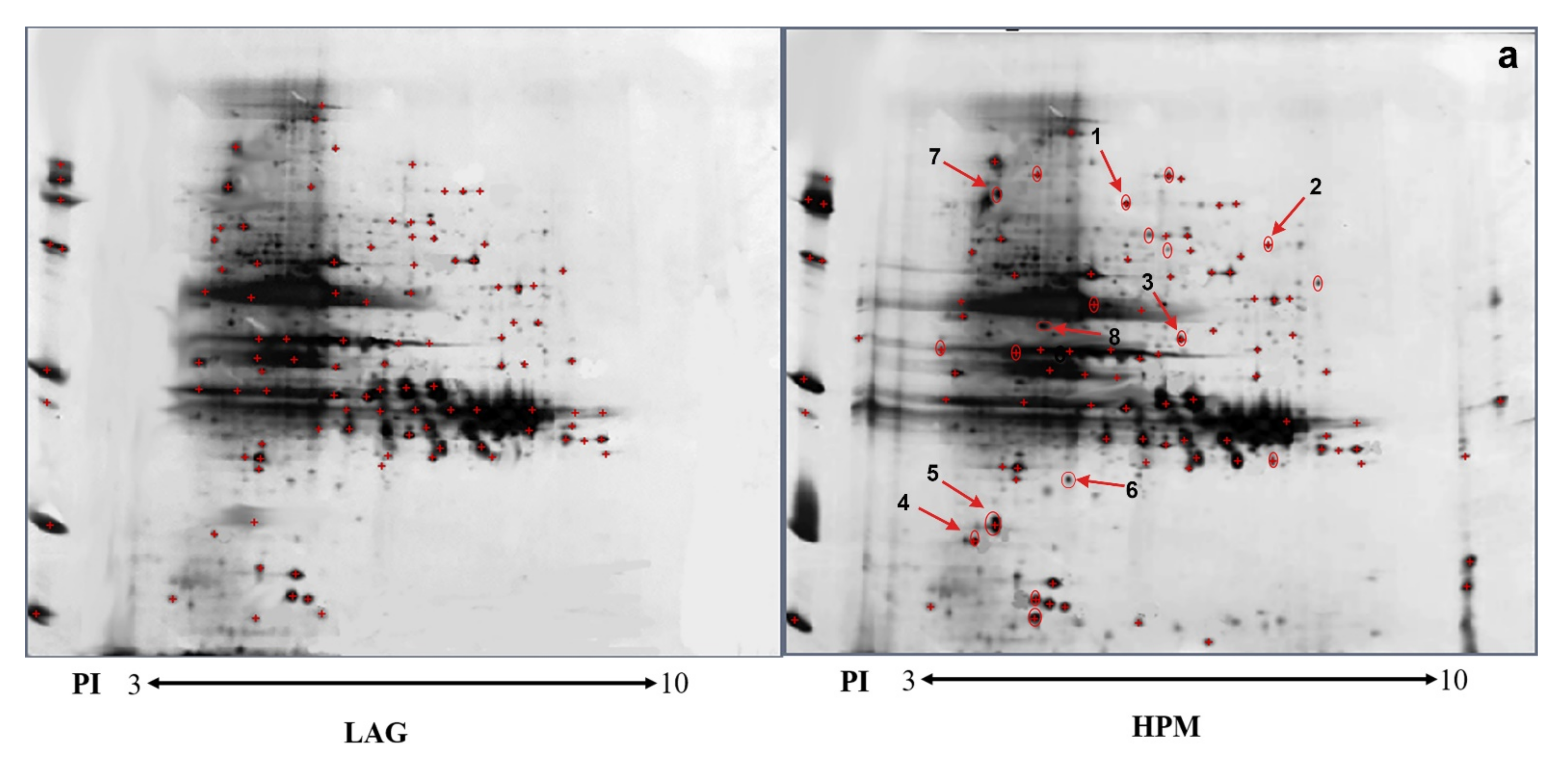


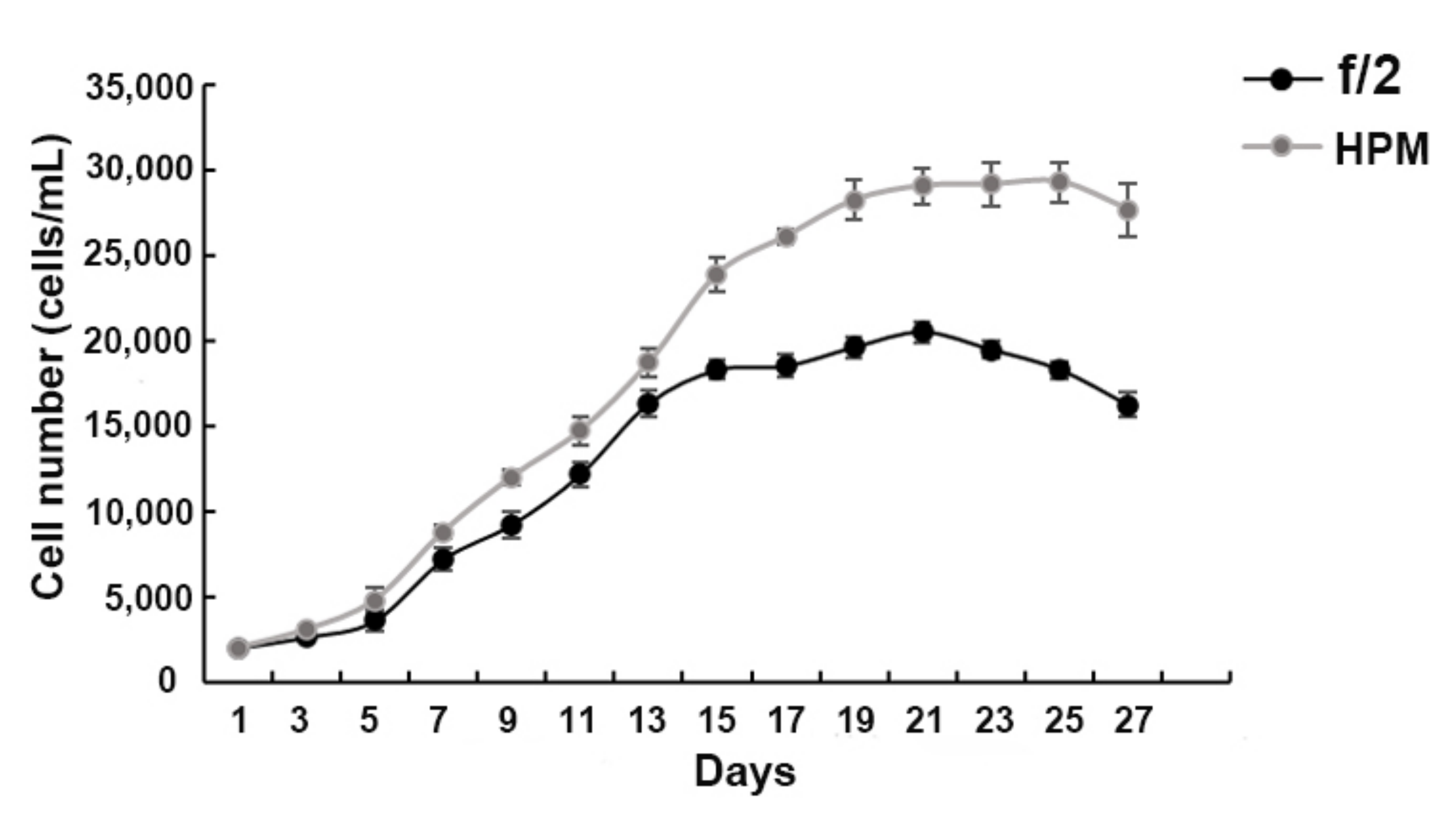
| Spot ID | Accession Number | Protein Score | Protein Score CI% | Peptide Count | MW/pI | Protein Description | HPM vs. LAG | |
|---|---|---|---|---|---|---|---|---|
| Fold Change | p-Value | |||||||
| 1 | GL433858.1 | 145 | 100 | 3 | 74.39/5.81 | Calcium/calmodulin-dependent protein kinase 2B | ||
| 3 | CAA48538.1 | 121 | 99,248 | 4 | 47.30/6.79 | Serine/threonine kinase | ||
| 6 | NM001003039.1 | 116 | 99,997 | 5 | 80.08/4.12 | Annexin A4 | 2.5 | 0.003 |
| 8 | XM001703196.1 | 150 | 100 | 3 | 80.52/5.54 | Inositol-3-phosphate synthase | 2.2 | 0.007 |
| Name | Sequence 5′→3′ | Application | Size (bp) |
|---|---|---|---|
| cam-f | CGGGATCCATGGCTGACCAGCTCACG | Complete cDNA sequence of CaM cloning | 450 |
| cam-r | CACGCGTCGACTCACTTTGCCATCATCATCTTCAC | ||
| CAMK2B-f | AAAATCTGTGATCCACACTTGACT | Ca2+/calmodulin-dependent protein kinase (CAMK2B) cloning and qPCR | 114 |
| CAMK2B-r | CTTACAGTTCTTCCCCAGGACATT | ||
| STK-f | GCACTTGCCGACACGCTTACAT | Serine/threonine-protein kinase (STK) cloning and qPCR | 170 |
| STK-r | GTGAGCCGACTGGGTTTTCCTT | ||
| MIPS3-f | CAACCACTTGGGGAACAATGATGG | Myo-inositol-3-phosphate synthase(MIPS3) cloning and qPCR | 138 |
| MIPS3-r | AATAGTGTTCGTTCCCCCCATAAC | ||
| ANNEXIN-f | CCTCTCTTGGTCGCTTTACTTGTC | ANNEXIN cloning and qPCR | 182 |
| ANNEXIN-r | TGACAACTTGACGCTTTGGACC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zhu, Z.; Sui, Z.; Liu, H.; Riaz, S. Calmodulin and Its Interactive Proteins Participate in Regulating the Explosive Growth of Alexandrium pacificum (Dinoflagellate). Int. J. Mol. Sci. 2022, 23, 145. https://doi.org/10.3390/ijms23010145
Liu Y, Zhu Z, Sui Z, Liu H, Riaz S. Calmodulin and Its Interactive Proteins Participate in Regulating the Explosive Growth of Alexandrium pacificum (Dinoflagellate). International Journal of Molecular Sciences. 2022; 23(1):145. https://doi.org/10.3390/ijms23010145
Chicago/Turabian StyleLiu, Yuan, Zhimei Zhu, Zhenghong Sui, Haoxin Liu, and Sadaf Riaz. 2022. "Calmodulin and Its Interactive Proteins Participate in Regulating the Explosive Growth of Alexandrium pacificum (Dinoflagellate)" International Journal of Molecular Sciences 23, no. 1: 145. https://doi.org/10.3390/ijms23010145






