Molecular Mechanisms of Cardiac Amyloidosis
Abstract
1. Introduction
2. Cardiac Amyloidosis
3. Molecular Mechanisms of Amyloidosis
3.1. ATTR Amyloidosis
3.1.1. Wild-Type ATTR Amyloidosis
3.1.2. Hereditary ATTR Amyloidosis
3.1.3. Cardiac Deposition of ATTR
3.1.4. Toxicity of ATTR to Cardiomyocytes
3.1.5. Effects of ATTR on Endothelial Cells
3.2. AL Amyloidosis
3.2.1. Toxicity of LC on Cardiac Cells
3.2.2. Extracellular Matrix Proteolysis in AL Cardiomyopathy
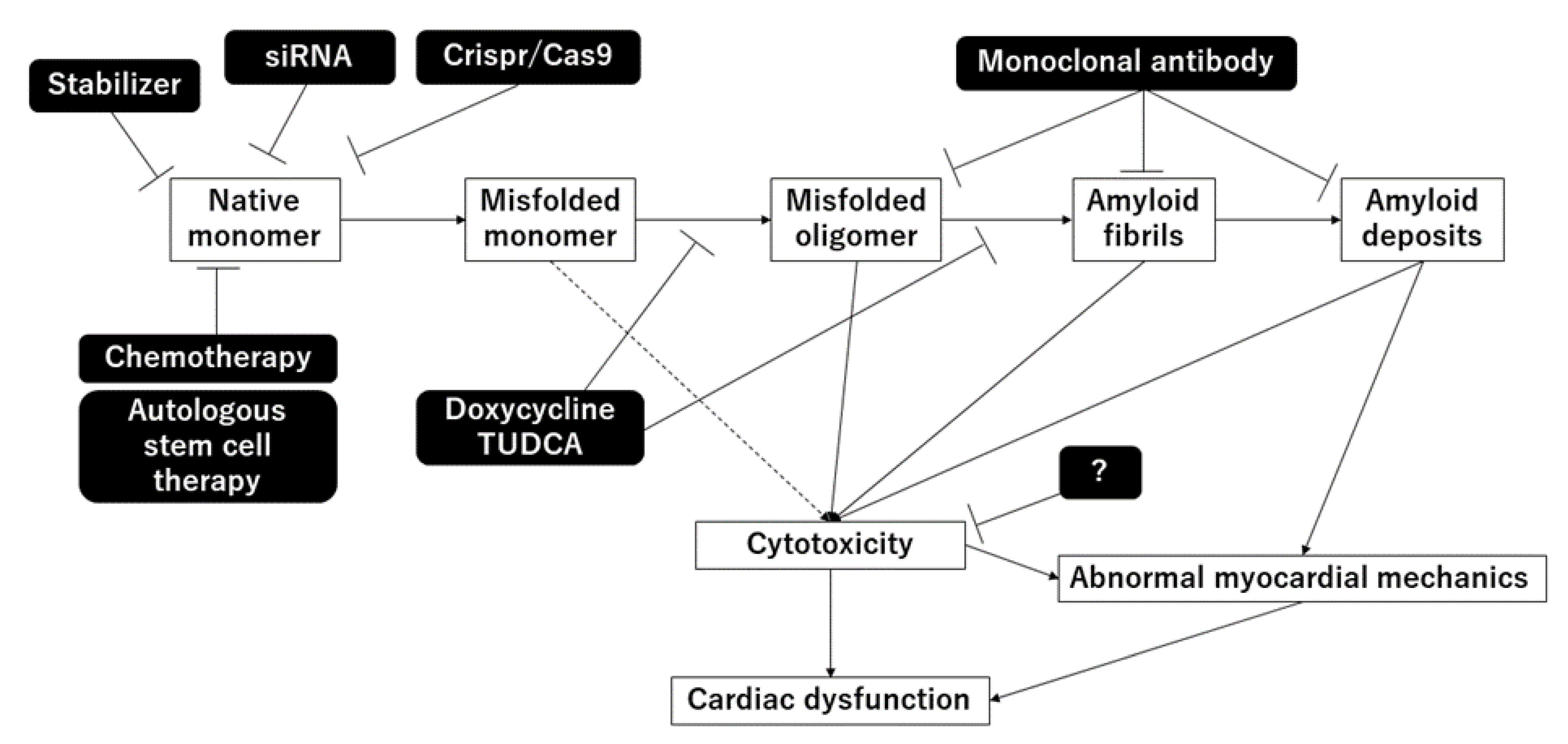
4. Treatment for Amyloidosis
4.1. Treatment for ATTR
4.1.1. Disruption of TTR Aggregation
4.1.2. Stabilization of TTR Tetramers
4.1.3. Suppression of TTR Synthesis
4.1.4. Removal of TTR Aggregates
4.1.5. Anti-Seeding Therapy
4.2. Treatment for AL Amyloidosis
4.2.1. Elimination of Light Chain Sources
4.2.2. Disruption of Light Chain Aggregation
4.2.3. Removal of Amyloid Deposits
4.2.4. Stabilization of Amyloidogenic LC
5. Summary
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | amyloid A |
| AL | amyloid light-chain |
| ATTR amyloidosis | transthyretin amyloidosis |
| ATTRv | hereditary ATTR amyloidosis |
| ATTRwt | wild-type ATTR amyloidosis |
| siRNA | small interfering RNA |
| TTR | transthyretin |
References
- Merlini, G.; Bellotti, V. Molecular mechanisms of amyloidosis. N. Engl. J. Med. 2003, 349, 583–596. [Google Scholar] [CrossRef]
- Grogan, M.; Scott, C.G.; Kyle, R.A.; Zeldenrust, S.R.; Gertz, M.A.; Lin, G.; Klarich, K.W.; Miller, W.L.; Maleszewski, J.J.; Dispenzieri, A. Natural History of Wild-Type Transthyretin Cardiac Amyloidosis and Risk Stratification Using a Novel Staging System. J. Am. Coll. Cardiol. 2016, 68, 1014–1020. [Google Scholar] [CrossRef]
- Fontana, M.; Ćorović, A.; Scully, P.; Moon, J.C. Myocardial Amyloidosis: The Exemplar Interstitial Disease. JACC Cardiovasc. Imaging 2019, 12, 2345–2356. [Google Scholar] [CrossRef]
- Kyle, R.A.; Linos, A.; Beard, C.M.; Linke, R.P.; Gertz, M.A.; O’Fallon, W.M.; Kurland, L.T. Incidence and natural history of primary systemic amyloidosis in Olmsted County, Minnesota, 1950 through 1989. Blood 1992, 79, 1817–1822. [Google Scholar] [CrossRef]
- Madan, S.; Kumar, S.K.; Dispenzieri, A.; Lacy, M.Q.; Hayman, S.R.; Buadi, F.K.; Dingli, D.; Rajkumar, S.V.; Hogan, W.J.; Leung, N.; et al. High-dose melphalan and peripheral blood stem cell transplantation for light-chain amyloidosis with cardiac involvement. Blood 2012, 119, 1117–1122. [Google Scholar] [CrossRef]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef]
- Benson, M.D.; Waddington-Cruz, M.; Berk, J.L.; Polydefkis, M.; Dyck, P.J.; Wang, A.K.; Plante-Bordeneuve, V.; Barroso, F.A.; Merlini, G.; Obici, L.; et al. Inotersen Treatment for Patients with Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 22–31. [Google Scholar] [CrossRef]
- Maurer, M.S.; Schwartz, J.H.; Gundapaneni, B.; Elliott, P.M.; Merlini, G.; Waddington-Cruz, M.; Kristen, A.V.; Grogan, M.; Witteles, R.; Damy, T.; et al. Tafamidis Treatment for Patients with Transthyretin Amyloid Cardiomyopathy. N. Engl. J. Med. 2018, 379, 1007–1016. [Google Scholar] [CrossRef]
- Gillmore, J.D.; Gane, E.; Taubel, J.; Kao, J.; Fontana, M.; Maitland, M.L.; Seitzer, J.; O’Connell, D.; Walsh, K.R.; Wood, K.; et al. CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N. Engl. J. Med. 2021, 385, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, T.; Mizuguchi, M. Transthyretin Amyloidogenesis Inhibitors: From Discovery to Current Developments. J. Med. Chem. 2020, 63, 14228–14242. [Google Scholar] [CrossRef]
- Benson, M.D.; Buxbaum, J.N.; Eisenberg, D.S.; Merlini, G.; Saraiva, M.J.M.; Sekijima, Y.; Sipe, J.D.; Westermark, P. Amyloid nomenclature 2020: Update and recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid 2020, 27, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Theis, J.D.; Vrana, J.A.; Rech, K.L.; Dao, L.N.; Howard, M.T.; Dispenzieri, A.; Gertz, M.A.; Hasadsri, L.; Highsmith, W.E.; et al. Amyloid Typing by Mass Spectrometry in Clinical Practice: A Comprehensive Review of 16,175 Samples. Mayo Clin. Proc. 2020, 95, 1852–1864. [Google Scholar] [CrossRef]
- Rognoni, P.; Mazzini, G.; Caminito, S.; Palladini, G.; Lavatelli, F. Dissecting the Molecular Features of Systemic Light Chain (AL) Amyloidosis: Contributions from Proteomics. Medicina 2021, 57, 916. [Google Scholar] [CrossRef]
- Larsen, B.T.; Mereuta, O.M.; Dasari, S.; Fayyaz, A.U.; Theis, J.D.; Vrana, J.A.; Grogan, M.; Dogan, A.; Dispenzieri, A.; Edwards, W.D.; et al. Correlation of histomorphological pattern of cardiac amyloid deposition with amyloid type: A histological and proteomic analysis of 108 cases. Histopathology 2016, 68, 648–656. [Google Scholar] [CrossRef]
- Mueller, P.S.; Edwards, W.D.; Gertz, M.A. Symptomatic ischemic heart disease resulting from obstructive intramural coronary amyloidosis. Am. J. Med. 2000, 109, 181–188. [Google Scholar] [CrossRef]
- Neben-Wittich, M.A.; Wittich, C.M.; Mueller, P.S.; Larson, D.R.; Gertz, M.A.; Edwards, W.D. Obstructive intramural coronary amyloidosis and myocardial ischemia are common in primary amyloidosis. Am. J. Med. 2005, 118, 1287. [Google Scholar] [CrossRef]
- Wittich, C.M.; Neben-Wittich, M.A.; Mueller, P.S.; Gertz, M.A.; Edwards, W.D. Deposition of amyloid proteins in the epicardial coronary arteries of 58 patients with primary systemic amyloidosis. Cardiovasc. Pathol. 2007, 16, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Witteles, R.M.; Liedtke, M. ALAmyloidosis for the Cardiologist and Oncologist: Epidemiology, Diagnosis, and Management. JACC Cardio Oncol. 2019, 1, 117–130. [Google Scholar] [CrossRef]
- Koike, H.; Katsuno, M. The Ultrastructure of Tissue Damage by Amyloid Fibrils. Molecules 2021, 26, 4611. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.R.; Saraiva, M.J. Clearance of extracellular misfolded proteins in systemic amyloidosis: Experience with transthyretin. FEBS Lett. 2012, 586, 2891–2896. [Google Scholar] [CrossRef]
- Westermark, P.; Sletten, K.; Johansson, B.; Cornwell, G.G., 3rd. Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc. Natl. Acad. Sci. USA 1990, 87, 2843–2845. [Google Scholar] [CrossRef]
- Palha, J.A. Transthyretin as a thyroid hormone carrier: Function revisited. Clin. Chem. Lab. Med. 2002, 40, 1292–1300. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, T.; Mizuguchi, M.; Nabeshima, Y.; Kusaka, K.; Yamada, T.; Hosoya, T.; Ohhara, T.; Kurihara, K.; Tomoyori, K.; Tanaka, I.; et al. Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin. J. Struct. Biol. 2012, 177, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Blake, C.C.; Geisow, M.J.; Oatley, S.J.; Rérat, B.; Rérat, C. Structure of prealbumin: Secondary, tertiary and quaternary interactions determined by Fourier refinement at 1.8 A. J. Mol. Biol. 1978, 121, 339–356. [Google Scholar] [CrossRef]
- Foss, T.R.; Wiseman, R.L.; Kelly, J.W. The pathway by which the tetrameric protein transthyretin dissociates. Biochemistry 2005, 44, 15525–15533. [Google Scholar] [CrossRef]
- McCutchen, S.L.; Colon, W.; Kelly, J.W. Transthyretin mutation Leu-55-Pro significantly alters tetramer stability and increases amyloidogenicity. Biochemistry 1993, 32, 12119–12127. [Google Scholar] [CrossRef]
- Quintas, A.; Saraiva, M.J.; Brito, R.M. The tetrameric protein transthyretin dissociates to a non-native monomer in solution. A novel model for amyloidogenesis. J. Biol. Chem. 1999, 274, 32943–32949. [Google Scholar] [CrossRef]
- Armen, R.S.; Alonso, D.O.; Daggett, V. Anatomy of an amyloidogenic intermediate: Conversion of beta-sheet to alpha-sheet structure in transthyretin at acidic pH. Structure 2004, 12, 1847–1863. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Kim, B.S.; Muniyappan, S.; Lee, Y.H.; Kim, J.H.; Yu, W. Aggregation-Prone Structural Ensembles of Transthyretin Collected With Regression Analysis for NMR Chemical Shift. Front. Mol. Biosci. 2021, 8, 766830. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Buxbaum, J.N.; Reixach, N. Age-related oxidative modifications of transthyretin modulate its amyloidogenicity. Biochemistry 2013, 52, 1913–1926. [Google Scholar] [CrossRef]
- Buxbaum, J.N.; Tagoe, C.; Gallo, G.; Walker, J.R.; Kurian, S.; Salomon, D.R. Why are some amyloidoses systemic? Does hepatic “chaperoning at a distance” prevent cardiac deposition in a transgenic model of human senile systemic (transthyretin) amyloidosis? FASEB J. 2012, 26, 2283–2293. [Google Scholar] [CrossRef]
- Stadtman, E.R. Protein oxidation and aging. Free Radic Res. 2006, 40, 1250–1258. [Google Scholar] [CrossRef] [PubMed]
- Vatner, S.F.; Zhang, J.; Oydanich, M.; Berkman, T.; Naftalovich, R.; Vatner, D.E. Healthful aging mediated by inhibition of oxidative stress. Ageing Res. Rev. 2020, 64, 101194. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Colón, W.; Kelly, J.W. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry 1996, 35, 6470–6482. [Google Scholar] [CrossRef] [PubMed]
- Palaninathan, S.K.; Mohamedmohaideen, N.N.; Snee, W.C.; Kelly, J.W.; Sacchettini, J.C. Structural insight into pH-induced conformational changes within the native human transthyretin tetramer. J. Mol. Biol. 2008, 382, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Si, J.B.; Kim, B.; Kim, J.H. Transthyretin Misfolding, A Fatal Structural Pathogenesis Mechanism. Int. J. Mol. Sci. 2021, 22, 4429. [Google Scholar] [CrossRef]
- Ihse, E.; Suhr, O.B.; Hellman, U.; Westermark, P. Variation in amount of wild-type transthyretin in different fibril and tissue types in ATTR amyloidosis. J. Mol. Med. 2011, 89, 171–180. [Google Scholar] [CrossRef]
- Bergström, J.; Gustavsson, A.; Hellman, U.; Sletten, K.; Murphy, C.L.; Weiss, D.T.; Solomon, A.; Olofsson, B.O.; Westermark, P. Amyloid deposits in transthyretin-derived amyloidosis: Cleaved transthyretin is associated with distinct amyloid morphology. J. Pathol. 2005, 206, 224–232. [Google Scholar] [CrossRef]
- Connors, L.H.; Sam, F.; Skinner, M.; Salinaro, F.; Sun, F.; Ruberg, F.L.; Berk, J.L.; Seldin, D.C. Heart Failure Resulting From Age-Related Cardiac Amyloid Disease Associated With Wild-Type Transthyretin: A Prospective, Observational Cohort Study. Circulation 2016, 133, 282–290. [Google Scholar] [CrossRef]
- González-López, E.; Gagliardi, C.; Dominguez, F.; Quarta, C.C.; de Haro-Del Moral, F.J.; Milandri, A.; Salas, C.; Cinelli, M.; Cobo-Marcos, M.; Lorenzini, M.; et al. Clinical characteristics of wild-type transthyretin cardiac amyloidosis: Disproving myths. Eur. Heart J. 2017, 38, 1895–1904. [Google Scholar] [CrossRef]
- Costa, P.P.; Figueira, A.S.; Bravo, F.R. Amyloid fibril protein related to prealbumin in familial amyloidotic polyneuropathy. Proc. Natl. Acad. Sci. USA 1978, 75, 4499–4503. [Google Scholar] [CrossRef] [PubMed]
- Rowczenio, D.M.; Noor, I.; Gillmore, J.D.; Lachmann, H.J.; Whelan, C.; Hawkins, P.N.; Obici, L.; Westermark, P.; Grateau, G.; Wechalekar, A.D. Online registry for mutations in hereditary amyloidosis including nomenclature recommendations. Hum. Mutat. 2014, 35, E2403–E2412. [Google Scholar] [CrossRef]
- Quintas, A.; Saraiva, M.J.; Brito, R.M. The amyloidogenic potential of transthyretin variants correlates with their tendency to aggregate in solution. FEBS Lett. 1997, 418, 297–300. [Google Scholar] [CrossRef]
- Hammarström, P.; Sekijima, Y.; White, J.T.; Wiseman, R.L.; Lim, A.; Costello, C.E.; Altland, K.; Garzuly, F.; Budka, H.; Kelly, J.W. D18G transthyretin is monomeric, aggregation prone, and not detectable in plasma and cerebrospinal fluid: A prescription for central nervous system amyloidosis? Biochemistry 2003, 42, 6656–6663. [Google Scholar] [CrossRef] [PubMed]
- Niraula, T.N.; Haraoka, K.; Ando, Y.; Li, H.; Yamada, H.; Akasaka, K. Decreased thermodynamic stability as a crucial factor for familial amyloidotic polyneuropathy. J. Mol. Biol. 2002, 320, 333–342. [Google Scholar] [CrossRef]
- Shinohara, Y.; Mizuguchi, M.; Matsubara, K.; Takeuchi, M.; Matsuura, A.; Aoki, T.; Igarashi, K.; Nagadome, H.; Terada, Y.; Kawano, K. Biophysical analyses of the transthyretin variants, Tyr114His and Tyr116Ser, associated with familial amyloidotic polyneuropathy. Biochemistry 2003, 42, 15053–15060. [Google Scholar] [CrossRef]
- Jiang, X.; Buxbaum, J.N.; Kelly, J.W. The V122I cardiomyopathy variant of transthyretin increases the velocity of rate-limiting tetramer dissociation, resulting in accelerated amyloidosis. Proc. Natl. Acad. Sci. USA 2001, 98, 14943–14948. [Google Scholar] [CrossRef]
- Dasari, A.K.R.; Hughes, R.M.; Wi, S.; Hung, I.; Gan, Z.; Kelly, J.W.; Lim, K.H. Transthyretin Aggregation Pathway toward the Formation of Distinct Cytotoxic Oligomers. Sci. Rep. 2019, 9, 33. [Google Scholar] [CrossRef]
- Frangolho, A.; Correia, B.E.; Vaz, D.C.; Almeida, Z.L.; Brito, R.M.M. Oligomerization Profile of Human Transthyretin Variants with Distinct Amyloidogenicity. Molecules 2020, 25, 5698. [Google Scholar] [CrossRef]
- Sekijima, Y.; Wiseman, R.L.; Matteson, J.; Hammarström, P.; Miller, S.R.; Sawkar, A.R.; Balch, W.E.; Kelly, J.W. The biological and chemical basis for tissue-selective amyloid disease. Cell 2005, 121, 73–85. [Google Scholar] [CrossRef]
- Rapezzi, C.; Quarta, C.C.; Obici, L.; Perfetto, F.; Longhi, S.; Salvi, F.; Biagini, E.; Lorenzini, M.; Grigioni, F.; Leone, O.; et al. Disease profile and differential diagnosis of hereditary transthyretin-related amyloidosis with exclusively cardiac phenotype: An Italian perspective. Eur. Heart J. 2013, 34, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, D.R.; Pastore, R.D.; Yaghoubian, R.; Kane, I.; Gallo, G.; Buck, F.S.; Buxbaum, J.N. Variant-sequence transthyretin (isoleucine 122) in late-onset cardiac amyloidosis in black Americans. N. Engl. J. Med. 1997, 336, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Mangione, P.P.; Porcari, R.; Gillmore, J.D.; Pucci, P.; Monti, M.; Porcari, M.; Giorgetti, S.; Marchese, L.; Raimondi, S.; Serpell, L.C.; et al. Proteolytic cleavage of Ser52Pro variant transthyretin triggers its amyloid fibrillogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 1539–1544. [Google Scholar] [CrossRef]
- Thylén, C.; Wahlqvist, J.; Haettner, E.; Sandgren, O.; Holmgren, G.; Lundgren, E. Modifications of transthyretin in amyloid fibrils: Analysis of amyloid from homozygous and heterozygous individuals with the Met30 mutation. EMBO J. 1993, 12, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Marcoux, J.; Mangione, P.P.; Porcari, R.; Degiacomi, M.T.; Verona, G.; Taylor, G.W.; Giorgetti, S.; Raimondi, S.; Sanglier-Cianférani, S.; Benesch, J.L.; et al. Anovel mechano-enzymatic cleavage mechanism underlies transthyretin amyloidogenesis. EMBO Mol. Med. 2015, 7, 1337–1349. [Google Scholar] [CrossRef]
- Mangione, P.P.; Verona, G.; Corazza, A.; Marcoux, J.; Canetti, D.; Giorgetti, S.; Raimondi, S.; Stoppini, M.; Esposito, M.; Relini, A.; et al. Plasminogen activation triggers transthyretin amyloidogenesis in vitro. J. Biol. Chem. 2018, 293, 14192–14199. [Google Scholar] [CrossRef]
- Bezerra, F.; Niemietz, C.; Schmidt, H.H.J.; Zibert, A.; Guo, S.; Monia, B.P.; Gonçalves, P.; Saraiva, M.J.; Almeida, M.R. In Vitro and In Vivo Effects of SerpinA1 on the Modulation of Transthyretin Proteolysis. Int. J. Mol. Sci. 2021, 22, 9488. [Google Scholar] [CrossRef] [PubMed]
- Sant’Anna, R.; Almeida, M.R.; Varejāo, N.; Gallego, P.; Esperante, S.; Ferreira, P.; Pereira-Henriques, A.; Palhano, F.L.; de Carvalho, M.; Foguel, D.; et al. Cavity filling mutations at the thyroxine-binding site dramatically increase transthyretin stability and prevent its aggregation. Sci. Rep. 2017, 7, 44709. [Google Scholar] [CrossRef] [PubMed]
- Sekijima, Y.; Dendle, M.T.; Wiseman, R.L.; White, J.T.; D’Haeze, W.; Kelly, J.W. R104H may suppress transthyretin amyloidogenesis by thermodynamic stabilization, but not by the kinetic mechanism characterizing T119 interallelic trans-suppression. Amyloid 2006, 13, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Hammarström, P.; Wiseman, R.L.; Powers, E.T.; Kelly, J.W. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science 2003, 299, 713–716. [Google Scholar] [CrossRef]
- Hammarström, P.; Jiang, X.; Hurshman, A.R.; Powers, E.T.; Kelly, J.W. Sequence-dependent denaturation energetics: A major determinant in amyloid disease diversity. Proc. Natl. Acad. Sci. USA 2002, 99 (Suppl. 4), 16427–16432. [Google Scholar] [CrossRef]
- Yee, A.W.; Aldeghi, M.; Blakeley, M.P.; Ostermann, A.; Mas, P.J.; Moulin, M.; de Sanctis, D.; Bowler, M.W.; Mueller-Dieckmann, C.; Mitchell, E.P.; et al. Amolecular mechanism for transthyretin amyloidogenesis. Nat. Commun. 2019, 10, 925. [Google Scholar] [CrossRef] [PubMed]
- Hornstrup, L.S.; Frikke-Schmidt, R.; Nordestgaard, B.G.; Tybjærg-Hansen, A. Genetic stabilization of transthyretin, cerebrovascular disease, and life expectancy. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1441–1447. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.M.; Ticau, S.; Butler, J.; Erbe, D.; Merkel, M.; Aldinc, E.; Hinkle, G.; Nioi, P. Transthyretin-stabilising mutation T119M is not associated with protection against vascular disease or death in the UK Biobank. Amyloid 2020, 27, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Misumi, Y.; Ando, Y.; Ueda, M.; Obayashi, K.; Jono, H.; Su, Y.; Yamashita, T.; Uchino, M. Chain reaction of amyloid fibril formation with induction of basement membrane in familial amyloidotic polyneuropathy. J. Pathol. 2009, 219, 481–490. [Google Scholar] [CrossRef]
- Bourgault, S.; Choi, S.; Buxbaum, J.N.; Kelly, J.W.; Price, J.L.; Reixach, N. Mechanisms of transthyretin cardiomyocyte toxicity inhibition by resveratrol analogs. Biochem. Biophys. Res. Commun. 2011, 410, 707–713. [Google Scholar] [CrossRef]
- Manral, P.; Reixach, N. Amyloidogenic and non-amyloidogenic transthyretin variants interact differently with human cardiomyocytes: Insights into early events of non-fibrillar tissue damage. Biosci. Rep. 2015, 35, e00172. [Google Scholar] [CrossRef] [PubMed]
- Sartiani, L.; Bucciantini, M.; Spinelli, V.; Leri, M.; Natalello, A.; Nosi, D.; Maria Doglia, S.; Relini, A.; Penco, A.; Giorgetti, S.; et al. Biochemical and Electrophysiological Modification of Amyloid Transthyretin on Cardiomyocytes. Biophys. J. 2016, 111, 2024–2038. [Google Scholar] [CrossRef] [PubMed]
- Hein, S.; Furkel, J.; Knoll, M.; Aus dem Siepen, F.; Schönland, S.; Hegenbart, U.; Katus, H.A.; Kristen, A.V.; Konstandin, M.H. Impaired in vitro growth response of plasma-treated cardiomyocytes predicts poor outcome in patients with transthyretin amyloidosis. Clin. Res. Cardiol. 2021, 110, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Bispo, M.; Marcelino, P.; Freire, A.; Martins, A.; Mourão, L.; Barroso, E. High incidence of thrombotic complications early after liver transplantation for familial amyloidotic polyneuropathy. Transpl. Int. 2009, 22, 165–171. [Google Scholar] [CrossRef]
- Nunes, R.J.; de Oliveira, P.; Lages, A.; Becker, J.D.; Marcelino, P.; Barroso, E.; Perdigoto, R.; Kelly, J.W.; Quintas, A.; Santos, S.C. Transthyretin proteins regulate angiogenesis by conferring different molecular identities to endothelial cells. J. Biol. Chem. 2013, 288, 31752–31760. [Google Scholar] [CrossRef] [PubMed]
- Padlan, E.A. Anatomy of the antibody molecule. Mol. Immunol. 1994, 31, 169–217. [Google Scholar] [CrossRef]
- Bourne, P.C.; Ramsland, P.A.; Shan, L.; Fan, Z.C.; DeWitt, C.R.; Shultz, B.B.; Terzyan, S.S.; Moomaw, C.R.; Slaughter, C.A.; Guddat, L.W.; et al. Three-dimensional structure of an immunoglobulin light-chain dimer with amyloidogenic properties. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Merlini, G.; Dispenzieri, A.; Sanchorawala, V.; Schönland, S.O.; Palladini, G.; Hawkins, P.N.; Gertz, M.A. Systemic immunoglobulin light chain amyloidosis. Nat. Rev. Dis. Primers 2018, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Swuec, P.; Lavatelli, F.; Tasaki, M.; Paissoni, C.; Rognoni, P.; Maritan, M.; Brambilla, F.; Milani, P.; Mauri, P.; Camilloni, C.; et al. Cryo-EM structure of cardiac amyloid fibrils from an immunoglobulin light chain AL amyloidosis patient. Nat. Commun. 2019, 10, 1269. [Google Scholar] [CrossRef] [PubMed]
- Radamaker, L.; Lin, Y.H.; Annamalai, K.; Huhn, S.; Hegenbart, U.; Schönland, S.O.; Fritz, G.; Schmidt, M.; Fändrich, M. Cryo-EM structure of a light chain-derived amyloid fibril from a patient with systemic AL amyloidosis. Nat. Commun. 2019, 10, 1103. [Google Scholar] [CrossRef]
- Radamaker, L.; Baur, J.; Huhn, S.; Haupt, C.; Hegenbart, U.; Schönland, S.; Bansal, A.; Schmidt, M.; Fändrich, M. Cryo-EM reveals structural breaks in a patient-derived amyloid fibril from systemic AL amyloidosis. Nat. Commun. 2021, 12, 875. [Google Scholar] [CrossRef] [PubMed]
- Kazman, P.; Absmeier, R.M.; Engelhardt, H.; Buchner, J. Dissection of the amyloid formation pathway in AL amyloidosis. Nat. Commun. 2021, 12, 6516. [Google Scholar] [CrossRef] [PubMed]
- Kazman, P.; Vielberg, M.T.; Pulido Cendales, M.D.; Hunziger, L.; Weber, B.; Hegenbart, U.; Zacharias, M.; Köhler, R.; Schönland, S.; Groll, M.; et al. Fatal amyloid formation in a patient’s antibody light chain is caused by a single point mutation. Elife 2020, 9, e52200. [Google Scholar] [CrossRef] [PubMed]
- Liao, R.; Jain, M.; Teller, P.; Connors, L.H.; Ngoy, S.; Skinner, M.; Falk, R.H.; Apstein, C.S. Infusion of light chains from patients with cardiac amyloidosis causes diastolic dysfunction in isolated mouse hearts. Circulation 2001, 104, 1594–1597. [Google Scholar] [CrossRef]
- Lavatelli, F.; Imperlini, E.; Orrù, S.; Rognoni, P.; Sarnataro, D.; Palladini, G.; Malpasso, G.; Soriano, M.E.; Di Fonzo, A.; Valentini, V.; et al. Novel mitochondrial protein interactors of immunoglobulin light chains causing heart amyloidosis. FASEB J. 2015, 29, 4614–4628. [Google Scholar] [CrossRef]
- Imperlini, E.; Gnecchi, M.; Rognoni, P.; Sabidò, E.; Ciuffreda, M.C.; Palladini, G.; Espadas, G.; Mancuso, F.M.; Bozzola, M.; Malpasso, G.; et al. Proteotoxicity in cardiac amyloidosis: Amyloidogenic light chains affect the levels of intracellular proteins in human heart cells. Sci. Rep. 2017, 7, 15661. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Mishra, S.; Shi, J.; Plovie, E.; Qiu, Y.; Cao, X.; Gianni, D.; Jiang, B.; Del Monte, F.; Connors, L.H.; et al. Stanniocalcin1 is a key mediator of amyloidogenic light chain induced cardiotoxicity. Basic Res. Cardiol. 2013, 108, 378. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.A.; Jain, M.; Pimentel, D.R.; Wang, B.; Connors, L.H.; Skinner, M.; Apstein, C.S.; Liao, R. Human amyloidogenic light chains directly impair cardiomyocyte function through an increase in cellular oxidant stress. Circ. Res. 2004, 94, 1008–1010. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Guan, J.; Jiang, B.; Brenner, D.A.; Del Monte, F.; Ward, J.E.; Connors, L.H.; Sawyer, D.B.; Semigran, M.J.; Macgillivray, T.E.; et al. Amyloidogenic light chains induce cardiomyocyte contractile dysfunction and apoptosis via a non-canonical p38alpha MAPK pathway. Proc. Natl. Acad. Sci. USA 2010, 107, 4188–4193. [Google Scholar] [CrossRef]
- McWilliams-Koeppen, H.P.; Foster, J.S.; Hackenbrack, N.; Ramirez-Alvarado, M.; Donohoe, D.; Williams, A.; Macy, S.; Wooliver, C.; Wortham, D.; Morrell-Falvey, J.; et al. Light Chain Amyloid Fibrils Cause Metabolic Dysfunction in Human Cardiomyocytes. PLoS ONE 2015, 10, e0137716. [Google Scholar] [CrossRef] [PubMed]
- Jordan, T.L.; Maar, K.; Redhage, K.R.; Misra, P.; Blancas-Mejia, L.M.; Dick, C.J.; Wall, J.S.; Williams, A.; Dietz, A.B.; van Wijnen, A.J.; et al. Light chain amyloidosis induced inflammatory changes in cardiomyocytes and adipose-derived mesenchymal stromal cells. Leukemia 2020, 34, 1383–1393. [Google Scholar] [CrossRef] [PubMed]
- Biolo, A.; Ramamurthy, S.; Connors, L.H.; O’Hara, C.J.; Meier-Ewert, H.K.; Soo Hoo, P.T.; Sawyer, D.B.; Seldin, D.C.; Sam, F. Matrix metalloproteinases and their tissue inhibitors in cardiac amyloidosis: Relationship to structural, functional myocardial changes and to light chain amyloid deposition. Circ. Heart Fail. 2008, 1, 249–257. [Google Scholar] [CrossRef]
- Macedo, B.; Batista, A.R.; Ferreira, N.; Almeida, M.R.; Saraiva, M.J. Anti-apoptotic treatment reduces transthyretin deposition in a transgenic mouse model of Familial Amyloidotic Polyneuropathy. Biochim. Biophys. Acta-Mol. Basis Dis. 2008, 1782, 517–522. [Google Scholar] [CrossRef]
- Cardoso, I.; Saraiva, M.J. Doxycycline disrupts transthyretin amyloid: Evidence from studies in a FAP transgenic mice model. Faseb J. 2006, 20, 234–239. [Google Scholar] [CrossRef]
- Cardoso, I.; Martins, D.; Ribeiro, T.; Merlini, G.; Saraiva, M.J. Synergy of combined doxycycline/TUDCA treatment in lowering Transthyretin deposition and associated biomarkers: Studies in FAP mouse models. J. Transl. Med. 2010, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Obici, L.; Cortese, A.; Lozza, A.; Lucchetti, J.; Gobbi, M.; Palladini, G.; Perlini, S.; Saraiva, M.J.; Merlini, G. Doxycycline plus tauroursodeoxycholic acid for transthyretin amyloidosis: A phase II study. Amyloid 2012, 19 (Suppl. 1), 34–36. [Google Scholar] [CrossRef] [PubMed]
- Karlstedt, E.; Jimenez-Zepeda, V.; Howlett, J.G.; White, J.A.; Fine, N.M. Clinical Experience With the Use of Doxycycline and Ursodeoxycholic Acid for the Treatment of Transthyretin Cardiac Amyloidosis. J. Card. Fail. 2019, 25, 147–153. [Google Scholar] [CrossRef]
- Sinha, S.; Lopes, D.H.; Du, Z.; Pang, E.S.; Shanmugam, A.; Lomakin, A.; Talbiersky, P.; Tennstaedt, A.; McDaniel, K.; Bakshi, R.; et al. Lysine-specific molecular tweezers are broad-spectrum inhibitors of assembly and toxicity of amyloid proteins. J. Am. Chem. Soc. 2011, 133, 16958–16969. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, N.; Pereira-Henriques, A.; Attar, A.; Klärner, F.G.; Schrader, T.; Bitan, G.; Gales, L.; Saraiva, M.J.; Almeida, M.R. Molecular tweezers targeting transthyretin amyloidosis. Neurotherapeutics 2014, 11, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, N.; Saraiva, M.J.; Almeida, M.R. Natural polyphenols inhibit different steps of the process of transthyretin (TTR) amyloid fibril formation. FEBS Lett. 2011, 585, 2424–2430. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, N.; Santos, S.A.; Domingues, M.R.; Saraiva, M.J.; Almeida, M.R. Dietary curcumin counteracts extracellular transthyretin deposition: Insights on the mechanism of amyloid inhibition. Biochim. Biophys. Acta 2013, 1832, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, N.; Saraiva, M.J.; Almeida, M.R. Epigallocatechin-3-gallate as a potential therapeutic drug for TTR-related amyloidosis: “in vivo” evidence from FAP mice models. PLoS ONE 2012, 7, e29933. [Google Scholar]
- Ferreira, N.; Gonçalves, N.P.; Saraiva, M.J.; Almeida, M.R. Curcumin: A multi-target disease-modifying agent for late-stage transthyretin amyloidosis. Sci. Rep. 2016, 6, 26623. [Google Scholar] [CrossRef]
- Miroy, G.J.; Lai, Z.; Lashuel, H.A.; Peterson, S.A.; Strang, C.; Kelly, J.W. Inhibiting transthyretin amyloid fibril formation via protein stabilization. Proc. Natl. Acad. Sci. USA 1996, 93, 15051–15056. [Google Scholar] [CrossRef]
- Johnson, S.M.; Wiseman, R.L.; Sekijima, Y.; Green, N.S.; Adamski-Werner, S.L.; Kelly, J.W. Native state kinetic stabilization as a strategy to ameliorate protein misfolding diseases: A focus on the transthyretin amyloidoses. Acc. Chem. Res. 2005, 38, 911–921. [Google Scholar] [CrossRef]
- Baures, P.W.; Peterson, S.A.; Kelly, J.W. Discovering transthyretin amyloid fibril inhibitors by limited screening. Bioorganic Med. Chem. 1998, 6, 1389–1401. [Google Scholar] [CrossRef]
- McCammon, M.G.; Scott, D.J.; Keetch, C.A.; Greene, L.H.; Purkey, H.E.; Petrassi, H.M.; Kelly, J.W.; Robinson, C.V. Screening transthyretin amyloid fibril inhibitors: Characterization of novel multiprotein, multiligand complexes by mass spectrometry. Structure 2002, 10, 851–863. [Google Scholar] [CrossRef]
- Reixach, N.; Adamski-Werner, S.L.; Kelly, J.W.; Koziol, J.; Buxbaum, J.N. Cell based screening of inhibitors of transthyretin aggregation. Biochem. Biophys. Res. Commun. 2006, 348, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Kelly, J.W. Acompetition assay to identify amyloidogenesis inhibitors by monitoring the fluorescence emitted by the covalent attachment of a stilbene derivative to transthyretin. Bioorganic Med. Chem. 2011, 19, 1505–1514. [Google Scholar] [CrossRef] [PubMed]
- Alhamadsheh, M.M.; Connelly, S.; Cho, A.; Reixach, N.; Powers, E.T.; Pan, D.W.; Wilson, I.A.; Kelly, J.W.; Graef, I.A. Potent kinetic stabilizers that prevent transthyretin-mediated cardiomyocyte proteotoxicity. Sci. Transl. Med. 2011, 3, 97ra81. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.R.; Sekijima, Y.; Kelly, J.W. Native state stabilization by NSAIDs inhibits transthyretin amyloidogenesis from the most common familial disease variants. Lab. Invest. 2004, 84, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Tojo, K.; Sekijima, Y.; Kelly, J.W.; Ikeda, S. Diflunisal stabilizes familial amyloid polyneuropathy-associated transthyretin variant tetramers in serum against dissociation required for amyloidogenesis. Neurosci. Res. 2006, 56, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Sekijima, Y.; Dendle, M.A.; Kelly, J.W. Orally administered diflunisal stabilizes transthyretin against dissociation required for amyloidogenesis. Amyloid 2006, 13, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Sekijima, Y.; Tojo, K.; Morita, H.; Koyama, J.; Ikeda, S. Safety and efficacy of long-term diflunisal administration in hereditary transthyretin (ATTR) amyloidosis. Amyloid 2015, 22, 79–83. [Google Scholar] [CrossRef]
- Castaño, A.; Helmke, S.; Alvarez, J.; Delisle, S.; Maurer, M.S. Diflunisal for ATTR cardiac amyloidosis. Congest. Heart Fail. 2012, 18, 315–319. [Google Scholar] [CrossRef]
- Miller, M.; Pal, A.; Albusairi, W.; Joo, H.; Pappas, B.; Haque Tuhin, M.T.; Liang, D.; Jampala, R.; Liu, F.; Khan, J.; et al. Enthalpy-Driven Stabilization of Transthyretin by AG10 Mimics a Naturally Occurring Genetic Variant That Protects from Transthyretin Amyloidosis. J. Med. Chem. 2018, 61, 7862–7876. [Google Scholar] [CrossRef]
- Penchala, S.C.; Connelly, S.; Wang, Y.; Park, M.S.; Zhao, L.; Baranczak, A.; Rappley, I.; Vogel, H.; Liedtke, M.; Witteles, R.M.; et al. AG10 inhibits amyloidogenesis and cellular toxicity of the familial amyloid cardiomyopathy-associated V122I transthyretin. Proc. Natl. Acad. Sci. USA 2013, 110, 9992–9997. [Google Scholar] [CrossRef] [PubMed]
- Judge, D.P.; Heitner, S.B.; Falk, R.H.; Maurer, M.S.; Shah, S.J.; Witteles, R.M.; Grogan, M.; Selby, V.N.; Jacoby, D.; Hanna, M.; et al. Transthyretin Stabilization by AG10 in Symptomatic Transthyretin Amyloid Cardiomyopathy. J. Am. Coll. Cardiol. 2019, 74, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Sant’Anna, R.; Gallego, P.; Robinson, L.Z.; Pereira-Henriques, A.; Ferreira, N.; Pinheiro, F.; Esperante, S.; Pallares, I.; Huertas, O.; Almeida, M.R.; et al. Repositioning tolcapone as a potent inhibitor of transthyretin amyloidogenesis and associated cellular toxicity. Nat. Commun. 2016, 7, 10787. [Google Scholar] [CrossRef]
- Pinheiro, F.; Varejão, N.; Esperante, S.; Santos, J.; Velázquez-Campoy, A.; Reverter, D.; Pallarès, I.; Ventura, S. Tolcapone, a potent aggregation inhibitor for the treatment of familial leptomeningeal amyloidosis. FEBS J. 2021, 288, 310–324. [Google Scholar] [CrossRef] [PubMed]
- Gamez, J.; Salvadó, M.; Reig, N.; Suñé, P.; Casasnovas, C.; Rojas-Garcia, R.; Insa, R. Transthyretin stabilization activity of the catechol-O-methyltransferase inhibitor tolcapone (SOM0226) in hereditary ATTR amyloidosis patients and asymptomatic carriers: Proof-of-concept study. Amyloid 2019, 26, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.M.; Connelly, S.; Fearns, C.; Powers, E.T.; Kelly, J.W. The transthyretin amyloidoses: From delineating the molecular mechanism of aggregation linked to pathology to a regulatory-agency-approved drug. J. Mol. Biol. 2012, 421, 185–203. [Google Scholar] [CrossRef]
- Tsuchiya, A.; Yazaki, M.; Kametani, F.; Takei, Y.; Ikeda, S. Marked regression of abdominal fat amyloid in patients with familial amyloid polyneuropathy during long-term follow-up after liver transplantation. Liver Transpl. 2008, 14, 563–570. [Google Scholar] [CrossRef]
- Groothof, D.; Nienhuis, H.L.A.; Bijzet, J.; Houwerzijl, E.J.; van den Berg, M.P.; Glaudemans, A.; Slart, R.; Hazenberg, B.P.C. Regression of Bone-Tracer Uptake in Cardiac Transthyretin Amyloidosis. Mayo Clin. Proc. 2020, 95, 417–418. [Google Scholar] [CrossRef] [PubMed]
- Finn, J.D.; Smith, A.R.; Patel, M.C.; Shaw, L.; Youniss, M.R.; van Heteren, J.; Dirstine, T.; Ciullo, C.; Lescarbeau, R.; Seitzer, J.; et al. Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018, 22, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Episkopou, V.; Maeda, S.; Nishiguchi, S.; Shimada, K.; Gaitanaris, G.A.; Gottesman, M.E.; Robertson, E.J. Disruption of the transthyretin gene results in mice with depressed levels of plasma retinol and thyroid hormone. Proc. Natl. Acad. Sci. USA 1993, 90, 2375–2379. [Google Scholar] [CrossRef] [PubMed]
- Liz, M.A.; Faro, C.J.; Saraiva, M.J.; Sousa, M.M. Transthyretin, a new cryptic protease. J. Biol. Chem. 2004, 279, 21431–21438. [Google Scholar] [CrossRef] [PubMed]
- Liz, M.A.; Fleming, C.E.; Nunes, A.F.; Almeida, M.R.; Mar, F.M.; Choe, Y.; Craik, C.S.; Powers, J.C.; Bogyo, M.; Sousa, M.M. Substrate specificity of transthyretin: Identification of natural substrates in the nervous system. Biochem. J. 2009, 419, 467–474. [Google Scholar] [CrossRef]
- Liz, M.A.; Mar, F.M.; Franquinho, F.; Sousa, M.M. Aboard transthyretin: From transport to cleavage. IUBMB Life 2010, 62, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.D.; Lambertsen, K.L.; Clausen, B.H.; Akinc, A.; Alvarez, R.; Finsen, B.; Saraiva, M.J. CSFtransthyretin neuroprotection in a mouse model of brain ischemia. J. Neurochem. 2010, 115, 1434–1444. [Google Scholar] [CrossRef]
- Misumi, Y.; Ando, Y.; Gonçalves, N.P.; Saraiva, M.J. Fibroblasts endocytose and degrade transthyretin aggregates in transthyretin-related amyloidosis. Lab. Invest. 2013, 93, 911–920. [Google Scholar] [CrossRef]
- Hosoi, A.; Su, Y.; Torikai, M.; Jono, H.; Ishikawa, D.; Soejima, K.; Higuchi, H.; Guo, J.; Ueda, M.; Suenaga, G.; et al. Novel Antibody for the Treatment of Transthyretin Amyloidosis. J. Biol. Chem. 2016, 291, 25096–25105. [Google Scholar] [CrossRef]
- George, J.; Rappaport, M.; Shimoni, S.; Goland, S.; Voldarsky, I.; Fabricant, Y.; Edri, O.; Cuciuc, V.; Lifshitz, S.; Tshori, S.; et al. Anovel monoclonal antibody targeting aggregated transthyretin facilitates its removal and functional recovery in an experimental model. Eur. Heart J. 2020, 41, 1260–1270. [Google Scholar] [CrossRef]
- Michalon, A.; Hagenbuch, A.; Huy, C.; Varela, E.; Combaluzier, B.; Damy, T.; Suhr, O.B.; Saraiva, M.J.; Hock, C.; Nitsch, R.M.; et al. Ahuman antibody selective for transthyretin amyloid removes cardiac amyloid through phagocytic immune cells. Nat. Commun. 2021, 12, 3142. [Google Scholar] [CrossRef] [PubMed]
- Higaki, J.N.; Chakrabartty, A.; Galant, N.J.; Hadley, K.C.; Hammerson, B.; Nijjar, T.; Torres, R.; Tapia, J.R.; Salmans, J.; Barbour, R.; et al. Novel conformation-specific monoclonal antibodies against amyloidogenic forms of transthyretin. Amyloid 2016, 23, 86–97. [Google Scholar] [CrossRef]
- Goldsteins, G.; Persson, H.; Andersson, K.; Olofsson, A.; Dacklin, I.; Edvinsson, A.; Saraiva, M.J.; Lundgren, E. Exposure of cryptic epitopes on transthyretin only in amyloid and in amyloidogenic mutants. Proc. Natl. Acad. Sci. USA 1999, 96, 3108–3113. [Google Scholar] [CrossRef]
- Carvalho, A.; Rocha, A.; Lobato, L. Liver transplantation in transthyretin amyloidosis: Issues and challenges. Liver Transpl. 2015, 21, 282–292. [Google Scholar] [CrossRef]
- Gustafsson, S.; Ihse, E.; Henein, M.Y.; Westermark, P.; Lindqvist, P.; Suhr, O.B. Amyloid fibril composition as a predictor of development of cardiomyopathy after liver transplantation for hereditary transthyretin amyloidosis. Transplantation 2012, 93, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Stangou, A.J.; Hawkins, P.N.; Heaton, N.D.; Rela, M.; Monaghan, M.; Nihoyannopoulos, P.; O’Grady, J.; Pepys, M.B.; Williams, R. Progressive cardiac amyloidosis following liver transplantation for familial amyloid polyneuropathy: Implications for amyloid fibrillogenesis. Transplantation 1998, 66, 229–233. [Google Scholar] [CrossRef]
- Yazaki, M.; Tokuda, T.; Nakamura, A.; Higashikata, T.; Koyama, J.; Higuchi, K.; Harihara, Y.; Baba, S.; Kametani, F.; Ikeda, S. Cardiac amyloid in patients with familial amyloid polyneuropathy consists of abundant wild-type transthyretin. Biochem. Biophys. Res. Commun. 2000, 274, 702–706. [Google Scholar] [CrossRef] [PubMed]
- Saelices, L.; Chung, K.; Lee, J.H.; Cohn, W.; Whitelegge, J.P.; Benson, M.D.; Eisenberg, D.S. Amyloid seeding of transthyretin by ex vivo cardiac fibrils and its inhibition. Proc. Natl. Acad. Sci. USA 2018, 115, E6741–E6750. [Google Scholar] [CrossRef]
- Saelices, L.; Nguyen, B.A.; Chung, K.; Wang, Y.; Ortega, A.; Lee, J.H.; Coelho, T.; Bijzet, J.; Benson, M.D.; Eisenberg, D.S. Apair of peptides inhibits seeding of the hormone transporter transthyretin into amyloid fibrils. J. Biol. Chem. 2019, 294, 6130–6141. [Google Scholar] [CrossRef] [PubMed]
- Jaccard, A.; Moreau, P.; Leblond, V.; Leleu, X.; Benboubker, L.; Hermine, O.; Recher, C.; Asli, B.; Lioure, B.; Royer, B.; et al. High-dose melphalan versus melphalan plus dexamethasone for AL amyloidosis. N. Engl. J. Med. 2007, 357, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Cibeira, M.T.; Sanchorawala, V.; Seldin, D.C.; Quillen, K.; Berk, J.L.; Dember, L.M.; Segal, A.; Ruberg, F.; Meier-Ewert, H.; Andrea, N.T.; et al. Outcome of AL amyloidosis after high-dose melphalan and autologous stem cell transplantation: Long-term results in a series of 421 patients. Blood 2011, 118, 4346–4352. [Google Scholar] [CrossRef]
- Wechalekar, A.D.; Gillmore, J.D.; Bird, J.; Cavenagh, J.; Hawkins, S.; Kazmi, M.; Lachmann, H.J.; Hawkins, P.N.; Pratt, G. Guidelines on the management of AL amyloidosis. Br. J. Haematol. 2015, 168, 186–206. [Google Scholar] [CrossRef]
- Bianchi, G.; Zhang, Y.; Comenzo, R.L. ALAmyloidosis: Current Chemotherapy and Immune Therapy Treatment Strategies: JACC: CardioOncology State-of-the-Art Review. JACC Cardio Oncol. 2021, 3, 467–487. [Google Scholar] [CrossRef]
- Seckinger, A.; Hillengass, J.; Emde, M.; Beck, S.; Kimmich, C.; Dittrich, T.; Hundemer, M.; Jauch, A.; Hegenbart, U.; Raab, M.S.; et al. CD38 as Immunotherapeutic Target in Light Chain Amyloidosis and Multiple Myeloma-Association With Molecular Entities, Risk, Survival, and Mechanisms of Upfront Resistance. Front. Immunol. 2018, 9, 1676. [Google Scholar] [CrossRef]
- Sanchez, L.; Wang, Y.; Siegel, D.S.; Wang, M.L. Daratumumab: A first-in-class CD38 monoclonal antibody for the treatment of multiple myeloma. J. Hematol. Oncol. 2016, 9, 51. [Google Scholar] [CrossRef]
- Lin, P.; Owens, R.; Tricot, G.; Wilson, C.S. Flow cytometric immunophenotypic analysis of 306 cases of multiple myeloma. Am. J. Clin. Pathol. 2004, 121, 482–488. [Google Scholar] [CrossRef]
- van Gameren, I.I.; van Rijswijk, M.H.; Bijzet, J.; Vellenga, E.; Hazenberg, B.P. Histological regression of amyloid in AL amyloidosis is exclusively seen after normalization of serum free light chain. Haematologica 2009, 94, 1094–1100. [Google Scholar] [CrossRef]
- Brahmanandam, V.; McGraw, S.; Mirza, O.; Desai, A.A.; Farzaneh-Far, A. Regression of cardiac amyloidosis after stem cell transplantation assessed by cardiovascular magnetic resonance imaging. Circulation 2014, 129, 2326–2328. [Google Scholar] [CrossRef]
- Katoh, N.; Matsushima, A.; Kurozumi, M.; Matsuda, M.; Ikeda, S. Marked and rapid regression of hepatic amyloid deposition in a patient with systemic light chain (AL) amyloidosis after high-dose melphalan therapy with stem cell transplantation. Intern. Med. 2014, 53, 1991–1995. [Google Scholar] [CrossRef]
- Martinez-Naharro, A.; Abdel-Gadir, A.; Treibel, T.A.; Zumbo, G.; Knight, D.S.; Rosmini, S.; Lane, T.; Mahmood, S.; Sachchithanantham, S.; Whelan, C.J.; et al. CMR-Verified Regression of Cardiac AL Amyloid After Chemotherapy. JACC Cardiovasc. Imaging 2018, 11, 152–154. [Google Scholar] [CrossRef]
- Ward, J.E.; Ren, R.; Toraldo, G.; Soohoo, P.; Guan, J.; O’Hara, C.; Jasuja, R.; Trinkaus-Randall, V.; Liao, R.; Connors, L.H.; et al. Doxycycline reduces fibril formation in a transgenic mouse model of AL amyloidosis. Blood 2011, 118, 6610–6617. [Google Scholar] [CrossRef]
- Kumar, S.K.; Dispenzieri, A.; Lacy, M.Q.; Hayman, S.R.; Buadi, F.K.; Dingli, D.; Zeldenrust, S.R.; Ramirez-Alvarado, M.; Kapoor, P.; Hogan, W.; et al. Doxycycline Used As Post Transplant Antibacterial Prophylaxis Improves Survival in Patients with Light Chain Amyloidosis Undergoing Autologous Stem Cell Transplantation. Blood 2012, 120, 3138. [Google Scholar] [CrossRef]
- Wechalekar, A.; Whelan, C.; Sachchithanantham, S.; Fontana, M.; Mahmood, S.; Foard, D.; Lane, T.; Lachmann, H.J.; Gillmore, J.D.; Hawkins, P.N. AMatched Case Control Study of Doxycycline Added to Chemotherapy for Reducing Early Mortality in Patients with Advanced Cardiac AL Amyloidosis from the Alchemy Study Cohort. Blood 2014, 124, 3485. [Google Scholar] [CrossRef]
- D’Souza, A.; Szabo, A.; Flynn, K.E.; Dhakal, B.; Chhabra, S.; Pasquini, M.C.; Weihrauch, D.; Hari, P.N. Adjuvant doxycycline to enhance anti-amyloid effects: Results from the dual phase 2 trial. EClinicalMedicine 2020, 23, 100361. [Google Scholar] [CrossRef] [PubMed]
- Bodin, K.; Ellmerich, S.; Kahan, M.C.; Tennent, G.A.; Loesch, A.; Gilbertson, J.A.; Hutchinson, W.L.; Mangione, P.P.; Gallimore, J.R.; Millar, D.J.; et al. Antibodies to human serum amyloid P component eliminate visceral amyloid deposits. Nature 2010, 468, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Richards, D.B.; Cookson, L.M.; Berges, A.C.; Barton, S.V.; Lane, T.; Ritter, J.M.; Fontana, M.; Moon, J.C.; Pinzani, M.; Gillmore, J.D.; et al. Therapeutic Clearance of Amyloid by Antibodies to Serum Amyloid P Component. N. Engl. J. Med. 2015, 373, 1106–1114. [Google Scholar] [CrossRef]
- Gertz, M.A.; Landau, H.; Comenzo, R.L.; Seldin, D.; Weiss, B.; Zonder, J.; Merlini, G.; Schönland, S.; Walling, J.; Kinney, G.G.; et al. First-in-Human Phase I/II Study of NEOD001 in Patients With Light Chain Amyloidosis and Persistent Organ Dysfunction. J. Clin. Oncol. 2016, 34, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Renz, M.; Torres, R.; Dolan, P.J.; Tam, S.J.; Tapia, J.R.; Li, L.; Salmans, J.R.; Barbour, R.M.; Shughrue, P.J.; Nijjar, T.; et al. 2A4 binds soluble and insoluble light chain aggregates from AL amyloidosis patients and promotes clearance of amyloid deposits by phagocytosis. Amyloid 2016, 23, 168–177. [Google Scholar] [CrossRef]
- Varga, C.; Lentzsch, S.; Comenzo, R.L. Beyond NEOD001 for systemic light-chain amyloidosis. Blood 2018, 132, 1992–1993. [Google Scholar] [CrossRef] [PubMed]
- Hrncic, R.; Wall, J.; Wolfenbarger, D.A.; Murphy, C.L.; Schell, M.; Weiss, D.T.; Solomon, A. Antibody-mediated resolution of light chain-associated amyloid deposits. Am. J. Pathol. 2000, 157, 1239–1246. [Google Scholar] [CrossRef]
- Edwards, C.V.; Gould, J.; Langer, A.L.; Mapara, M.Y.; Radhakrishnan, J.; Maurer, M.S.; Raza, S.; Mears, J.G.; Leng, S.; Wall, J.S.; et al. Final Analysis of the Phase 1a/b Study of Chimeric Fibril-Reactive Monoclonal Antibody 11-1F4 in Patients with Relapsed or Refractory AL Amyloidosis. Blood 2017, 130 (Suppl. 1), 509. [Google Scholar]
- Bhutani, D.; Leng, S.; Eisenberger, A.; Maurer, M.S.; Shames, S.; Goldsmith, J.; Lentzsch, S. Improvement in Global Longitudinal Strain (GLS) Correlates with NT-Probnp Response in Patients with Cardiac Amyloidosis Treated on a Phase 1b Study of Anti-Amyloid Mab Cael-101. Blood 2018, 132 (Suppl. 1), 958. [Google Scholar] [CrossRef]
- Pepys, M.B.; Dyck, R.F.; de Beer, F.C.; Skinner, M.; Cohen, A.S. Binding of serum amyloid P-component (SAP) by amyloid fibrils. Clin. Exp. Immunol. 1979, 38, 284–293. [Google Scholar] [PubMed]
- Morgan, G.J.; Yan, N.L.; Mortenson, D.E.; Rennella, E.; Blundon, J.M.; Gwin, R.M.; Lin, C.Y.; Stanfield, R.L.; Brown, S.J.; Rosen, H.; et al. Stabilization of amyloidogenic immunoglobulin light chains by small molecules. Proc. Natl. Acad. Sci. USA 2019, 116, 8360–8369. [Google Scholar] [CrossRef] [PubMed]
| Precursor Protein | Underlying Condition | Cardiac Involvement | Treatment | |
|---|---|---|---|---|
| ATTRwt | Wild-type transthyretin | Aging | ++ | TTR tetramer stabilizer |
| ATTRv | Abnormal transthyretin | TTR gene variant | + | Liver transplantation Transthyretin tetramer stabilizer siRNA/antisense oligomer |
| AL | Immunoglobulin light chain | Plasma cell abnormality | ++ | Chemotherapy Autologous stem cell transplantation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saito, Y.; Nakamura, K.; Ito, H. Molecular Mechanisms of Cardiac Amyloidosis. Int. J. Mol. Sci. 2022, 23, 25. https://doi.org/10.3390/ijms23010025
Saito Y, Nakamura K, Ito H. Molecular Mechanisms of Cardiac Amyloidosis. International Journal of Molecular Sciences. 2022; 23(1):25. https://doi.org/10.3390/ijms23010025
Chicago/Turabian StyleSaito, Yukihiro, Kazufumi Nakamura, and Hiroshi Ito. 2022. "Molecular Mechanisms of Cardiac Amyloidosis" International Journal of Molecular Sciences 23, no. 1: 25. https://doi.org/10.3390/ijms23010025
APA StyleSaito, Y., Nakamura, K., & Ito, H. (2022). Molecular Mechanisms of Cardiac Amyloidosis. International Journal of Molecular Sciences, 23(1), 25. https://doi.org/10.3390/ijms23010025







