Search for Structural Basis of Interactions of Biogenic Amines with Human TAAR1 and TAAR6 Receptors
Abstract
:1. Introduction
2. Results
2.1. Bioinformatics Analysis of Amino Acid Sequences of TAAR Proteins in Different Animal Species
2.2. Amino Acid Sequence Identity of Human, Mouse, and Fish TAARs
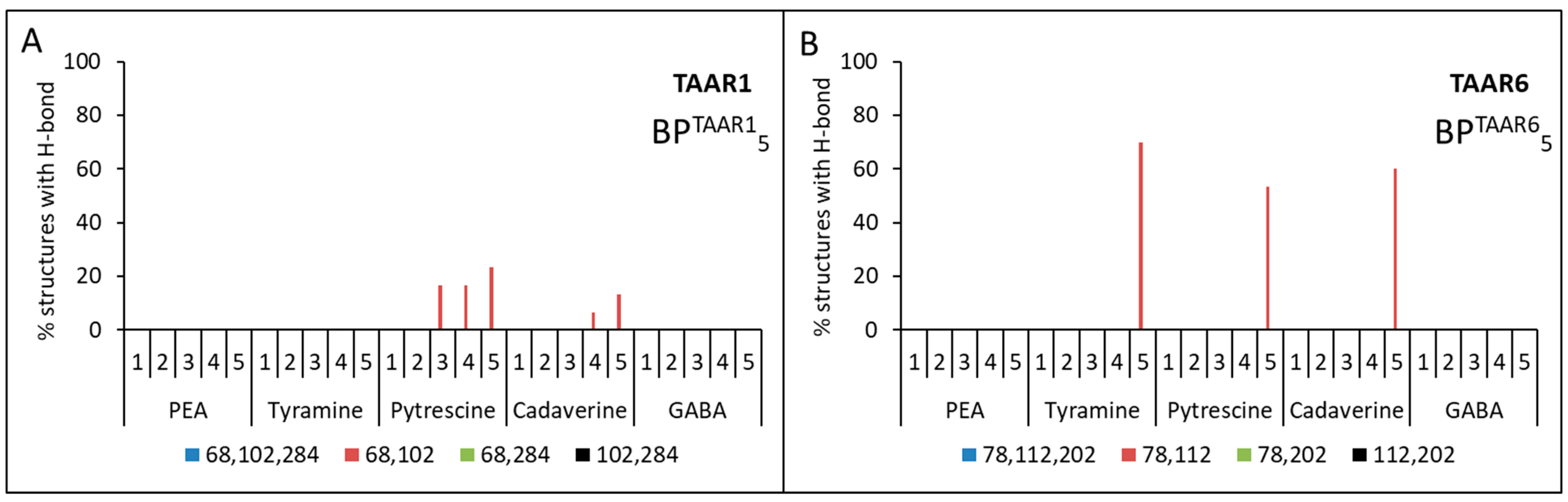
2.3. Docking of Ligands with Human TAAR1 and TAAR6 Receptors
3. Discussion
4. Materials and Methods
4.1. Analysis of the Sequences of Genes of the TAAR Family in Animals
4.2. Calculation of Amino Acid Sequence Identities for Human, Mouse, and Fish TAARs
4.3. Modeling the Interaction of Human TAAR1 and TAAR6 Receptors with Ligands
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TAARs | Trace amine-associated receptors |
| A-GPCR | class A G protein-coupled receptor family |
| PAM | positive allosteric modulators |
| NAM | negative allosteric modulators |
| GABA | gamma-aminobutyric acid |
References
- Moinard, C.; Cynober, L.; de Bandt, J.-P. Polyamines: Metabolism and Implications in Human Diseases. Clin. Nutr. 2005, 24, 184–197. [Google Scholar] [CrossRef]
- Sagar, N.A.; Tarafdar, S.; Agarwal, S.; Tarafdar, A.; Sharma, S. Polyamines: Functions, Metabolism, and Role in Human Disease Management. Med. Sci. 2021, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.D. Mammalian Central Nervous System Trace Amines. Pharmacologic Amphetamines, Physiologic Neuromodulators. J. Neurochem. 2004, 90, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Z.; Nawaz, W. The Emerging Roles of Human Trace Amines and Human Trace Amine-Associated Receptors (HTAARs) in Central Nervous System. Biomed. Pharmacother. 2016, 83, 439–449. [Google Scholar] [CrossRef]
- Gainetdinov, R.R.; Hoener, M.C.; Berry, M.D. Trace Amines and Their Receptors. Pharmacol. Rev. 2018, 70, 549–620. [Google Scholar] [CrossRef] [Green Version]
- Grandy, D.K. Trace Amine-Associated Receptor 1-Family Archetype or Iconoclast? Pharmacol. Ther. 2007, 116, 355–390. [Google Scholar] [CrossRef] [Green Version]
- Espinoza, S.; Sukhanov, I.; Efimova, E.V.; Kozlova, A.; Antonova, K.A.; Illiano, P.; Leo, D.; Merkulyeva, N.; Kalinina, D.; Musienko, P.; et al. Trace Amine-Associated Receptor 5 Provides Olfactory Input Into Limbic Brain Areas and Modulates Emotional Behaviors and Serotonin Transmission. Front. Mol. Neurosci. 2020, 13, 18. [Google Scholar] [CrossRef] [PubMed]
- Efimova, E.V.; Kozlova, A.A.; Razenkova, V.; Katolikova, N.V.; Antonova, K.A.; Sotnikova, T.D.; Merkulyeva, N.S.; Veshchitskii, A.S.; Kalinina, D.S.; Korzhevskii, D.E.; et al. Increased Dopamine Transmission and Adult Neurogenesis in Trace Amine-Associated Receptor 5 (TAAR5) Knockout Mice. Neuropharmacology 2021, 182, 108373. [Google Scholar] [CrossRef] [PubMed]
- Panas, M.W.; Xie, Z.; Panas, H.N.; Hoener, M.C.; Vallender, E.J.; Miller, G.M. Trace Amine Associated Receptor 1 Signaling in Activated Lymphocytes. J. Neuroimmune Pharmacol. 2012, 7, 866–876. [Google Scholar] [CrossRef] [Green Version]
- Khuhawar, M.Y.; Memon, A.A.; Jaipal, P.D.; Bhanger, M.I. Capillary Gas Chromatographic Determination of Putrescine and Cadaverine in Serum of Cancer Patients Using Trifluoroacetylacetone as Derivatizing Reagent. J. Chromatogr. B Biomed. Sci. Appl. 1999, 723, 17–24. [Google Scholar] [CrossRef]
- Le Gall, G.; Noor, S.O.; Ridgway, K.; Scovell, L.; Jamieson, C.; Johnson, I.T.; Colquhoun, I.J.; Kemsley, E.K.; Narbad, A. Metabolomics of Fecal Extracts Detects Altered Metabolic Activity of Gut Microbiota in Ulcerative Colitis and Irritable Bowel Syndrome. J. Proteome Res. 2011, 10, 4208–4218. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Suh, J.W.; Chung, B.C.; Kim, S.O. Polyamine Profiles in the Urine of Patients with Leukemia. Cancer Lett. 1998, 122, 1–8. [Google Scholar] [CrossRef]
- Lee, Y.R.; Lee, J.W.; Hong, J.; Chung, B.C. Simultaneous Determination of Polyamines and Steroids in Human Serum from Breast Cancer Patients Using Liquid Chromatography-Tandem Mass Spectrometry. Molecules 2021, 26, 1153. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, P.; Bi, C.W.; Ma, R.; Yin, Y.; Bi, K.; Li, Q. Plasma N-Acetylputrescine, Cadaverine and 1,3-Diaminopropane: Potential Biomarkers of Lung Cancer Used to Evaluate the Efficacy of Anticancer Drugs. Oncotarget 2017, 8, 88575–88585. [Google Scholar] [CrossRef] [Green Version]
- Löser, C.; Fölsch, U.R.; Paprotny, C.; Creutzfeldt, W. Polyamine Concentrations in Pancreatic Tissue, Serum, and Urine of Patients with Pancreatic Cancer. Pancreas 1990, 5, 119–127. [Google Scholar] [CrossRef]
- Song, J.; Shan, Z.; Mao, J.; Teng, W. Serum Polyamine Metabolic Profile in Autoimmune Thyroid Disease Patients. Clin. Endocrinol. 2019, 90, 727–736. [Google Scholar] [CrossRef]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary Electrophoresis Mass Spectrometry-Based Saliva Metabolomics Identified Oral, Breast and Pancreatic Cancer-Specific Profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [Green Version]
- Kovács, T.; Mikó, E.; Vida, A.; Sebő, É.; Toth, J.; Csonka, T.; Boratkó, A.; Ujlaki, G.; Lente, G.; Kovács, P.; et al. Cadaverine, a Metabolite of the Microbiome, Reduces Breast Cancer Aggressiveness through Trace Amino Acid Receptors. Sci. Rep. 2019, 9, 1300. [Google Scholar] [CrossRef] [Green Version]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef]
- Pegg, A.E.; McGill, S. Decarboxylation of Ornithine and Lysine in Rat Tissues. Biochim. Biophys. Acta 1979, 568, 416–427. [Google Scholar] [CrossRef]
- Alhonen-Hongisto, L.; Jänne, J. Polyamine Depletion Induces Enhanced Synthesis and Accumulation of Cadaverine in Cultured Ehrlich Ascites Carcinoma Cells. Biochem. Biophys. Res. Commun. 1980, 93, 1005–1013. [Google Scholar] [CrossRef]
- Hesterberg, R.S.; Cleveland, J.L.; Epling-Burnette, P.K. Role of Polyamines in Immune Cell Functions. Med. Sci. 2018, 6, 22. [Google Scholar] [CrossRef] [Green Version]
- Soda, K. The Mechanisms by Which Polyamines Accelerate Tumor Spread. J. Exp. Clin. Cancer Res. 2011, 30, 95. [Google Scholar] [CrossRef] [Green Version]
- Babusyte, A.; Kotthoff, M.; Fiedler, J.; Krautwurst, D. Biogenic Amines Activate Blood Leukocytes via Trace Amine-Associated Receptors TAAR1 and TAAR2. J. Leukoc. Biol. 2013, 93, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Jun, J.J.; Cuyler, J.; Xie, X.-Q. Covalent Allosteric Modulation: An Emerging Strategy for GPCRs Drug Discovery. Eur. J. Med. Chem. 2020, 206, 112690. [Google Scholar] [CrossRef] [PubMed]
- Conn, P.J.; Christopoulos, A.; Lindsley, C.W. Allosteric Modulators of GPCRs: A Novel Approach for the Treatment of CNS Disorders. Nat. Rev. Drug Discov. 2009, 8, 41–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, A.; Saraiva, L.R.; Ferrero, D.M.; Ahuja, G.; Krishna, V.S.; Liberles, S.D.; Korsching, S.I. High-Affinity Olfactory Receptor for the Death-Associated Odor Cadaverine. Proc. Natl. Acad. Sci. USA 2013, 110, 19579–19584. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Tachie-Baffour, Y.; Liu, Z.; Baldwin, M.W.; Kruse, A.C.; Liberles, S.D. Non-Classical Amine Recognition Evolved in a Large Clade of Olfactory Receptors. Elife 2015, 4, e10441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, M.D.; Gainetdinov, R.R.; Hoener, M.C.; Shahid, M. Pharmacology of Human Trace Amine-Associated Receptors: Therapeutic Opportunities and Challenges. Pharmacol. Ther. 2017, 180, 161–180. [Google Scholar] [CrossRef]
- Almeida-Santos, D.; Duarte, A.C.; Gonçalves, I.; Ferreira, C.L.; Ferrer, I.; Ishikawa, H.; Schwerk, C.; Schroten, H.; Santos, C.R.A. Cadaverine and Spermine Elicit Ca2+ Uptake in Human CP Cells via a Trace Amine-Associated Receptor 1 Dependent Pathway. J. Mol. Neurosci. 2021, 71, 625–637. [Google Scholar] [CrossRef]
- Izquierdo, C.; Gómez-Tamayo, J.C.; Nebel, J.-C.; Pardo, L.; Gonzalez, A. Identifying Human Diamine Sensors for Death Related Putrescine and Cadaverine Molecules. PLoS Comput. Biol. 2018, 14, e1005945. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Yang, J.; Jang, R.; Zhang, Y. GPCR-I-TASSER: A Hybrid Approach to G Protein-Coupled Receptor Structure Modeling and the Application to the Human Genome. Structure 2015, 23, 1538–1549. [Google Scholar] [CrossRef] [Green Version]
- van der Westhuizen, E.T.; Valant, C.; Sexton, P.M.; Christopoulos, A. Endogenous Allosteric Modulators of G Protein-Coupled Receptors. J. Pharmacol. Exp. Ther. 2015, 353, 246–260. [Google Scholar] [CrossRef] [Green Version]
- Eyun, S.-I.; Moriyama, H.; Hoffmann, F.G.; Moriyama, E.N. Molecular Evolution and Functional Divergence of Trace Amine-Associated Receptors. PLoS ONE 2016, 11, e0151023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liberles, S.D.; Buck, L.B. A Second Class of Chemosensory Receptors in the Olfactory Epithelium. Nature 2006, 442, 645–650. [Google Scholar] [CrossRef]
- Liberles, S.D. Trace Amine-Associated Receptors: Ligands, Neural Circuits, and Behaviors. Curr. Opin. Neurobiol. 2015, 34, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashiguchi, Y.; Nishida, M. Evolution of Trace Amine Associated Receptor (TAAR) Gene Family in Vertebrates: Lineage-Specific Expansions and Degradations of a Second Class of Vertebrate Chemosensory Receptors Expressed in the Olfactory Epithelium. Mol. Biol. Evol. 2007, 24, 2099–2107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saraiva, L.R.; Ibarra-Soria, X.; Khan, M.; Omura, M.; Scialdone, A.; Mombaerts, P.; Marioni, J.C.; Logan, D.W. Hierarchical Deconstruction of Mouse Olfactory Sensory Neurons: From Whole Mucosa to Single-Cell RNA-Seq. Sci. Rep. 2015, 5, 18178. [Google Scholar] [CrossRef]
- Saraiva, L.R.; Riveros-McKay, F.; Mezzavilla, M.; Abou-Moussa, E.H.; Arayata, C.J.; Makhlouf, M.; Trimmer, C.; Ibarra-Soria, X.; Khan, M.; Van Gerven, L.; et al. A Transcriptomic Atlas of Mammalian Olfactory Mucosae Reveals an Evolutionary Influence on Food Odor Detection in Humans. Sci. Adv. 2019, 5, eaax0396. [Google Scholar] [CrossRef] [Green Version]
- Hussain, A.; Saraiva, L.R.; Korsching, S.I. Positive Darwinian Selection and the Birth of an Olfactory Receptor Clade in Teleosts. Proc. Natl. Acad. Sci. USA 2009, 106, 4313–4318. [Google Scholar] [CrossRef] [Green Version]
- Ferrero, D.M.; Wacker, D.; Roque, M.A.; Baldwin, M.W.; Stevens, R.C.; Liberles, S.D. Agonists for 13 Trace Amine-Associated Receptors Provide Insight into the Molecular Basis of Odor Selectivity. ACS Chem. Biol. 2012, 7, 1184–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewan, A. Olfactory Signaling via Trace Amine-Associated Receptors. Cell Tissue Res. 2021, 383, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, M.Y.; Galzitskaya, O.V. The Ising Model for Prediction of Disordered Residues from Protein Sequence Alone. Phys. Biol. 2011, 8, 035004. [Google Scholar] [CrossRef]
- Ko, J.; Park, H.; Heo, L.; Seok, C. GalaxyWEB Server for Protein Structure Prediction and Refinement. Nucleic Acids Res. 2012, 40, W294–W297. [Google Scholar] [CrossRef]
- Gurevich, V.V.; Gurevich, E.V. How and Why Do GPCRs Dimerize? Trends Pharmacol. Sci. 2008, 29, 234–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiller, C.; Kühhorn, J.; Gmeiner, P. Class A G-Protein-Coupled Receptor (GPCR) Dimers and Bivalent Ligands. J. Med. Chem. 2013, 56, 6542–6559. [Google Scholar] [CrossRef]
- Feng, Y.H.; Saad, Y.; Karnik, S.S. Reversible Inactivation of AT(2) Angiotensin II Receptor from Cysteine-Disulfide Bond Exchange. FEBS Lett. 2000, 484, 133–138. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Huang, X.; Zhang, J.; Gui, J.; Zhang, Q. Subcellular Localization and Characterization of G Protein-Coupled Receptor Homolog from Lymphocystis Disease Virus Isolated in China. Viral. Immunol. 2007, 20, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Konvicka, K.; Guarnieri, F.; Ballesteros, J.A.; Weinstein, H. A Proposed Structure for Transmembrane Segment 7 of G Protein-Coupled Receptors Incorporating an Asn-Pro/Asp-Pro Motif. Biophys. J. 1998, 75, 601–611. [Google Scholar] [CrossRef] [Green Version]






| Name | TAAR Conservative Motifs | ||
|---|---|---|---|
| Fingerprint 1 | Fingerprint 2 | Fingerprint 3 | |
| TAAR1 H. sapiens | SISHFKQLHTP * | SMVRSAEHCWYFG | DPFLHYIIPPTLND |
| TAAR1 M. musculus | SISHFKQLHTP | SMVRTVERCWYFG | DPFLGYVIPPSLND |
| TAAR2 H. sapiens | SISYFKQLHTP | SMIRSVENCWYFG | DPFLNFSTPVVLFD |
| TAAR2 M. musculus | SISYFKQLHTP | SMVRSVENCWYFG | DPFLNFSTPAVLFD |
| TAAR5 H. sapiens | AVSYFKALHTP | STIRSVESCWFFG | DSLLHFITPPLVFD |
| TAAR5 M. musculus | AVSYFKVLHTP | STVRSVESCWFFG | DSLLNFITPPLVFD |
| TAAR6 H. sapiens | SILHFKQLHSP | SMVRTVESCWYFG | DAFMGFITPACIYE |
| TAAR6 M. musculus | SILHFKQLHSP | SMVRSIESCWYFG | DAFMGFITPAYIYE |
| TAAR8 H. sapiens | SVLHFKQLHSP | SMVRTVESCWYFG | DAFMGFLTPAYIYE |
| TAAR8 M. musculus | SVLHFKQLHSP | SMVRSIESCWYFG | DAFMGFITPAYVYE |
| TAAR8b M. musculus | SVLHFKQLHSP | SMVRSIESCWYFG | DAFVGFITPAYVYE |
| TAAR8c M. musculus | SVLHFKQLHSP | SMVRSIESCWYFG | DAFMGFITPAYVYE |
| TAAR9 H. sapiens | AILHFKQLHTP | STVRSVESCWYFG | DAYMNFITPPYVYE |
| TAAR9 M. musculus | AILHFKQLHTP | STVRSVESCWYFG | DAYMNFITPAYVYE |
| TAAR13c D. rerio | SIAHFKQLQTP | SMIRSVDGCWYYG | DPYINFSTPYALFD |
| Human | TAAR1 | TAAR2 | TAAR5 | TAAR6 | TAAR8 | TAAR9 |
|---|---|---|---|---|---|---|
| TAAR1 | 100 | 51 | 39 | 41 | 39 | 43 |
| TAAR2 | 100 | 43 | 39 | 37 | 40 | |
| TAAR5 | 100 | 44 | 44 | 45 | ||
| TAAR6 | 100 | 80 | 69 | |||
| TAAR8 | 100 | 68 | ||||
| TAAR9 | 100 |
| TAAR1 Mouse | TAAR6 Mouse | TAAR13c Fish | |
|---|---|---|---|
| TAAR1 human | 75 | 41 | 45 |
| TAAR6 human | 40 | 89 | 43 |
| TAAR13c fish | 44 | 42 |
| Ligand | TAAR1 | TAAR6 |
|---|---|---|
| cadaverine | 1. Asp68(2.50) + Asp102(3.32) | 1. Asp78(2.50) + Asp112(3.32) 2. Asp112(3.32) + Asp202(5.42) |
| putrescine | 1. Asp68(2.50) + Asp102(3.32) | 1. Asp78(2.50) + Asp112(3.32) 2. Asp112(3.32) + Asp202(5.42) |
| tyramine | - | 1. Asp78(2.50) + Asp112(3.32) 2. Asp112(3.32) + Asp202(5.42) |
| β-phenylethylamine | - | - |
| GABA | - | - |
| TAAR1 | TAAR6 | ||
|---|---|---|---|
| Asp68 (helix 2) | 12 | Asp78 (helix 2) | 19 |
| Asp102 (helix 3) | 21 | Asp112 (helix 3) | 21 |
| Asp284 (helix 7) | 1 | Asp202 (helix 5) | 6 |
| Ser106 (helix 3) | 9 | Ser205 (helix 5) | 4 |
| Trp261 (helix 6) | 4 | Trp271 (helix 6) | 6 |
| Asn293 (helix 7) | 6 | Asn303 (helix 7) | 9 |
| Ser294 (helix 7) | 4 | Ser304 (helix 7) | 5 |
| TAAR1 (TAAR6) | Model 1 | Model 2 | Model 3 | Model 4 | Model 5 |
|---|---|---|---|---|---|
| Model 1 | 0 | 4.7 (4.4) | 3 (1.7) | 4.5 (2.5) | 2 (4) |
| Model 2 | 0 | 0 | 4.8 (5) | 2.1 (5.2) | 5.3 (6.2) |
| Model 3 | 0 | 0 | 0 | 5.6 (3.7) | 2.2 (3.8) |
| Model 4 | 0 | 0 | 0 | 0 | 5.5 (4.9) |
| Model 5 | 0 | 0 | 0 | 0 | 0 |
| Ligand Type Name | Structure | |
|---|---|---|
| aromatic | β-phenylethylamine |  |
| tyramine |  | |
| aliphatic | putrescine |  |
| cadaverine |  | |
| gamma-aminobutyric acid (GABA) |  | |
| Calculation Option for Each Model | Aspartic Acid Residue Numbers | |
|---|---|---|
| TAAR1 | TAAR6 | |
| 1 | BPTAAR11{Asp68(2.50), Asp102(3.32), Asp284 (6.61)} | BPTAAR61{Asp78(2.50), Asp112(3.32), Asp202(5.42)} |
| 2 | BPTAAR12{Asp68 (2.50), Asp102(3.32)} | BPTAAR62{Asp78(2.50), Asp112(3.32)} |
| 3 | BPTAAR13{Asp102(3.32), Asp284 (6.61)} | BPTAAR63{Asp112(3.32), Asp202(5.42)} |
| 4 | BPTAAR14{Asp68(2.50), Asp284 (6.61)} | BPTAAR64{Asp78(2.50), Asp202(5.42)} |
| 5 | BPTAAR15{–} 1 | BPTAAR65{–} 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glyakina, A.V.; Pavlov, C.D.; Sopova, J.V.; Gainetdinov, R.R.; Leonova, E.I.; Galzitskaya, O.V. Search for Structural Basis of Interactions of Biogenic Amines with Human TAAR1 and TAAR6 Receptors. Int. J. Mol. Sci. 2022, 23, 209. https://doi.org/10.3390/ijms23010209
Glyakina AV, Pavlov CD, Sopova JV, Gainetdinov RR, Leonova EI, Galzitskaya OV. Search for Structural Basis of Interactions of Biogenic Amines with Human TAAR1 and TAAR6 Receptors. International Journal of Molecular Sciences. 2022; 23(1):209. https://doi.org/10.3390/ijms23010209
Chicago/Turabian StyleGlyakina, Anna V., Constantine D. Pavlov, Julia V. Sopova, Raul R. Gainetdinov, Elena I. Leonova, and Oxana V. Galzitskaya. 2022. "Search for Structural Basis of Interactions of Biogenic Amines with Human TAAR1 and TAAR6 Receptors" International Journal of Molecular Sciences 23, no. 1: 209. https://doi.org/10.3390/ijms23010209
APA StyleGlyakina, A. V., Pavlov, C. D., Sopova, J. V., Gainetdinov, R. R., Leonova, E. I., & Galzitskaya, O. V. (2022). Search for Structural Basis of Interactions of Biogenic Amines with Human TAAR1 and TAAR6 Receptors. International Journal of Molecular Sciences, 23(1), 209. https://doi.org/10.3390/ijms23010209








