The Ceramide Synthase Subunit Lac1 Regulates Cell Growth and Size in Fission Yeast
Abstract
1. Introduction
2. Results
2.1. Fission Yeast Ceramide Synthases Are Conserved and Localize at the ER
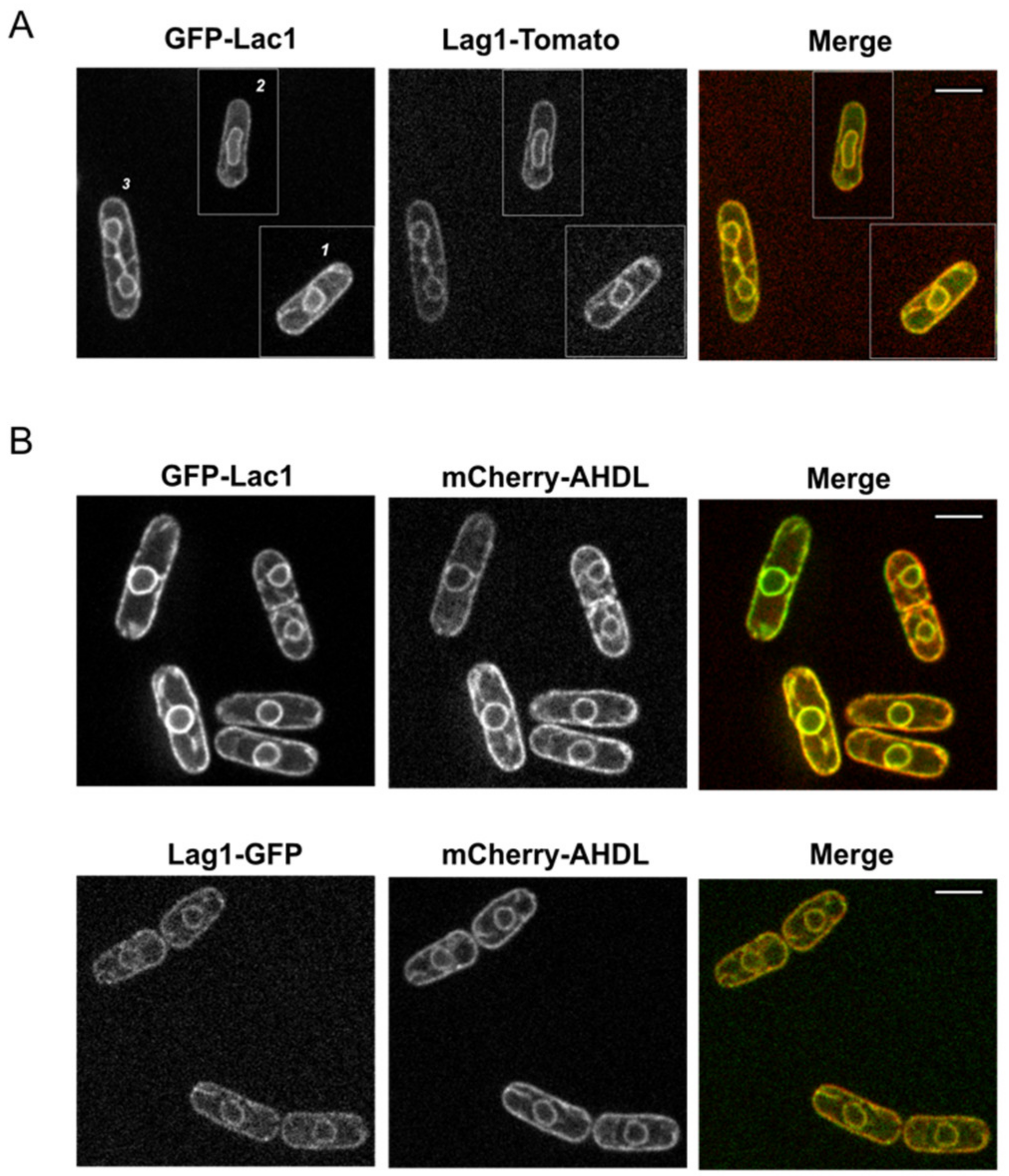
2.2. Fission Yeast Lac1 and Lag1 Co-Localize at the Endoplasmic Reticulum
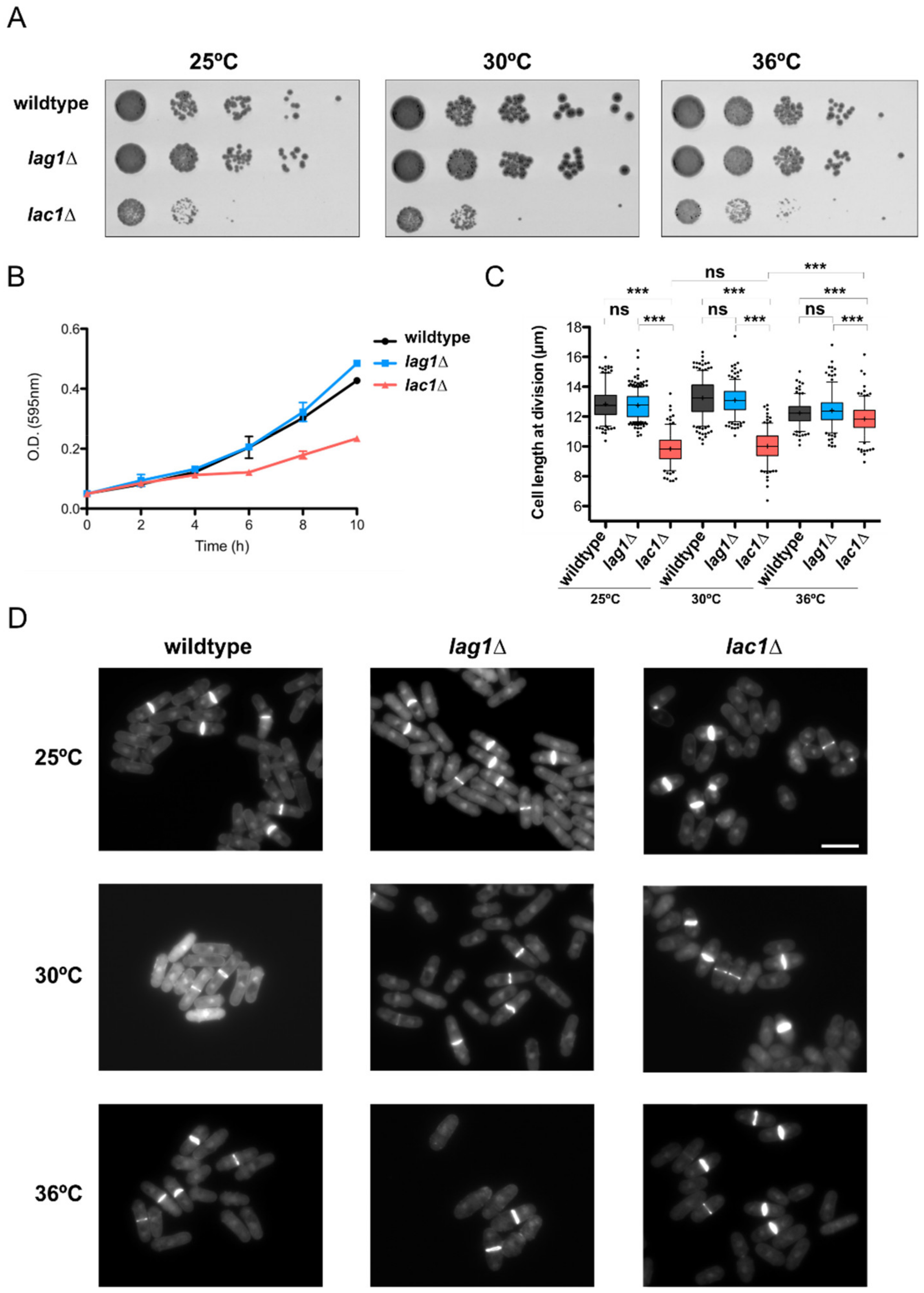
2.3. Lac1 Is Implicated in Control of Cell Growth and Size in Fission Yeast
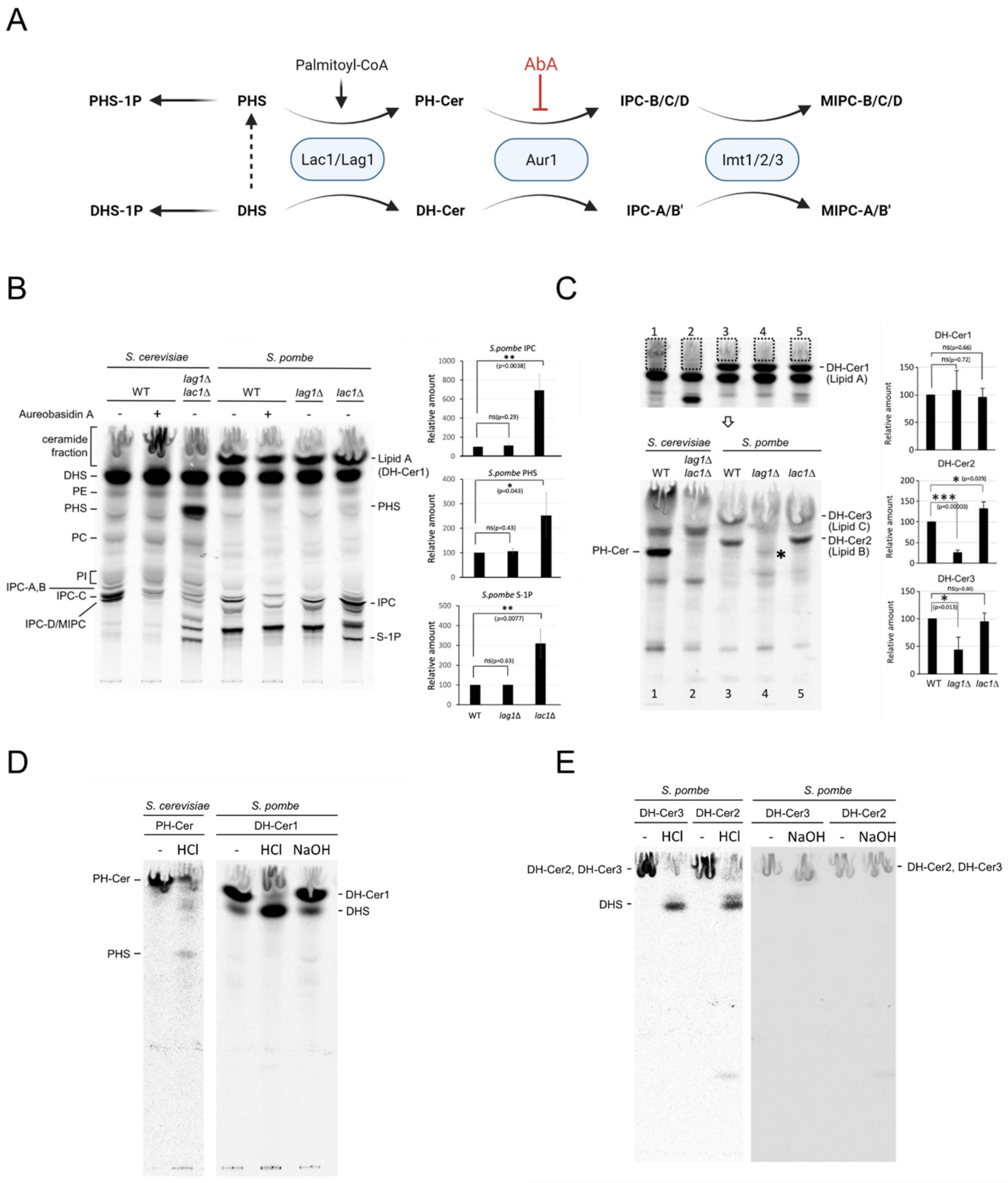
2.4. Lac1 and Lag1 Generate Different Species of Ceramides and Complex Sphingolipids
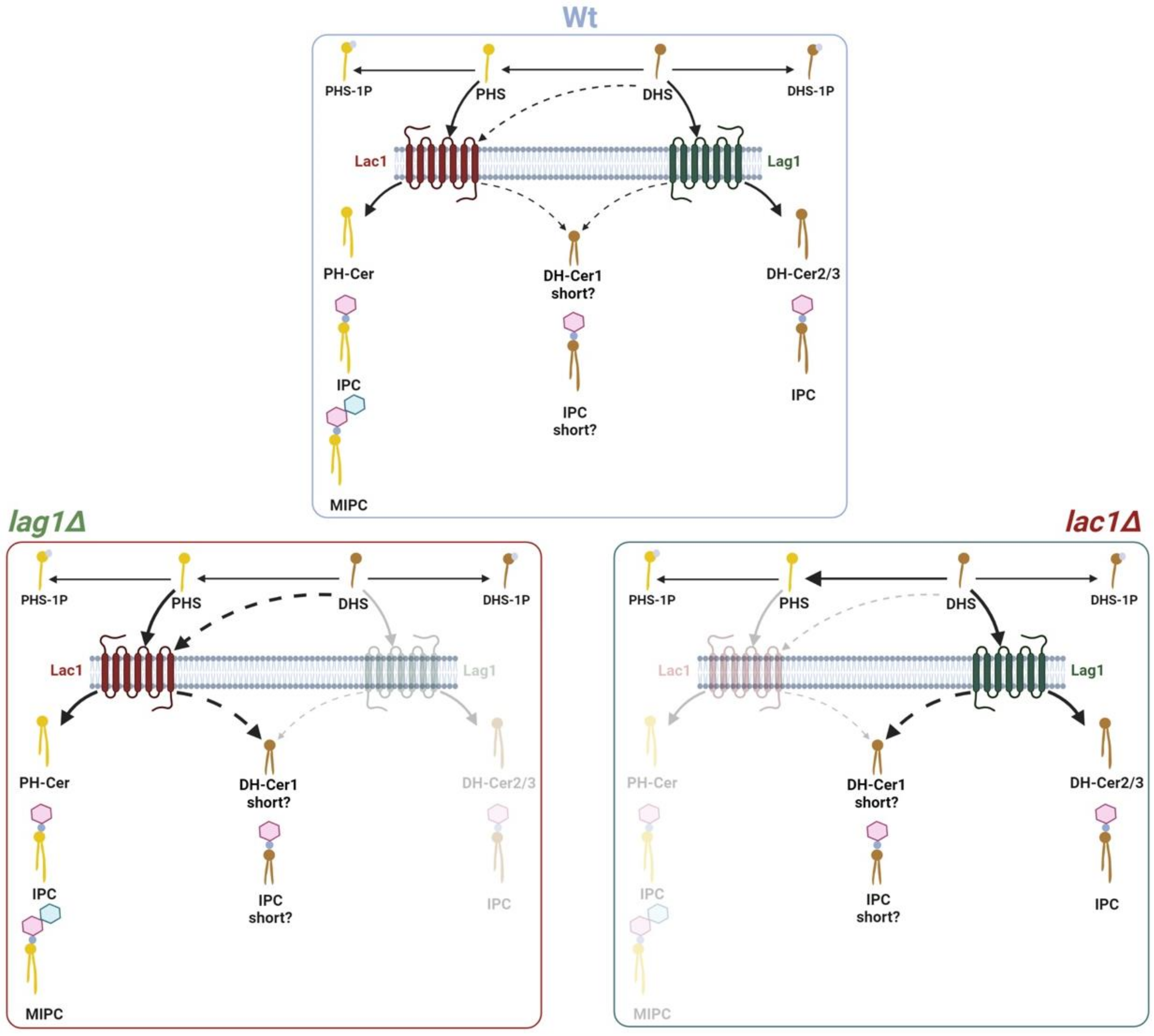
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids and Media
4.2. Sequence Alignment
4.3. Microscopy Analysis
4.4. Native Co-Immunoprecipitation
4.5. Lipid Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schaechter, M.; Maaløe, O.; Kjeldgaard, N.O. Dependency on Medium and Temperature of Cell Size and Chemical Composition during Balanced Growth of Salmonella Typhimurium. J. Gen. Microbiol. 1958, 19, 592–606. [Google Scholar] [CrossRef]
- Johnston, G.C.; Pringle, J.R.; Hartwell, L.H. Coordination of Growth with Cell Division in the Yeast Saccharomyces cerevisiae. Exp. Cell Res. 1977, 105, 79–98. [Google Scholar] [CrossRef]
- Hirsch, J.; Han, P.W. Effects of Growth, Starvation, and Obesity. Lipids 1969, 10, 77–82. [Google Scholar]
- Fantes, P.; Nurse, P. Control of Cell Size at Division in Fission Yeast by a Growth-Modulated Size Control over Nuclear Division. Exp. Cell Res. 1977, 107, 377–386. [Google Scholar] [CrossRef]
- Lucena, R.; Alcaide-Gavilán, M.; Schubert, K.; He, M.; Domnauer, M.G.; Marquer, C.; Klose, C.; Surma, M.A.; Kellogg, D.R. Cell Size and Growth Rate Are Modulated by TORC2-Dependent Signals. Curr. Biol. 2018, 28, 196–210. [Google Scholar] [CrossRef]
- Dickson, R.C. Thematic Review Series: Sphingolipids. New Insights into Sphingolipid Metabolism and Function in Budding Yeast. J. Lipid Res. 2008, 49, 909–921. [Google Scholar] [CrossRef]
- Megyeri, M.; Riezman, H.; Schuldiner, M.; Futerman, A.H. Making Sense of the Yeast Sphingolipid Pathway. J. Mol. Biol. 2016, 428, 4765–4775. [Google Scholar] [CrossRef]
- Guillas, I.; Kirchman, P.A.; Chuard, R.; Pfefferli, M.; Jiang, J.C.; Jazwinski, S.M.; Conzelmann, A. C26-CoA-Dependent Ceramide Synthesis of Saccharomyces cerevisiae Is Operated by Lag1p and Lac1p. EMBO J. 2001, 20, 2655–2665. [Google Scholar] [CrossRef]
- Schorling, S.; Vallee, B.; Barz, W.P.; Riezman, H.; Oesterhelt, D. Lag1p and Lac1p Are Essential for the Acyl-CoA-Dependent Ceramide Synthase Reaction in Saccharomyces cerevisae. Mol. Biol. Cell 2001, 12, 3417–3427. [Google Scholar] [CrossRef]
- Vallée, B.; Riezman, H. Lip1p: A Novel Subunit of Acyl-CoA Ceramide Synthase. EMBO J. 2005, 24, 730–741. [Google Scholar] [CrossRef]
- Funato, K.; Riezman, H. Vesicular and Nonvesicular Transport of Ceramide from ER to the Golgi Apparatus in Yeast. J. Cell Biol. 2001, 155, 949–959. [Google Scholar] [CrossRef]
- Jiang, J.C.; Kirchman, P.A.; Zagulski, M.; Hunt, J.; Jazwinski, S.M. Homologs of the Yeast Longevity Gene LAG1 in Caenorhabditis Elegans and Human. Genome Res. 1998, 8, 1259–1272. [Google Scholar] [CrossRef]
- Megyeri, M.; Prasad, R.; Volpert, G.; Sliwa-Gonzalez, A.; Haribowo, A.G.; Aguilera-Romero, A.; Riezman, H.; Barral, Y.; Futerman, A.H.; Schuldiner, M. Yeast Ceramide Synthases, Lag1 and Lac1, Have Distinct Substrate Specificity. J. Cell Sci. 2019, 132, jcs228411. [Google Scholar] [CrossRef]
- Mitchison, J.M.; Nurse, P. Growth in Cell Length in the Fission Yeast Schizosaccharomyces Pombe. J. Cell Sci. 1985, 75, 357–376. [Google Scholar] [CrossRef] [PubMed]
- Wood, E.; Nurse, P. Sizing Up to Divide: Mitotic Cell Size Control in Fission Yeast. Annu. Rev. Cell Dev. Biol. 2014, 31, 11–29. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.G.; Berthelot-grosjean, M. Polar Gradients of the DYRK-Family Kinase Pom1 Couple Cell Length with the Cell Cycle. Nature 2009, 459, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Moseley, J.B.; Mayeux, A.; Paoletti, A.; Nurse, P. A Spatial Gradient Coordinates Cell Size and Mitotic Entry in Fission Yeast. Nature 2009, 459, 857–860. [Google Scholar] [CrossRef]
- Pan, K.Z.; Saunders, T.E.; Flor-Parra, I.; Howard, M.; Chang, F. Cortical Regulation of Cell Size by a Sizer Cdr2p. eLife 2014, 3, e02040. [Google Scholar] [CrossRef]
- Wood, E.; Nurse, P. Pom1 and Cell Size Homeostasis in Fission Yeast. Cell Cycle 2013, 12, 3228–3236. [Google Scholar] [CrossRef]
- Keifenheim, D.; Sun, X.-M.; D’Souza, E.; Ohira, M.J.; Magner, M.; Mayhew, M.B.; Marguerat, S.; Rhind, N. Size-Dependent Expression of the Mitotic Activator Cdc25 Suggests a Mechanism of Size Control in Fission Yeast. Curr. Biol. 2017, 27, 1491–1497.e4. [Google Scholar] [CrossRef]
- Facchetti, G.; Knapp, B.; Flor-Parra, I.; Chang, F.; Correspondence, M.H. Reprogramming Cdr2-Dependent Geometry-Based Cell Size Control in Fission Yeast. Curr. Biol. 2018, 29, 350–358.e4. [Google Scholar] [CrossRef] [PubMed]
- Mandon, E.C.; Ehses, I.; Rother, J.; van Echten, G.; Sandhoff, K. Subcellular Localization and Membrane Topology of Serine Palmitoyltransferase, 3-Dehydrosphinganine Reductase, and Sphinganine N-Acyltransferase in Mouse Liver. J. Biol. Chem. 1992, 267, 11144–11148. [Google Scholar] [CrossRef]
- Barz, W.P.; Walter, P. Two Endoplasmic Reticulum (ER) Membrane Proteins That Facilitate ER-to-Golgi Transport of Glycosylphosphatidylinositol-Anchored Proteins. Mol. Biol. Cell 1999, 10, 1043–1059. [Google Scholar] [CrossRef] [PubMed]
- Fresques, T.; Niles, B.; Aronova, S.; Mogri, H.; Rakhshandehroo, T.; Powers, T. Regulation of Ceramide Synthase by Casein Kinase 2-Dependent Phosphorylation in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 1395–1403. [Google Scholar] [CrossRef]
- Forsburg, S.L. Comparison of Schizosaccharomyces Pombe Expression Systems. Nucleic Acids Res. 1993, 21, 2955–2956. [Google Scholar] [CrossRef]
- Zhang, D.; Vjestica, A.; Oliferenko, S. The Cortical ER Network Limits the Permissive Zone for Actomyosin Ring Assembly. Curr. Biol. 2010, 20, 1029–1034. [Google Scholar] [CrossRef]
- Dawson, K.; Toone, W.M.; Jones, N.; Wilkinson, C.R.M. Loss of Regulators of Vacuolar ATPase Function and Ceramide Synthesis Results in Multidrug Sensitivity in Schizosaccharomyces Pombe. Eukaryot. Cell 2008, 7, 926–937. [Google Scholar] [CrossRef]
- Mao, C.; Wadleigh, M.; Jenkins, G.M.; Hannun, Y.A.; Obeid, L.M. Identification and Characterization of Saccharomyces cerevisiae Dihydrosphingosine-1-Phosphate Phosphatase. J. Biol. Chem. 1997, 272, 28690–28694. [Google Scholar] [CrossRef]
- Mandala, S.M.; Thornton, R.; Tu, Z.; Kurtz, M.B.; Nickels, J.; Broach, J.; Menzeleev, R.; Spiegel, S. Sphingoid Base 1-Phosphate Phosphatase: A Key Regulator of Sphingolipid Metabolism and Stress Response. Proc. Natl. Acad. Sci. USA 1998, 95, 150–155. [Google Scholar] [CrossRef]
- Kolaczkowski, M.; Kolaczkowska, A.; Gaigg, B.; Schneiter, R.; Moye-Rowley, W.S. Differential Regulation of Ceramide Synthase Components LAC1 and LAG1 in Saccharomyces cerevisiae. Eukaryot. Cell 2004, 3, 880–892. [Google Scholar] [CrossRef]
- Muir, A.; Ramachandran, S.; Roelants, F.M.; Timmons, G.; Thorner, J. TORC2-Dependent Protein Kinase Ypk1 Phosphorylates Ceramide Synthase to Stimulate Synthesis of Complex Sphingolipids. eLife 2014, 3, e03779. [Google Scholar] [CrossRef]
- Wells, G.B.; Dickson, R.C.; Lester, R.L. Heat-Induced Elevation of Ceramide in Saccharomyces cerevisiae via De Novo Synthesis. J. Biol. Chem. 1998, 273, 7235–7243. [Google Scholar] [CrossRef]
- Klose, C.; Surma, M.A.; Gerl, M.J.; Meyenhofer, F.; Shevchenko, A.; Simons, K. Flexibility of a Eukaryotic Lipidome—Insights from Yeast Lipidomics. PLoS ONE 2012, 7, e35063. [Google Scholar] [CrossRef] [PubMed]
- Epstein, S.; Castillon, G.A.; Qin, Y.; Riezman, H. An Essential Function of Sphingolipids in Yeast Cell Division. Mol. Microbiol. 2012, 84, 1018–1032. [Google Scholar] [CrossRef]
- Shui, G.; Guan, X.L.; Low, C.P.; Chua, G.H.; Goh, J.S.Y.; Yang, H.; Wenk, M.R. Toward One Step Analysis of Cellular Lipidomes Using Liquid Chromatography Coupled with Mass Spectrometry: Application to Saccharomyces cerevisiae and Schizosaccharomyces pombe Lipidomics. Mol. Biosyst. 2010, 6, 1008–1017. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, T. Sphingolipids from the Human Fungal Pathogen Aspergillus fumigatus. Biochimie 2017, 141, 9–15. [Google Scholar] [CrossRef]
- Fernandes, C.M.; Goldman, G.H.; Del Poeta, M. Biological Roles Played by Sphingolipids in Dimorphic and Filamentous Fungi. mBio 2018, 9, e00642-18. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.; Klar, A.; Nurse, P. Guide to Yeast Genetics and Molecular Biology. Methods Enzymol. 1991, 194, 795–823. [Google Scholar] [CrossRef] [PubMed]
- Bähler, J.; Wu, J.-Q.; Longtine, M.S.; Shah, N.G.; Mckenzie, A., III; Steever, A.B.; Wach, A.; Philippsen, P.; Pringle, J.R. Heterologous Modules for Efficient and Versatile PCR-Based Gene Targeting InSchizosaccharomyces Pombe. Yeast 1998, 14, 943–951. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; The UniProt Consortium; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering Key Features in Protein Structures with the New ENDscript Server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.M.; Hong, E.L.; Amundsen, C.; Balakrishnan, R.; Binkley, G.; Chan, E.T.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; et al. Saccharomyces Genome Database: The Genomics Resource of Budding Yeast. Nucleic Acids Res. 2012, 40, D700–D705. [Google Scholar] [CrossRef] [PubMed]
- Lock, A.; Rutherford, K.; Harris, M.A.; Hayles, J.; Oliver, S.G.; Bähler, J.; Wood, V. PomBase 2018: User-Driven Reimplementation of the Fission Yeast Database Provides Rapid and Intuitive Access to Diverse, Interconnected Information. Nucleic Acids Res. 2019, 47, D821–D827. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Hagan, I.M. Chromatin and Cell Wall Staining of Schizosaccharomyces pombe. Cold Spring Harb. Protoc. 2016, 2016, 549–553. [Google Scholar] [CrossRef]
- Rodriguez-Gallardo, S.; Kurokawa, K.; Sabido-Bozo, S.; Cortes-Gomez, A.; Ikeda, A.; Zoni, V.; Aguilera-Romero, A.; Perez-Linero, A.M.; Lopez, S.; Waga, M.; et al. Ceramide Chain Length–Dependent Protein Sorting into Selective Endoplasmic Reticulum Exit Sites. Sci. Adv. 2020, 6, eaba8237. [Google Scholar] [CrossRef]
- Lucena, R.; Alcaide-Gavilán, M.; Anastasia, S.D.; Kellogg, D.R. Wee1 and Cdc25 Are Controlled by Conserved PP2A-Dependent Mechanisms in Fission Yeast. Cell Cycle 2017, 16, 428–435. [Google Scholar] [CrossRef]
- Ikeda, A.; Hanaoka, K.; Funato, K. Protocol for Measuring Sphingolipid Metabolism in Budding Yeast. STAR Protoc. 2021, 2, 100412. [Google Scholar] [CrossRef]


| Strain | Genotype | Source |
|---|---|---|
| NJ1 | h+ his7-366, ade6-M210 | Nick Jones lab |
| NJ4 | h+ lag1::KanMX his7-366, ade6-M210 | Nick Jones lab |
| NJ5 | h- lac1::KanMX his7-366, ade6-M210 | Nick Jones lab |
| FYL2 | h- ade6-M210 leu1-32 ura4-D18 | This study |
| FYL19 | h+ lag1-GFP:KanMX lac1::KanMX leu1-32 ura4-D18 | This study |
| FYL27 | h+ lag1::HphMX his7-366, ade6-M210 | This study |
| FYL42 | h- lag1-3xHA:KanMX ade6-M210 leu1-32 ura4-D18 | This study |
| FYL46 | h- lac1::KanMX leu1-32 ura4-D18 | This study |
| FYL48 | h- lag1::NatMX leu1-32 ura4-D18 | This study |
| FYL51 | h+ KanMX:nmt1-GFP-lac1 leu1-32 ura4-D18 | This study |
| FYL52 | h- lag1-GFP:KanMX pBip1-mCherry-AHDL:Leu1 ade6-M210 leu1-32 ura4-D18 | This study |
| FYL65 | h- KanMX:nmt1-GFP-lac1 lag1-mTomato:NatMX ade6-M210 leu1-32 ura4-D18 | This study |
| FYL67 | h- KanMX:nmt1-GFP-lac1 pBip1-mCherry-AHDL:Leu1 ade6-M210 leu1-32 ura4-D18 | This study |
| FYL69 | h- KanMX:nmt1-GFP-lac1 lag1-3xHA:KanMX leu1-32 ura4-D18 | This study |
| FYL71 | h- KanMX:nmt1-GFP-lac1 lag1::HphMX leu1-32 ura4-D18 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flor-Parra, I.; Sabido-Bozo, S.; Ikeda, A.; Hanaoka, K.; Aguilera-Romero, A.; Funato, K.; Muñiz, M.; Lucena, R. The Ceramide Synthase Subunit Lac1 Regulates Cell Growth and Size in Fission Yeast. Int. J. Mol. Sci. 2022, 23, 303. https://doi.org/10.3390/ijms23010303
Flor-Parra I, Sabido-Bozo S, Ikeda A, Hanaoka K, Aguilera-Romero A, Funato K, Muñiz M, Lucena R. The Ceramide Synthase Subunit Lac1 Regulates Cell Growth and Size in Fission Yeast. International Journal of Molecular Sciences. 2022; 23(1):303. https://doi.org/10.3390/ijms23010303
Chicago/Turabian StyleFlor-Parra, Ignacio, Susana Sabido-Bozo, Atsuko Ikeda, Kazuki Hanaoka, Auxiliadora Aguilera-Romero, Kouichi Funato, Manuel Muñiz, and Rafael Lucena. 2022. "The Ceramide Synthase Subunit Lac1 Regulates Cell Growth and Size in Fission Yeast" International Journal of Molecular Sciences 23, no. 1: 303. https://doi.org/10.3390/ijms23010303
APA StyleFlor-Parra, I., Sabido-Bozo, S., Ikeda, A., Hanaoka, K., Aguilera-Romero, A., Funato, K., Muñiz, M., & Lucena, R. (2022). The Ceramide Synthase Subunit Lac1 Regulates Cell Growth and Size in Fission Yeast. International Journal of Molecular Sciences, 23(1), 303. https://doi.org/10.3390/ijms23010303






