Enzyme Inhibitors: The Best Strategy to Tackle Superbug NDM-1 and Its Variants
Abstract
1. Introduction
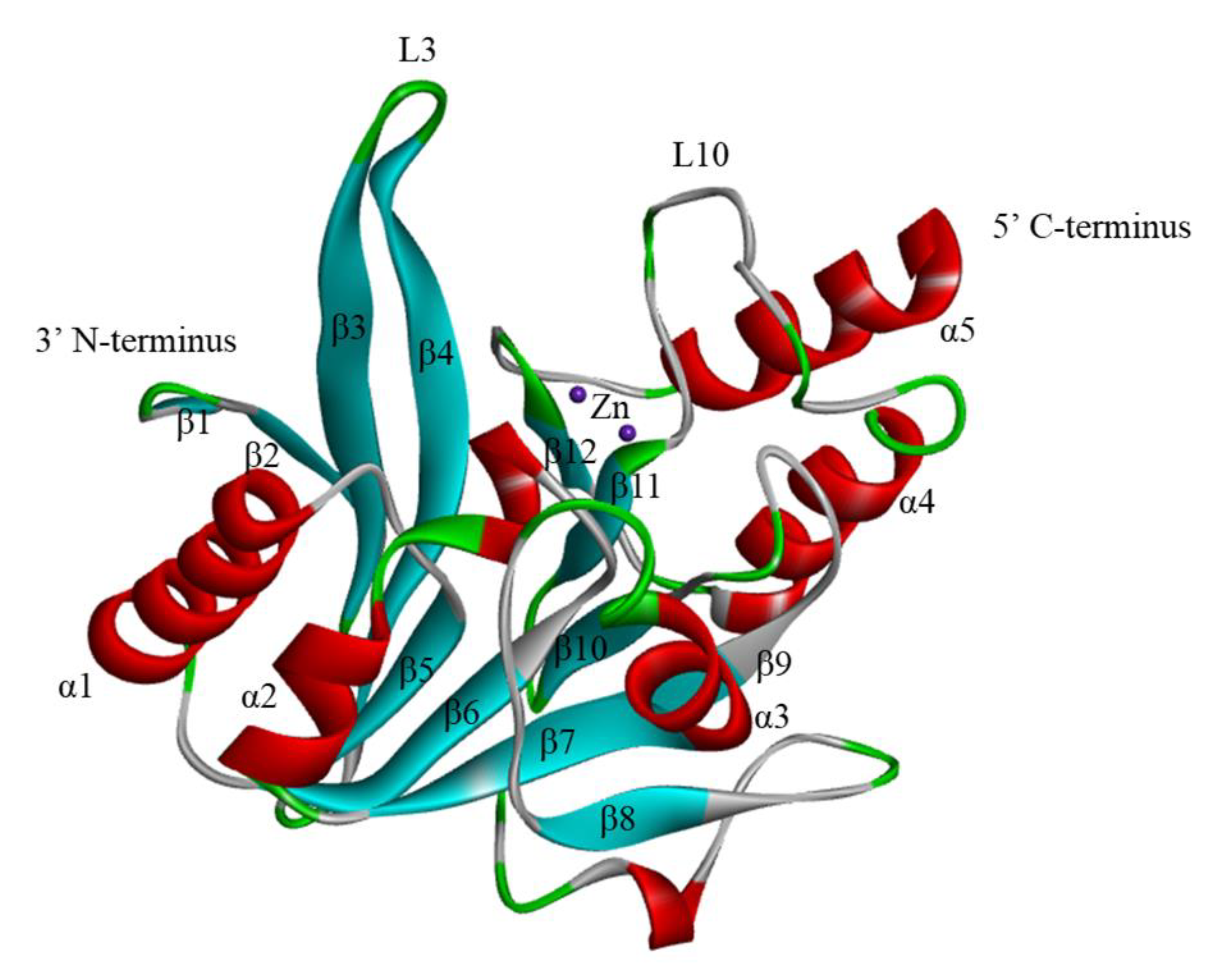
2. The Structure of NDM-1
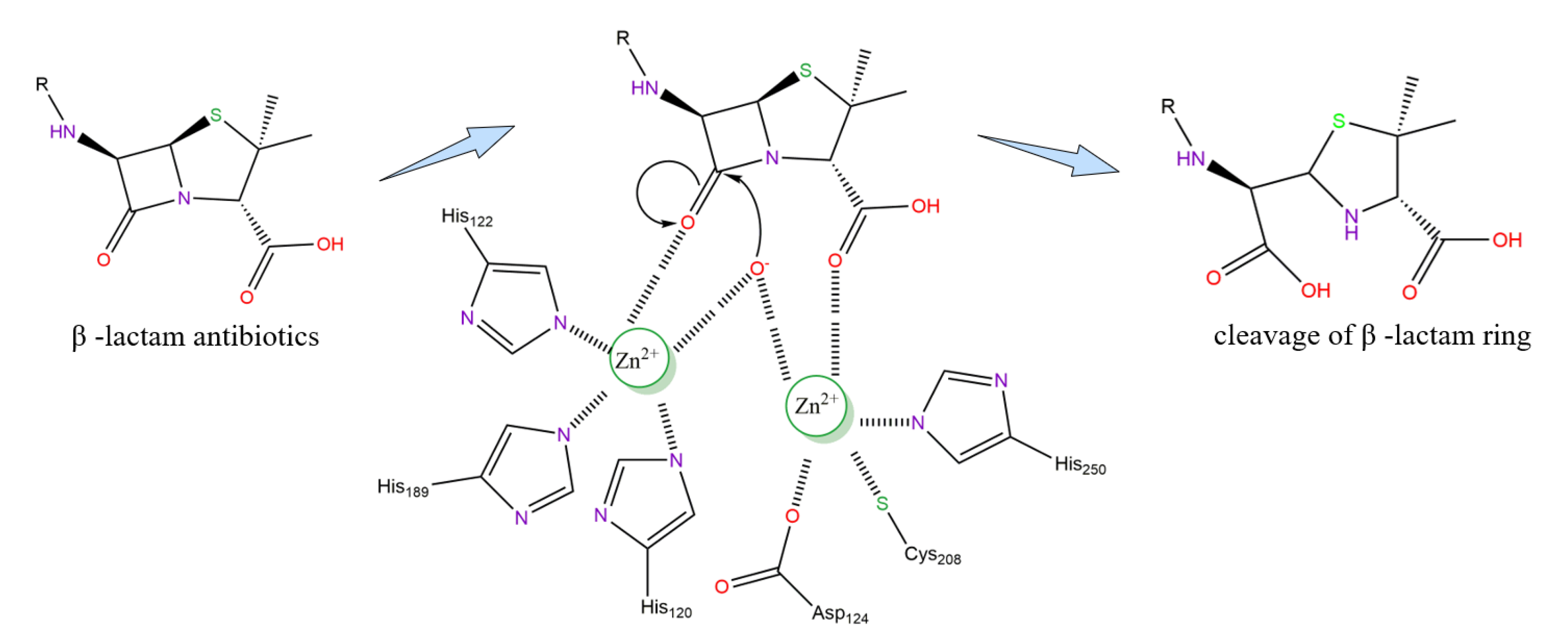
3. Active Site and Hydrolysis Mechanism of NDM-1
4. NDM-1 Variants and Global Distribution
5. NDM-1 Inhibitors: Discovery and Advances
5.1. Noncovalently-Bound Inhibitors
5.1.1. Zinc-Binding Inhibitors
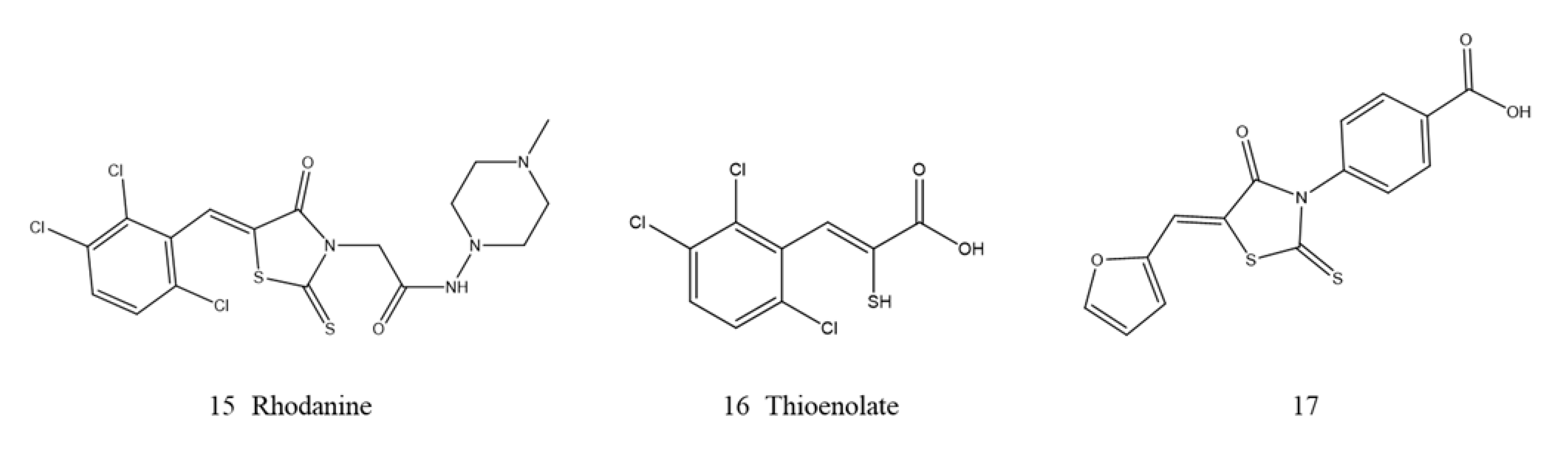
Thiol-Based Derivatives
Rhodanine
Thiadiazoles
Isatin-β-thiosemicarbazones (IBT)
Pyridine Dicarboxylic Acid Derivatives
Triazole-Thione Derivatives
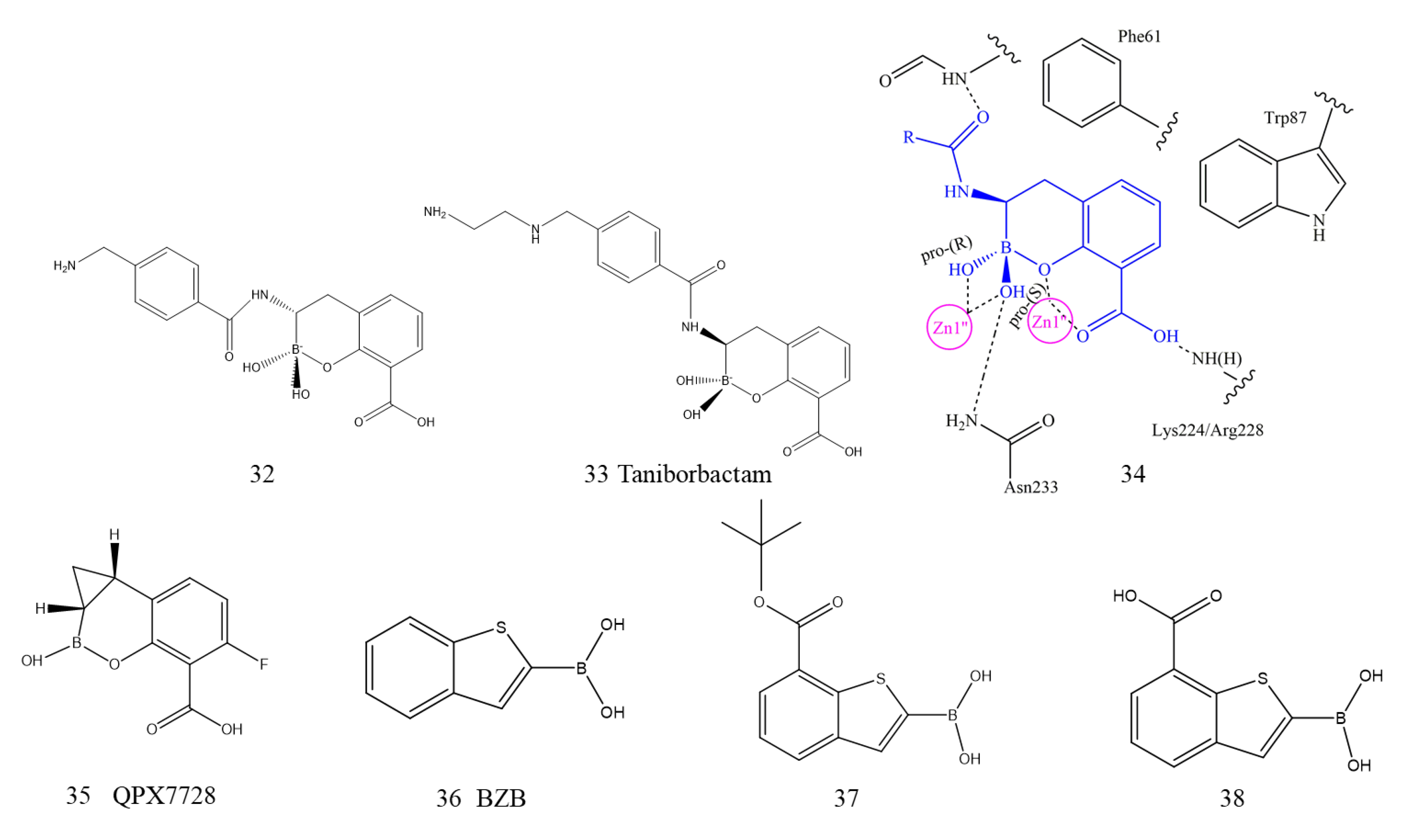
5.1.2. Boronic Acid Derivatives
5.1.3. Metal Chelating Inhibitors
5.2. Covalently Bound Inhibitors
5.3. Inhibitors with Other Mechanisms
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- De Graaf, M.; van Beek, J.; Koopmans, M.P. Human norovirus transmission and evolution in a changing world. Nat. Rev. Microbiol. 2016, 14, 421–433. [Google Scholar] [CrossRef]
- Hu, X.; Collier, M.G.; Xu, F. Hepatitis a outbreaks in developed countries: Detection, control, and prevention. Foodborne Pathog. Dis. 2020, 17, 166–171. [Google Scholar] [CrossRef]
- Richter, M.F.; Hergenrother, P.J. The challenge of converting Gram-positive-only compounds into broad-spectrum antibiotics. Ann. N. Y. Acad. Sci. 2019, 1435, 18–38. [Google Scholar] [CrossRef] [PubMed]
- Cornejo-juárez, P.; Vilar-compte, D.; Pérez-jiménez, C.; Ñamendys-Silva, S.A.; Sandoval-Hernández, S.; Volkow-Fernández, P. The impact of hospital-acquired infections with multidrug-resistant bacteria in an oncology intensive care unit. Int. J. Infect. Dis. 2015, 31, 31–34. [Google Scholar] [CrossRef]
- Karaiskos, I.; Lagou, S.; Pontikis, K.; Rapti, V.; Poulakou, G. The “Old” and the “New” Antibiotics for MDR Gram-Negative Pathogens: For Whom, When, and How. Front. Public Health 2019, 7, 151–176. [Google Scholar] [CrossRef]
- De Kraker, M.E.; Stewardson, A.J.; Harbarth, S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase inhibitors: An overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef] [PubMed]
- King, D.T.; Sobhanifar, S.; Strynadka, N. The Mechanisms of Resistance to β-Lactam Antibiotics; Springer: New York, NY, USA, 2014. [Google Scholar]
- Essack, S.Y. The development of beta-lactam antibiotics in response to the evolution of beta-lactamases. Pharm. Res. 2001, 18, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. Past and present perspectives on β-lactamases. Antimicrob. Agents Chemother. 2018, 62, e01076-18. [Google Scholar] [CrossRef] [PubMed]
- Ambler, R.P. The structure of beta-lactamases. Philos. Trans. R. Soc. B. 1980, 289, 321–331. [Google Scholar]
- Al-sehlawi, Z.; Almohana, A.; Al-thahab, A. Ampc β-Lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar]
- Tondi, D.; Cross, S.; Venturelli, A.; Costi, M.P.; Cruciani, G.; Spyrakis, F. Decoding the structural basis for carbapenem hydrolysis by class a β-lactamases: Fishing for a pharmacophore. Curr. Drug Targets 2016, 17, 983–1005. [Google Scholar] [CrossRef]
- Docquier, J.D.; Mangani, S. Structure-function relationships of class d carbapenemases. Curr. Drug Targets 2016, 17, 1061–1071. [Google Scholar] [CrossRef]
- Shimada, A.; Ishikawa, H.; Nakagawa, N.; Kuramitsu, S.; Masui, R. The first crystal structure of an archaeal metallo-beta-lactamase superfamily protein; ST1585 from Sulfolobus tokodaii. Proteins 2010, 78, 2399–2402. [Google Scholar] [CrossRef] [PubMed]
- Bebrone, C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007, 74, 1686–1701. [Google Scholar] [CrossRef] [PubMed]
- Palzkill, T. Metallo-β-lactamase structure and function. Ann. N. Y. Acad. Sci. 2013, 1277, 91–104. [Google Scholar] [CrossRef]
- Garau, G.; García-Sáez, I.; Bebrone, C.; Anne, C.; Mercuri, P.; Galleni, M.; Frère, J.M.; Dideberg, O. Update of the standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 2004, 48, 2347–2349. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.R.; Weeks, J.; Livermore, D.M.; Toleman, M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect. Dis. 2011, 11, 355–362. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Rolain, J.M.; Parola, P.; Cornaglia, G. New Delhi metallo-beta-lactamase (NDM-1): Towards a new pandemia? Clin. Microbiol. Infect. 2010, 16, 1699–1701. [Google Scholar] [CrossRef] [PubMed]
- Cornaglia, G.; Giamarellou, H.; Rossolini, G.M. Metallo-β-lactamases: A last frontier for β-lactams? Lancet Infect. Dis. 2011, 11, 381–393. [Google Scholar] [CrossRef]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef]
- Moellering, R.C. NDM-1--a cause for worldwide concern. N. Engl. J. Med. 2010, 363, 2377–2379. [Google Scholar] [CrossRef] [PubMed]
- Linciano, P.; Cendron, L.; Gianquinto, E.; Spyrakis, F.; Tondi, D. Ten years with new delhi metallo-β-lactamase-1 (ndm-1): From structural insights to inhibitor design. ACS Infect. Dis. 2019, 5, 9–34. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Poirel, L.; Nordmann, P. Plasmid-mediated transfer of the blaNDM-1 gene in Gram-negative rods. Fems. Microbiol. Lett. 2011, 324, 111–116. [Google Scholar] [CrossRef]
- Zubair, M. Microbiology of diabetic foot ulcer with special reference to ESBL infections. Am. J. Clin. Exp. Med. 2015, 3, 6–23. [Google Scholar] [CrossRef][Green Version]
- Lubick, N. Antibiotic resistance shows up in India’s drinking water. Nature 2011, 218. [Google Scholar] [CrossRef]
- Pan, C.Y.; Chen, J.C.; Chen, T.L.; Wu, J.L.; Hui, C.F.; Chen, J.Y. Piscidin is highly active against carbapenem-resistant Acinetobacter baumannii and NDM-1-producing Klebsiella pneumonia in a systemic Septicaemia infection mouse model. Mar. Drugs 2015, 13, 2287–2305. [Google Scholar] [CrossRef]
- Albur, M.; Noel, A.; Bowker, K.; MacGowan, A. Bactericidal activity of multiple combinations of tigecycline and colistin against NDM-1-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2012, 56, 3441–3443. [Google Scholar] [CrossRef] [PubMed]
- Rimrang, B.; Chanawong, A.; Lulitanond, A.; Wilailuckana, C.; Charoensri, N.; Sribenjalux, P.; Phumsrikaew, W.; Wonglakorn, L.; Chetchotisakd, P. New Delhi metallo-β-lactamase-1 (NDM-1)-producing enterobacteriaceae: First report in Thailand. Int. J. Infect. Dis. 2012, 16, e431. [Google Scholar] [CrossRef][Green Version]
- Charan, J.; Mulla, S.; Ryavanki, S.; Kantharia, N. New Delhi Metallo-beta lactamase-1 containing enterobacteriaceae: Origin, diagnosis, treatment and public health concern. Pan. Afr. Med. J. 2012, 11, 22. [Google Scholar] [PubMed]
- Zhang, H.; Hao, Q. Crystal structure of NDM-1 reveals a common β-lactam hydrolysis mechanism. FASEB J. 2011, 25, 2574–2582. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, J.; Niu, G.; Shui, W.; Sun, Y.; Zhou, H.; Zhang, Y.; Yang, C.; Lou, Z.; Rao, Z. A structural view of the antibiotic degradation enzyme NDM-1 from a superbug. Protein Cell 2011, 2, 384–394. [Google Scholar] [CrossRef]
- Kim, Y.; Tesar, C.; Mire, J.; Jedrzejczak, R.; Binkowski, A.; Babnigg, G.; Sacchettini, J.; Joachimiak, A. Structure of apo- and monometalated forms of NDM-1—A highly potent carbapenem-hydrolyzing metallo-β-lactamase. PLoS ONE 2011, 6, e24621. [Google Scholar] [CrossRef] [PubMed]
- Chiou, J.; Leung, Y.C.; Chen, S. Molecular mechanisms of substrate recognition and specificity of New Delhi metallo-ß-lactamase. Antimicrob. Agents Chemother. 2014, 58, 5372–5378. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.W.; Zheng, M.; Wu, S.; Guo, H.; Liu, D.; Xu, D.; Fast, W. Characterization of purified New Delhi metallo-β-lactamase-1. Biochemistry 2011, 50, 10102–10113. [Google Scholar] [CrossRef]
- King, D.; Strynadka, N. Crystal structure of New Delhi metallo-β-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 2011, 20, 1484–1491. [Google Scholar] [CrossRef]
- Rose, P.W.; Bi, C.; Bluhm, W.F.; Christie, C.H.; Dimitropoulos, D.; Dutta, S.; Green, R.K.; Goodsell, D.S.; Prlic, A.; Quesada, M.; et al. The RCSB Protein Data Bank: New resources for research and education. Nucleic Acids Res. 2013, 41, D475–D482. [Google Scholar] [CrossRef]
- King, D.T.; Worrall, L.J.; Gruninger, R.; Strynadka, N.C. New Delhi metallo-β-lactamase: Structural insights into β-lactam recognition and inhibitio. J. Am. Chem. Soc. 2012, 134, 11362–11365. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Xu, D. New Delhi metallo-β-lactamase I: Substrate binding and catalytic mechanism. J. Phys. Chem. B. 2013, 117, 11596–11607. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Ding, J.; Zhu, D.; Liu, X.; Xu, X.; Zhang, Y.; Zang, S.; Wang, D.C.; Liu, W. Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins. J. Am. Chem. Soc. 2014, 136, 14694–14697. [Google Scholar] [CrossRef]
- González, M.M.; Kosmopoulou, M.; Mojica, M.F.; Castillo, V.; Hinchliffe, P.; Pettinati, I.; Brem, J.; Schofield, C.J.; Mahler, G.; Bonomo, R.A.; et al. Bisthiazolidines: A Substrate-Mimicking Scaffold as an Inhibitor of the NDM-1 Carbapenemase. ACS Infect. Dis. 2015, 1, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. The ABCD’s of β-lactamase nomenclature. J. Infect. Chemother. 2013, 19, 549–559. [Google Scholar] [CrossRef]
- Frère, J.M.; Galleni, M.; Bush, K.; Dideberg, O. Is it necessary to change the classification of {beta}-lactamases? J. Antimicrob. Chemother. 2005, 55, 1051–1053. [Google Scholar] [CrossRef]
- Green, V.L.; Verma, A.; Owens, R.J.; Phillips, S.E.; Carr, S.B. Structure of New Delhi metallo-β-lactamase 1 (NDM-1). Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 1160–1164. [Google Scholar] [CrossRef]
- Tripathi, R.; Nair, N.N. Mechanism of Meropenem Hydrolysis by New Delhi Metallo β-Lactamase. Acs Catal. 2015, 5, 2577–2586. [Google Scholar] [CrossRef]
- Kim, Y.; Cunningham, M.A.; Mire, J.; Tesar, C.; Sacchettini, J.; Joachimiak, A. NDM-1, the ultimate promiscuous enzyme: Substrate recognition and catalytic mechanism. FASEB J. 2013, 27, 1917–1927. [Google Scholar] [CrossRef]
- Zhu, K.; Lu, J.; Liang, Z.; Kong, X.; Ye, F.; Jin, L.; Geng, H.; Chen, Y.; Zheng, M.; Jiang, H.; et al. A quantum mechanics/molecular mechanics study on the hydrolysis mechanism of New Delhi metallo-β-lactamase-1. J. Comput. Aided Mol. Des. 2013, 27, 247–256. [Google Scholar] [CrossRef]
- Doi, Y.; Hazen, T.H.; Boitano, M.; Tsai, Y.C.; Clark, T.A.; Korlach, J.; Rasko, D.A. Whole-genome assembly of Klebsiella pneumoniae coproducing NDM-1 and OXA-232 carbapenemases using single-molecule, real-time sequencing. Antimicrob. Agents Chemother. 2014, 58, 5947–5953. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Aitha, M.; Hetrick, A.M.; Richmond, T.K.; Tierney, D.L.; Crowder, M.W. Mechanistic and Spectroscopic Studies of Metallo-β-lactamase NDM-1. Biochemistry 2012, 51, 3839–3847. [Google Scholar] [CrossRef]
- Jacquin, O.; Balbeur, D.; Damblon, C.; Marchot, P.; De Pauw, E.; Roberts, G.C.; Frère, J.M.; Matagne, A. Positively cooperative binding of zinc ions to Bacillus cereus 569/H/9 beta-lactamase II suggests that the binuclear enzyme is the only relevant form for catalysis. J. Mol. Biol. 2009, 392, 1278–1291. [Google Scholar] [CrossRef]
- Page, M.I.; Badarau, A. The mechanisms of catalysis by metallo beta-lactamases. Bioinorg. Chem. Appl. 2008, 2008, 576297. [Google Scholar] [CrossRef]
- Yang, H.; Aitha, M.; Marts, A.R.; Hetrick, A.; Bennett, B.; Crowder, M.W.; Tierney, D.L. Spectroscopic and mechanistic studies of heterodimetallic forms of metallo-β-lactamase NDM-1. J. Am. Chem. Soc. 2014, 136, 7273–7285. [Google Scholar] [CrossRef]
- Pal, A.; Tripathi, A. An in silico approach for understanding the molecular evolution of clinically important metallo-beta-lactamases. Infect. Genet. Evol. 2013, 20, 39–47. [Google Scholar] [CrossRef]
- Shi, M.; Xu, D.; Zeng, J. GPU accelerated quantum virtual screening: Application for the natural inhibitors of New Dehli metallo protein (NDM-1). Front. Chem. 2018, 6, 564–572. [Google Scholar] [CrossRef]
- Krauss, M.; Gresh, N.; Antony, J. Binding and hydrolysis of ampicillin in the active site of a zinc lactamase. J. Phys. Chem. B 2003, 107, 1215–1229. [Google Scholar] [CrossRef]
- Stewart, A.C.; Bethel, C.R.; VanPelt, J.; Bergstrom, A.; Cheng, Z.; Miller, C.G.; Williams, C.; Poth, R.; Morris, M.; Lahey, O. Clinical variants of New Delhi Metallo-β-Lactamase are evolving to overcome zinc scarcity. ACS Infect. Dis. 2017, 3, 927–940. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Thomas, P.W.; Ju, L.; Bergstrom, A.; Mason, K.; Clayton, D.; Miller, C.; Bethel, C.R.; VanPelt, J.; Tierney, D.L.; et al. Evolution of New Delhi metallo-β-lactamase (NDM) in the clinic: Effects of NDM mutations on stability, zinc affinity, and mono-zinc activity. J. Biol. Chem. 2018, 293, 12606–12618. [Google Scholar] [CrossRef]
- Mitra, S.D.; Sebastian, S.C.; Rekha, I.; Mudigonda, A.; Shome, B.R. Molecular detection of the new delhi metallo-β-lactamase clinical variant with double mutation-V88L and M154L in Escherichia coli isolates from South India. Gene Rep. 2020, 21, 100880. [Google Scholar] [CrossRef]
- Raczynska, J.E.; Imiolczyk, B.; Komorowska, M.; Sliwiak, J.; Czyrko-Horczak, J.; Brzezinski, K.; Jaskolski, M. Flexible loops of New Delhi metallo-β-lactamase modulate its activity towards different substrates. Int. J. Biol. Macromol. 2020, 158, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM Metallo-β-Lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef] [PubMed]
- Makena, A.; Brem, J.; Pfeffer, I.; Geffen, R.E.; Wilkins, S.E.; Tarhonskaya, H.; Flashman, E.; Phee, L.M.; Wareham, D.W.; Schofield, C.J. Biochemical characterization of New Delhi metallo-β-lactamase variants reveals differences in protein stability. J. Antimicrob. Chemother. 2015, 70, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, H.; Zhu, T.; Zhou, D.; Zhang, F.; Lao, X.; Zheng, H. Asp120Asn mutation impairs the catalytic activity of NDM-1 metallo-β-lactamase: Experimental and computational study. Phys. Chem. Chem. Phys. 2014, 16, 6709–6716. [Google Scholar] [CrossRef]
- Ali, A.; Gupta, D.; Srivastava, G.; Sharma, A.; Khan, A.U. Molecular and computational approaches to understand resistance of New Delhi metallo β-lactamase variants (NDM-1, NDM-4, NDM-5, NDM-6, NDM-7)-producing strains against carbapenems. J. Biomol. Struct. Dyn. 2019, 37, 2061–2071. [Google Scholar] [CrossRef]
- Ali, A.; Kumar, R.; Iquebal, M.A.; Jaiswal, S.; Kumar, D.; Khan, A.U. The role of conserved residues in the catalytic activity of NDM-1: An approach involving site directed mutagenesis and molecular dynamics. Phys. Chem. Chem. Phys. 2019, 21, 17821–17835. [Google Scholar] [CrossRef]
- Bahr, G.; Vitor-Horen, L.; Bethel, C.R.; Bonomo, R.A.; González, L.J.; Vila, A.J. Clinical evolution of new delhi metallo-β-lactamase (ndm) optimizes resistance under zn(ii) deprivation. Antimicrob. Agents Chemother. 2017, 62, e01849-17. [Google Scholar] [CrossRef]
- Patel, M.P.; Fryszczyn, B.G.; Palzkill, T. Characterization of the global stabilizing substitution A77V and its role in the evolution of CTX-M β-lactamases. Antimicrob. Agents Chemother. 2015, 59, 6741–6748. [Google Scholar] [CrossRef]
- Tewari, R.; Mitra, S.D.; Ganaie, F.; Venugopal, N.; Das, S.; Shome, B.R. Prevalence of extended spectrum β-lactamase, AmpC β-lactamase and metallo β-lactamase mediated resistance in Escherichia coli from diagnostic and tertiary healthcare centers in south Bangalore, India. INT J. Med. Res. Health 2018, 6, 1308–1313. [Google Scholar] [CrossRef][Green Version]
- Ganta, S.R.; Perumal, S.; Pagadala, S.R.; Samuelsen, O.; Spencer, J.; Pratt, R.F.; Buynak, J.D. Approaches to the simultaneous inactivation of metallo- and serine-beta-lactamases. Bioorg. Med. Chem. Lett. 2009, 19, 1618–1622. [Google Scholar] [CrossRef] [PubMed]
- Groundwater, P.W.; Xu, S.; Lai, F.; Váradi, L.; Tan, J.; Perry, J.D.; Hibbs, D.E. New Delhi metallo-β-lactamase-1: Structure, inhibitors and detection of producers. Future Med. Chem. 2016, 8, 993–1012. [Google Scholar] [CrossRef]
- Khan, A.U.; Rehman, M.T. Role of Non-Active-Site Residue Trp-93 in the Function and Stability of New Delhi Metallo-β-Lactamase 1. Antimicrob. Agents Chemother. 2015, 60, 356–360. [Google Scholar] [CrossRef]
- Aitha, M.; Moller, A.J.; Sahu, I.D.; Horitani, M.; Tierney, D.L.; Crowder, M.W. Investigating the position of the hairpin loop in New Delhi metallo-β-lactamase, NDM-1, during catalysis and inhibitor binding. J. Inorg. Biochem. 2016, 156, 35–39. [Google Scholar] [CrossRef]
- Khan, S.; Ali, A.; Khan, A.U. Structural and functional insight of New Delhi Metallo β-lactamase-1 variants. Future Med. Chem. 2018, 10, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Feng, Y.; McNally, A.; Zong, Z. blaNDM-21, a new variant of blaNDM in an Escherichia coli clinical isolate carrying blaCTX-M-55 and rmtB. J. Antimicrob. Chemother. 2018, 73, 2336–2339. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Y.; Walsh, T.R.; Liu, D.; Shen, Z.; Zhang, R.; Yin, W.; Yao, H.; Li, J.; Shen, J. Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob. Agents Chemother. 2018, 61, e02233-16. [Google Scholar]
- Hornsey, M.; Phee, L.; Wareham, D.W. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 2011, 55, 5952–5954. [Google Scholar] [CrossRef]
- Li, X.; Mu, X.; Zhang, P.; Zhao, D.; Ji, J.; Quan, J.; Zhu, Y.; Yu, Y. Detection and characterization of a clinical Escherichia coli ST3204 strain coproducing NDM-16 and MCR-1. Infect. Drug Resist. 2018, 11, 1189–1195. [Google Scholar] [CrossRef]
- Liu, Z.; Piccirilli, A.; Liu, D.; Wang, Y.; Shen, J. Deciphering the Role of V88l Substitution in NDM-24 metallo-β-lactamase. Catalysts 2019, 9, 744. [Google Scholar] [CrossRef]
- Liu, Z.; Li, J.; Wang, X.; Liu, D.; Ke, Y.; Wang, Y.; Shen, J. Novel variant of New Delhi Metallo-β-lactamase, NDM-20, in Escherichia coli. Front. Microbiol. 2018, 9, 248. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Huang, Y.; Zhao, X.; Liu, W.; Dong, D.; Li, H.; Wang, X.; Huang, S.; Wei, X.; Yan, X.; et al. A novel New Delhi Metallo-β-lactamase variant, NDM-14, isolated in a Chinese Hospital possesses increased enzymatic activity against carbapenems. Antimicrob. Agents Chemother. 2015, 59, 2450–2453. [Google Scholar] [CrossRef]
- Nordmann, P.; Boulanger, A.E.; Poirel, L. NDM-4 Metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob. Agents Chemother. 2012, 56, 2184–2186. [Google Scholar] [CrossRef] [PubMed]
- Tada, T.; Miyoshi-Akiyama, T.; Dahal, R.K.; Sah, M.K.; Ohara, H.; Kirikae, T.; Pokhrel, B.M. NDM-8 metallo-β-lactamase in a multidrug-resistant Escherichia coli strain isolated in Nepal. Antimicrob. Agents Chemother. 2013, 57, 2394–2396. [Google Scholar] [CrossRef]
- Göttig, S.; Hamprecht, A.G.; Christ, S.; Kempf, V.A.; Wichelhaus, T.A. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-β-lactamase with increased carbapenemase activity. J. Antimicrob. Chemother. 2013, 68, 1737–1740. [Google Scholar] [CrossRef] [PubMed]
- Pal, L.R.; Moult, J. Genetic basis of Common Human Disease: Insight into the role of nonsynonymous SNPs from genome-wide association studies. J. Mol. Biol. 2015, 427, 2271–2289. [Google Scholar] [CrossRef]
- Ali, A.; Azam, M.W.; Khan, A.U. Non-active site mutation (Q123A) in New Delhi metallo-β-lactamase (NDM-1) enhanced its enzyme activity. Int. J. Biol. Macromol. 2018, 112, 1272–1277. [Google Scholar] [CrossRef]
- Mancini, S.; Keller, P.M.; Greiner, M.; Bruderer, V.; Imkamp, F. Detection of NDM-19, a novel variant of the New Delhi metallo-β-lactamase with increased carbapenemase activity under zinc-limited conditions, in Switzerland. Diagn. Microbiol. Infect. Dis. 2019, 95, 114851. [Google Scholar] [CrossRef]
- Lascols, C.; Hackel, M.; Marshall, S.H.; Hujer, A.M.; Bouchillon, S.; Badal, R.; Hoban, D.; Bonomo, R.A. Increasing prevalence and dissemination of NDM-1 metallo-β-lactamase in India: Data from the SMART study (2009). J. Antimicrob. Chemother. 2011, 66, 1992–1997. [Google Scholar] [CrossRef]
- Bonnin, R.A.; Poirel, L.; Naas, T.; Pirs, M.; Seme, K.; Schrenzel, J.; Nordmann, P. Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 2012, 18, E362–E365. [Google Scholar] [CrossRef]
- Rahman, M.; Mukhopadhyay, C.; Rai, R.P.; Singh, S.; Gupta, S.; Singh, A.; Pathak, A.; Prasad, K.N. Novel variant NDM-11 and other NDM-1 variants in multidrug-resistant Escherichia coli from South India. J. Glob. Antimicrob. Resist. 2018, 14, 154–157. [Google Scholar] [CrossRef]
- Khan, A.U.; Nordmann, P. NDM-1-producing Enterobacter cloacae and Klebsiella pneumoniae from diabetic foot ulcers in India. J. Med. Microbiol. 2012, 61, 454–456. [Google Scholar] [CrossRef][Green Version]
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, genetics and worldwide spread of New Delhi Metallo-β-lactamase (NDM): A threat to public health. BMC Microbiol. 2017, 17, 101. [Google Scholar] [CrossRef] [PubMed]
- Espinal, P.; Fugazza, G.; López, Y.; Kasma, M.; Lerman, Y.; Malhotra-Kumar, S.; Goossens, H.; Carmeli, Y.; Vila, J. Dissemination of an NDM-2-producing Acinetobacter baumannii clone in an Israeli rehabilitation center. Antimicrob. Agents Chemother. 2011, 55, 5396–5398. [Google Scholar] [CrossRef]
- Cantón, R.; Akóva, M.; Carmeli, Y.; Giske, C.G.; Glupczynski, Y.; Gniadkowski, M.; Livermore, D.M.; Miriagou, V.; Naas, T.; Rossolini, G.M.; et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2012, 18, 413–431. [Google Scholar] [CrossRef] [PubMed]
- Eu, W. Updated ECDC Risk Assessment on the Spread of New Delhi Metallo-β-lactamase and Its Variants within Europe; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2011. [Google Scholar]
- Coppo, E.; Del Bono, V.; Ventura, F.; Camera, M.; Orengo, G.; Viscoli, C.; Marchese, A. Identification of a New Delhi metallo-β-lactamase-4 (NDM-4)-producing Escherichia coli in Italy. BMC Microbiol. 2014, 14, 148. [Google Scholar] [CrossRef]
- Sassi, A.; Loucif, L.; Gupta, S.K.; Dekhil, M.; Chettibi, H.; Rolain, J.M. NDM-5 carbapenemase-encoding gene in multidrug-resistant clinical isolates of Escherichia coli from Algeria. Antimicrob. Agents Chemother. 2014, 58, 5606–5608. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Poirel, L.; Lagrutta, E.; Taylor, P.; Pham, J.; Nordmann, P. Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 2010, 54, 4914–4916. [Google Scholar] [CrossRef]
- Tondi, D.; Venturelli, A.; Bonnet, R.; Pozzi, C.; Shoichet, B.K.; Costi, M.P. Targeting class A and C serine β-lactamases with a broad-spectrum boronic acid derivative. J. Med. Chem. 2014, 57, 5449–5458. [Google Scholar] [CrossRef]
- Focco, V.; Bonomo, R.A. Exploring additional dimensions of complexity in inhibitor design for serine β-lactamases: Mechanistic and intra- and inter-molecular chemistry approaches. Front. Microbiol. 2018, 9, 622–632. [Google Scholar]
- Liénard, B.M.; Garau, G.; Horsfall, L.; Karsisiotis, A.I.; Damblon, C.; Lassaux, P.; Papamicael, C.; Roberts, G.C.; Galleni, M.; Dideberg, O.; et al. Structural basis for the broad-spectrum inhibition of Metallo-β-lactamases by thiols. Org. Biomol. Chem. 2008, 6, 2282–2294. [Google Scholar] [CrossRef]
- King, A.M.; Reid-Yu, S.A.; Wang, W.; King, D.T.; De Pascale, G.; Strynadka, N.C.; Walsh, T.R.; Coombes, B.K.; Wright, G.D. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature 2014, 510, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Yoshizumi, A.; Ishii, Y.; Livermore, D.M.; Woodford, N.; Kimura, S.; Saga, T.; Harada, S.; Yamaguchi, K.; Tateda, K. Efficacies of calcium-EDTA in combination with imipenem in a murine model of sepsis caused by Escherichia coli with NDM-1 β-lactamase. J. Infect. Chemother. 2013, 19, 992–995. [Google Scholar] [CrossRef] [PubMed]
- Drawz, S.M.; Bonomo, R.A. Three decades of β-lactamase inhibitors. Clin. Microbiol. Rev. 2010, 23, 160–201. [Google Scholar] [CrossRef]
- Buynak, J.D. β-Lactamase inhibitors: A review of the patent literature (2010–2013). Expert Opin Ther Pat. 2013, 23, 1469–1481. [Google Scholar] [CrossRef]
- Livermore, D.M.; Mushtaq, S.; Morinaka, A.; Ida, T.; Maebashi, K.; Hope, R. Activity of carbapenems with ME1071 (disodium 2,3-diethylmaleate) against Enterobacteriaceae and Acinetobacter spp. with carbapenemases, including NDM enzymes. J. Antimicrob. Chemother. 2013, 68, 153–158. [Google Scholar] [CrossRef]
- Niskanen, L.; Hedner, T.; Hansson, L.; Lanke, J.; Niklason, A.C. Study Group. Reduced cardiovascular morbidity and mortality in hypertensive diabetic patients on first-line therapy with an ACE inhibitor compared with a diuretic/beta-blocker-based treatment regimen: A subanalysis of the Captopril Prevention Project. Diabetes Care. 2001, 24, 2091–2096. [Google Scholar] [CrossRef][Green Version]
- Yusof, Y.; Tan, D.T.C.; Arjomandi, O.K.; Schenk, G.; McGeary, R.P. Captopril analogues as metallo-β-lactamase inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 1589–1593. [Google Scholar] [CrossRef] [PubMed]
- Brem, J.; Berkel, S.S.; Zollman, D.; Lee, S.Y.; Gileadi, O.; McHugh, P.J.; Walsh, T.R.; McDonough, M.A. Schofield CJ. Structural Basis of Metallo-β-Lactamase Inhibition by Captopril Stereoisomers. Antimicrob. Agents Chemother. 2015, 60, 142–150. [Google Scholar] [CrossRef]
- Li, N.; Xu, Y.; Xia, Q.; Bai, C.; Wang, T.; Wang, L.; He, D.; Xie, N.; Li, L.; Wang, J.; et al. Simplified captopril analogues as NDM-1 inhibitors. Bioorg. Med. Chem. Lett. 2014, 24, 386–389. [Google Scholar] [CrossRef]
- García-Sáez, I.; Mercuri, P.S.; Papamicael, C.; Kahn, R.; Frère, J.M.; Galleni, M.; Rossolini, G.M.; Dideberg, O. Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril. J. Mol. Biol. 2003, 325, 651–660. [Google Scholar] [CrossRef]
- Skagseth, S.; Akhter, S.; Paulsen, M.H.; Muhammad, Z.; Lauksund, S.; Samuelsen, Ø.; Leiros, H.S.; Bayer, A. Metallo-β-lactamase inhibitors by bioisosteric replacement: Preparation, activity and binding. Eur. J. Med. Chem. 2017, 135, 159–173. [Google Scholar] [CrossRef]
- Yamada, K.; Yanagihara, K.; Kaku, N.; Harada, Y.; Migiyama, Y.; Nagaoka, K.; Morinaga, Y.; Nakamura, S.; Imamura, Y.; Miyazaki, T.; et al. In vivo efficacy of biapenem with ME1071, a novel metallo-β-lactamase (MBL) Inhibitor, in a murine model mimicking ventilator-associated pseumonia caused by MBL-producing Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 2013, 42, 238–243. [Google Scholar] [CrossRef]
- Klingler, F.M.; Wichelhaus, T.A.; Frank, D.; Cuesta-Bernal, J.; El-Delik, J.; Müller, H.F.; Sjuts, H.; Göttig, S.; Koenigs, A.; Pos, K.M.; et al. Approved drugs Containing thiols as inhibitors of Metallo-β-lactamases: Strategy to combat multidrug-resistant bacteria. J. Med. Chem. 2015, 58, 3626–3630. [Google Scholar] [CrossRef]
- Tehrani, K.H.M.E.; Martin, N.I. Thiol-containing Metallo-β-Lactamase inhibitors resensitize resistant gram-negative bacteria to meropenem. ACS Infect. Dis. 2017, 3, 711–717. [Google Scholar] [CrossRef]
- Hinchliffe, P.; González, M.M.; Mojica, M.F.; González, J.M.; Castillo, V.; Saiz, C.; Kosmopoulou, M.; Tooke, C.L.; Llarrull, L.I.; Mahler, G.; et al. Cross-class Metallo-β-lactamase inhibition by bisthiazolidines reveals multiple binding modes. Proc. Natl. Acad. Sci. USA 2016, 113, E3745–E3754. [Google Scholar] [CrossRef] [PubMed]
- Cain, R.; Brem, J.; Zollman, D.; McDonough, M.A.; Johnson, R.M.; Spencer, J.; Makena, A.; Abboud, M.I.; Cahill, S.; Lee, S.Y.; et al. In silico fragment-based design identifies subfamily B1 Metallo-β-lactamase inhibitors. J. Med. Chem. 2018, 61, 1255–1260. [Google Scholar] [CrossRef]
- Bauer, R. Binding of D- and L-captopril inhibitors to metallo-beta-lactamase studied by polarizable molecular mechanics and quantum mechanics. J. Comput. Chem. 2002, 23, 1281–1296. [Google Scholar]
- Antony, J.; Piquemal, J.P.; Gresh, N. Complexes of thiomandelate and captopril mercaptocarboxylate inhibitors to metallo-beta-lactamase by polarizable molecular mechanics. validation on model binding sites by quantum chemistry. J. Comput. Chem. 2005, 26, 1131–1147. [Google Scholar] [CrossRef] [PubMed]
- Hinchliffe, P.; Moreno, D.M.; Rossi, M.A.; Mojica, M.F.; Martinez, V.; Villamil, V.; Spellberg, B.; Drusano, G.L.; Banchio, C.; Mahler, G.; et al. 2-Mercaptomethyl thiazolidines (MMTZS) inhibit all metallo-β-lactamase classes by maintaining a conserved binding mode. ACS Infect. Dis. 2021, 7, 2697–2706. [Google Scholar] [CrossRef]
- Zervosen, A.; Sauvage, E.; Frère, J.M.; Charlier, P.; Luxen, A. Development of new drugs for an old target: The penicillin binding proteins. Molecules 2012, 17, 12478–12505. [Google Scholar] [CrossRef] [PubMed]
- Zervosen, A.; Lu, W.P.; Chen, Z.; White, R.E.; Demuth, T.P.; Frère, J.M. Interactions between penicillin-binding proteins (PBPs) and two novel classes of PBP inhibitors, arylalkylidene rhodanines and arylalkylidene iminothiazolidin-4-ones. Antimicrob. Agents Chemother. 2004, 48, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Brem, J.; van Berkel, S.S.; Aik, W.; Rydzik, A.M.; Avison, M.B.; Pettinati, I.; Umland, K.D.; Kawamura, A.; Spencer, J.; Claridge, T.D.; et al. Rhodanine hydrolysis leads to potent thioenolate mediated metallo-β-lactamase inhibition. Nat. Chem. 2014, 6, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Chen, C.; Wang, W.M.; Xu, L.W.; Yang, K.W.; Oelschlaeger, P.; He, Y. Rhodanine as a potent scaffold for the development of broad-spectrum Metallo-β-lactamase inhibitors. ACS Med. Chem Lett. 2018, 9, 359–364. [Google Scholar] [CrossRef]
- Falconer, S.B.; Wang, W.; Gehrke, S.S.; Cuneo, J.D.; Britten, J.F.; Wright, G.D.; Brown, E.D. Metal-induced isomerization yields an intracellular chelator that disrupts bacterial iron homeostasis. Chem. Biol. 2014, 21, 136–145. [Google Scholar] [CrossRef]
- Falconer, S.B.; Reid-Yu, S.A.; King, A.M.; Gehrke, S.S.; Wang, W.; Britten, J.F.; Coombes, B.K.; Wright, G.D.; Brown, E.D. Zinc chelation by a small-molecule adjuvant potentiates meropenem activity in vivo against NDM-1-producing klebsiella pneumoniae. ACS Infect. Dis. 2015, 1, 533–543. [Google Scholar] [CrossRef]
- Ai, X.; Liu, H.; Lu, C.; Liang, C.; Sun, Y.; Chen, S.; Sun, B.; Li, Y.; Liu, Y.; Zhang, Q.; et al. Phenytoin silver: A new nanocompound for nromoting dermal wound healing via comprehensive pharmacological action. Theranostics 2017, 7, 425–435. [Google Scholar] [CrossRef][Green Version]
- Pandeya, S.N.; Smitha, S.; Jyoti, M.; Sridhar, S.K. Biological activities of isatin and its derivatives. Acta Pharm. 2005, 55, 27–46. [Google Scholar]
- Song, G.; Wang, W.; Li, Z.; Wang, Y.; Wang, J. First identification of isatin-β-thiosemicarbazones as novel inhibitors of New Delhi Metallo-β-lactamase-1: Chemical synthesis, biological evaluation and molecular simulation. Chin. Chem. Lett. 2018, 29, 899–902. [Google Scholar] [CrossRef]
- Zhang, X.M.; Guo, H.; Li, Z.S.; Song, F.H.; Wang, W.M.; Dai, H.Q.; Zhang, L.X.; Wang, J.G. Synthesis and evaluation of isatin-β-thiosemicarbazones as novel agents against antibiotic-resistant Gram-positive bacterial species. Eur. J. Med. Chem. 2015, 101, 419–430. [Google Scholar] [CrossRef]
- Chen, A.Y.; Thomas, P.W.; Stewart, A.C.; Bergstrom, A.; Cheng, Z.; Miller, C.; Bethel, C.R.; Marshall, S.H.; Credille, C.V.; Riley, C.L.; et al. Dipicolinic acid derivatives as inhibitors of New Delhi Metallo-β-lactamase-1. J. Med. Chem. 2017, 60, 7267–7283. [Google Scholar] [CrossRef]
- Hinchliffe, P.; Tanner, C.A.; Krismanich, A.P.; Labbé, G.; Goodfellow, V.J.; Marrone, L.; Desoky, A.Y.; Calvopiña, K.; Whittle, E.E.; Zeng, F.; et al. Structural and kinetic studies of the potent inhibition of metallo-β-lactamases by 6-phosphonomethylpyridine-2-carboxylates. Biochemistry 2018, 57, 1880–1892. [Google Scholar] [CrossRef] [PubMed]
- Mavrova, A.; Wesselinova, D.; Tsenov, Y.A.; Denkova, P. Synthesis, cytotoxicity and effects of some 1,2,4-triazole and 1,3,4-thiadiazole derivatives on immunocompetent cells. Eur. J. Med. Chem. 2009, 44, 63–69. [Google Scholar] [CrossRef]
- Kwapien, K.; Damergi, M.; Nader, S.; El Khoury, L.; Hobaika, Z.; Maroun, R.G.; Piquemal, J.P.; Gavara, L.; Berthomieu, D.; Hernandez, J.F.; et al. Calibration of 1,2,4-triazole-3-thione, an original Zn-binding group of metallo-β-lactamase inhibitors. validation of a polarizable MM/MD potential by quantum chemistry. J. Phys. Chem. B 2017, 121, 6295–6312. [Google Scholar] [CrossRef]
- Sevaille, L.; Gavara, L.; Bebrone, C.; De Luca, F.; Nauton, L.; Achard, M.; Mercuri, P.; Tanfoni, S.; Borgianni, L.; Guyon, C.; et al. 1,2,4-Triazole-3-thione compounds as inhibitors of dizinc metallo-β-lactamases. Chemmedchem 2017, 12, 972–985. [Google Scholar] [CrossRef] [PubMed]
- Gavara, L.; Sevaille, L.; De Luca, F.; Mercuri, P.; Bebrone, C.; Feller, G.; Legru, A.; Cerboni, G.; Tanfoni, S.; Baud, D.; et al. 4-Amino-1,2,4-triazole-3-thione-derived Schiff bases as metallo-β-lactamase inhibitors. Eur. J. Med. Chem. 2020, 208, 112720. [Google Scholar] [CrossRef]
- Fu, H.; Fang, H.; Sun, J.; Wang, H.; Liu, A.; Sun, J.; Wu, Z. Boronic acid-based enzyme inhibitors: A review of recent progress. Curr. Med. Chem. 2014, 21, 3271–3280. [Google Scholar] [CrossRef] [PubMed]
- Brem, J.; Cain, R.; Cahill, S.; McDonough, M.A.; Clifton, I.J.; Jiménez-Castellanos, J.C.; Avison, M.B.; Spencer, J.; Fishwick, C.W.; Schofield, C.J. Structural basis of metallo-β-lactamase, serine-β-lactamase and penicillin-binding protein inhibition by cyclic boronates. Nat. Commun. 2016, 7, 12406. [Google Scholar] [CrossRef]
- Hecker, S.J.; Reddy, K.R.; Lomovskaya, O.; Griffith, D.C.; Rubio-Aparicio, D.; Nelson, K.; Tsivkovski, R.; Sun, D.; Sabet, M.; Tarazi, Z.; et al. Discovery of cyclic boronic acid Qpx7728, an ultra-broad-spectrum inhibitor of serine and metallo beta-lactamases. J. Med. Chem. 2020, 63, 7491–7507. [Google Scholar] [CrossRef] [PubMed]
- Santucci, M.; Spyrakis, F.; Cross, S.; Quotadamo, A.; Farina, D.; Tondi, D.; De Luca, F.; Docquier, J.D.; Prieto, A.I.; Ibacache, C.; et al. Computational and biological profile of boronic acids for the detection of bacterial serine- and metallo-β-lactamases. Sci. Rep. 2017, 7, 17716. [Google Scholar] [CrossRef]
- Krajnc, A.; Brem, J.; Hinchliffe, P.; Calvopiña, K.; Panduwawala, T.D.; Lang, P.A.; Kamps, J.J.A.G.; Tyrrell, J.M.; Widlake, E.; Saward, B.G.; et al. Bicyclic boronate VNRX-5133 inhibits metallo- and serine-β-lactamases. J. Med. Chem. 2019, 62, 8544–8556. [Google Scholar] [CrossRef] [PubMed]
- Aaseth, J.; Skaug, M.A.; Cao, Y.; Andersen, O. Chelation in metal intoxication--principles and paradigms. J. Trace. Elem. Med. Biol. 2015, 31, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; McLeod, S.; MacCormack, K.; Sriram, S.; Gao, N.; Breeze, A.L.; Hu, J. Real-time monitoring of New Delhi metallo-β-lactamase activity in living bacterial cells by 1HNMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2014, 53, 2130–2133. [Google Scholar] [CrossRef]
- Somboro, A.M.; Tiwari, D.; Bester, L.A.; Parboosing, R.; Chonco, L.; Kruger, H.G.; Arvidsson, P.I.; Govender, T.; Naicker, T.; Essack, S.Y. NOTA: A potent metallo-β-lactamase inhibitor. J. Antimicrob. Chemother. 2015, 70, 1594–1956. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Bertini, I. A bioinformatics view of zinc enzymes. J. Inorg. Biochem. 2012, 111, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Arai, K.; Ashikawa, N.; Nakakita, Y.; Matsuura, A.; Munekata, M. Aspergillomarasmine A and B, potent microbial inhibitors of endothelin-converting enzyme. Biosci. Biotech. Bioch. 2014, 57, 1944–1945. [Google Scholar] [CrossRef][Green Version]
- Mikami, Y.; Suzuki, T. Novel microbial inhibitors of angiotensin-converting enzyme, aspergillomarasmines A and B. J. Agric. Chem. Soc. Jpn. 1983, 47, 2693–2695. [Google Scholar]
- Koteva, K.; King, A.M.; Caprett, A.; Wright, G.D. Total synthesis and activity of the metallo-β-lactamase inhibitor Aspergillomarasmine A. Angew. Chem. 2016, 55, 2210–2212. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, S.; Wei, Q.; Guo, Q.; Bai, Y.; Yang, S.; Song, F.; Zhang, L.; Lei, X. Synthesis and biological evaluation of Aspergillomarasmine a derivatives as novel NDM-1 inhibitor to overcome antibiotics resistance. Bioorg. Med. Chem. 2017, 25, 5133–5141. [Google Scholar] [CrossRef]
- Thomas, P.W.; Spicer, T.; Cammarata, M.; Brodbelt, J.S.; Hodder, P.; Fast, W. An altered zinc-binding site confers resistance to a covalent inactivator of New Delhi metallo-beta-lactamase-1 (NDM-1) discovered by high-throughput screening. Bioorg. Med. Chem. 2013, 21, 3138–3146. [Google Scholar] [CrossRef][Green Version]
- Parnham, M.J.; Sies, H. The early research and development of ebselen. Biochem. Pharmacol. 2013, 86, 1248–1253. [Google Scholar] [CrossRef]
- Chiou, J.; Wan, S.; Chan, K.F.; So, P.K.; He, D.; Chan, E.W.; Chan, T.H.; Wong, K.Y.; Tao, J.; Chen, S. Ebselen as a potent covalent inhibitor of New Delhi metallo-β-lactamase (NDM-1). Chem. Commun. 2015, 51, 9543–9546. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Xu, C.; Cheng, Q.; Qi, X.; Gao, W.; Zheng, Z.; Chan, E.W.C.; Leung, Y.; Chan, T.; Wong, K.; et al. Investigation of synergistic antimicrobial effects of the drug combinations of meropenem and 1,2-benzisoselenazol-3(2H)-one derivatives on carbapenem-resistant Enterobacteriaceae producing NDM-1. Eur. J. Med. Chem. 2018, 155, 285–302. [Google Scholar] [CrossRef]
- Su, J.; Liu, J.; Chen, C.; Zhang, Y.; Yang, K. Ebsulfur as a potent scaffold for inhibition and labelling of New Delhi metallo-β-lactamase-1 in vitro and in vivo. Bioorg. Chem. 2019, 84, 192–201. [Google Scholar] [CrossRef]
- Darabedian, N.; Chen, T.C.; Molina, H.; Pratt, M.R.; Schönthal, A.H. Bioorthogonal profiling of a cancer cell proteome identifies a large set of 3-bromopyruvate targets beyond glycolysis. ACS Chem. Biol. 2018, 13, 3054–3058. [Google Scholar] [CrossRef]
- Kang, P.; Su, J.; Sun, L.; Gao, H.; Yang, K. 3-Bromopyruvate as a potent covalently reversible inhibitor of New Delhi metallo-β-lactamase-1 (NDM-1). Eur. J. Pharm. Sci. 2020, 142, 105161. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.W.; Cammarata, M.; Brodbelt, J.S.; Fast, W. Covalent inhibition of New Delhi metallo-β-lactamase-1 (NDM-1) by cefaclor. Chembiochem 2014, 15, 2541–2548. [Google Scholar] [CrossRef]
- Christopeit, T.; Albert, A.; Leiros, H.S. Discovery of a novel covalent non-β-lactam inhibitor of the metallo-β-lactamase NDM-1. Bioorg. Med. Chem. 2016, 24, 2947–2953. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.W.; Cammarata, M.; Brodbelt, J.S.; Monzingo, A.F.; Pratt, R.F.; Fast, W. A lysine-targeted affinity label for serine-β-lactamase also covalently modifies New Delhi Metallo-β-lactamase-1 (NDM-1). Biochemistry 2019, 58, 2834–2843. [Google Scholar] [CrossRef]
- Sully, E.K.; Geller, B.L.; Li, L.; Moody, C.M.; Bailey, S.M.; Moore, A.L.; Wong, M.; Nordmann, P.; Daly, S.M.; Sturge, C.R.; et al. Peptide-conjugated phosphorodiamidate morpholino oligomer (PPMO) restores carbapenem susceptibility to NDM-1-positive pathogens in vitro and in vivo. J. Antimicrob. Chemother. 2017, 72, 782–790. [Google Scholar]
- Chandar, B.; Poovitha, S.; Ilango, K.; MohanKumar, R.; Parani, M. Inhibition of New Delhi Metallo-β-Lactamase 1 (NDM-1) producing Escherichia coli IR-6 by selected plant extracts and their synergistic actions with antibiotics. Front. Microbiol. 2017, 8, 1580. [Google Scholar] [CrossRef] [PubMed]
- Everett, M.; Sprynski, N.; Coelho, A.; Castandet, J.; Lemonnier, M. Discovery of a novel metallo-ß-lactamase inhibitor, which can potentiate meropenem activity against carbapenem-resistant enterobacteriaceae. Antimicrob. Agents Chemother. 2018, 62, e00074-18. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, J.; Zhou, Y.; Hu, N.; Li, J.; Wang, Y.; Niu, X.; Deng, X.; Wang, J. Pterostilbene restores carbapenem susceptibility in New Delhi metallo-β-lactamase-producing isolates by inhibiting the activity of New Delhi metallo-β-lactamases. Br. J. Pharmacol. 2019, 176, 4548–4557. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhou, Y.; Niu, X.; Wang, T.; Li, J.; Liu, Z.; Wang, J.; Tang, S.; Wang, Y.; Deng, X. Magnolol restores the activity of meropenem against NDM-1-producing Escherichia coli by inhibiting the activity of metallo-beta-lactamase. Cell Death Discov. 2018, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Kazi, M.I.; Perry, B.W.; Card, D.C.; Schargel, R.D.; Ali, H.B.; Obuekwe, V.C.; Sapkota, M.; Kang, K.N.; Pellegrino, M.W.; Greenberg, D.E.; et al. Discovery and characterization of New Delhi metallo-β-lactamase-1 inhibitor peptides that potentiate meropenem-dependent killing of carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2020, 75, 2843–2851. [Google Scholar] [CrossRef]











| NDM-1 Variant | Location of Amino Acid(s) Substitution | Source Organism(s) | ||
|---|---|---|---|---|
| A-Helices | β-Strands | Loop | ||
| NDM-2 | - | - | P28A | A. baumannii |
| NDM-3 | D95N | - | - | E. coli |
| NDM-4 | - | - | M154L | E. coli |
| NDM-5 | - | V88L | M154L | E. coli |
| NDM-6 | A233V | - | - | E. coli |
| NDM-7 | D130N | - | M154L | E. coli |
| NDM-8 | D130G | - | M154L | E. coli |
| NDM-9 | - | - | E152K | K. pneumoniae |
| NDM-10 | - | A74T | R32S, G36D, G69S, G200R | K. pneumoniae |
| NDM-11 | - | - | M154V | E. coli |
| NDM-12 | G222D | - | M154L | E. coli |
| NDM-13 | D95N | - | M154L | E. coli |
| NDM-14 | D130G | - | - | A. lwoffii |
| NDM-15 | A233V | - | M154L | E. coli |
| NDM-16 | A233V | V88L | M154L | E. coli |
| NDM-17 | - | V88L | M154L, E170K | E. coli |
| NDM-18 | - | - | QRFGD (44-48) | P. rettgeri |
| NDM-19 | D130N | - | M154L, A233V | E. coli |
| NDM-20 | - | V88L | M154L, R270H | E. coli |
| NDM-21 | - | V88L | G69S, M154L | E. coli |
| NDM-22 | - | - | M248L | E. cloacae |
| NDM-23 | I101L | - | - | K. pneumoniae |
| NDM-24 | - | V88L | - | P. stuartii |
| NDM-25 | - | A55S | - | K. pneumoniae |
| NDM-26 | G222S | V88L | M154L | E. coli |
| NDM-27 | D95N, A233V | - | - | E. coli |
| NDM-28 | A266V | - | - | K. pneumoniae |
| NDM-29 | D130N | - | - | K. pneumoniae |
| NDM-30 | D223Y | - | - | K. oxytoca |
| NDM-31 | P171T | - | - | C. werkmanii |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhao, D.; Li, W.; Sun, J.; Zhang, X. Enzyme Inhibitors: The Best Strategy to Tackle Superbug NDM-1 and Its Variants. Int. J. Mol. Sci. 2022, 23, 197. https://doi.org/10.3390/ijms23010197
Li X, Zhao D, Li W, Sun J, Zhang X. Enzyme Inhibitors: The Best Strategy to Tackle Superbug NDM-1 and Its Variants. International Journal of Molecular Sciences. 2022; 23(1):197. https://doi.org/10.3390/ijms23010197
Chicago/Turabian StyleLi, Xiaoting, Dongmei Zhao, Weina Li, Jichao Sun, and Xiuying Zhang. 2022. "Enzyme Inhibitors: The Best Strategy to Tackle Superbug NDM-1 and Its Variants" International Journal of Molecular Sciences 23, no. 1: 197. https://doi.org/10.3390/ijms23010197
APA StyleLi, X., Zhao, D., Li, W., Sun, J., & Zhang, X. (2022). Enzyme Inhibitors: The Best Strategy to Tackle Superbug NDM-1 and Its Variants. International Journal of Molecular Sciences, 23(1), 197. https://doi.org/10.3390/ijms23010197






