Down-Regulation of miR-194-5p for Predicting Metastasis in Breast Cancer Cells
Abstract
:1. Introduction
2. Results
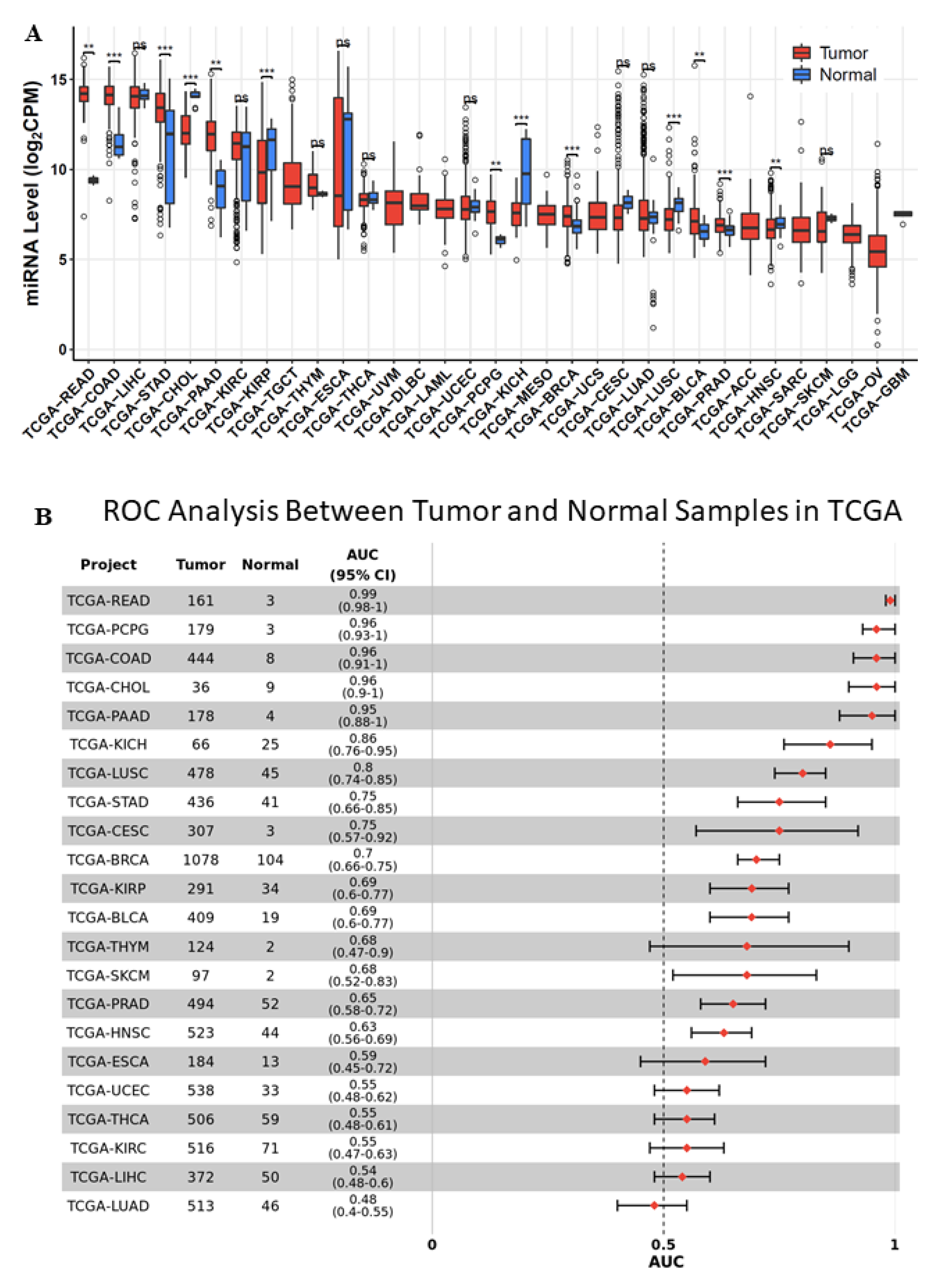
2.1. miR-194-5p Expression in TCGA-BRCA
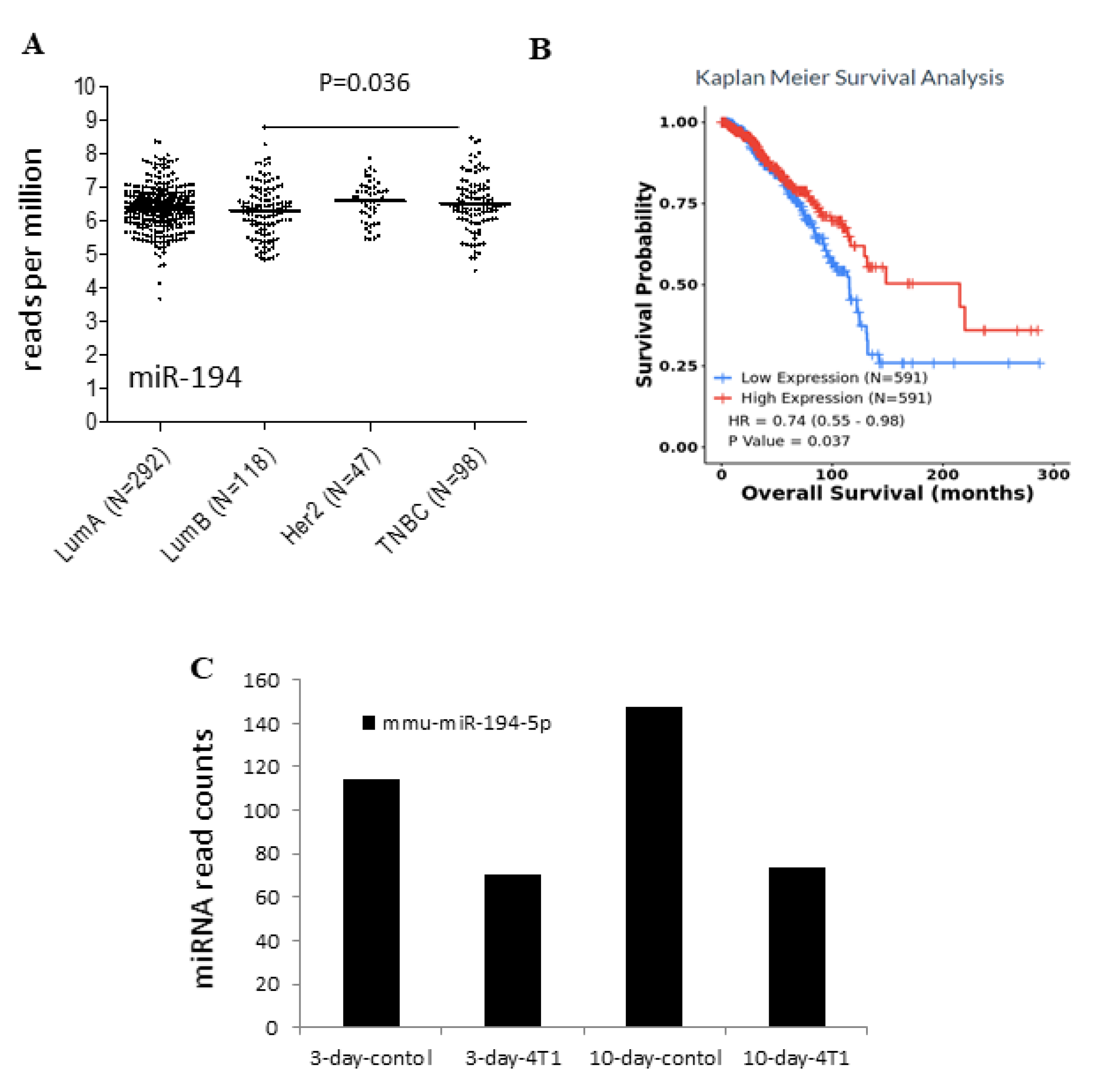
2.2. Low Expression of miR-194-5p in the Basal-like Group in Breast Cancer
2.3. Survival Analysis
2.4. Functional Enrichment Analysis of miR-194-5p in Breast Cancer Patients
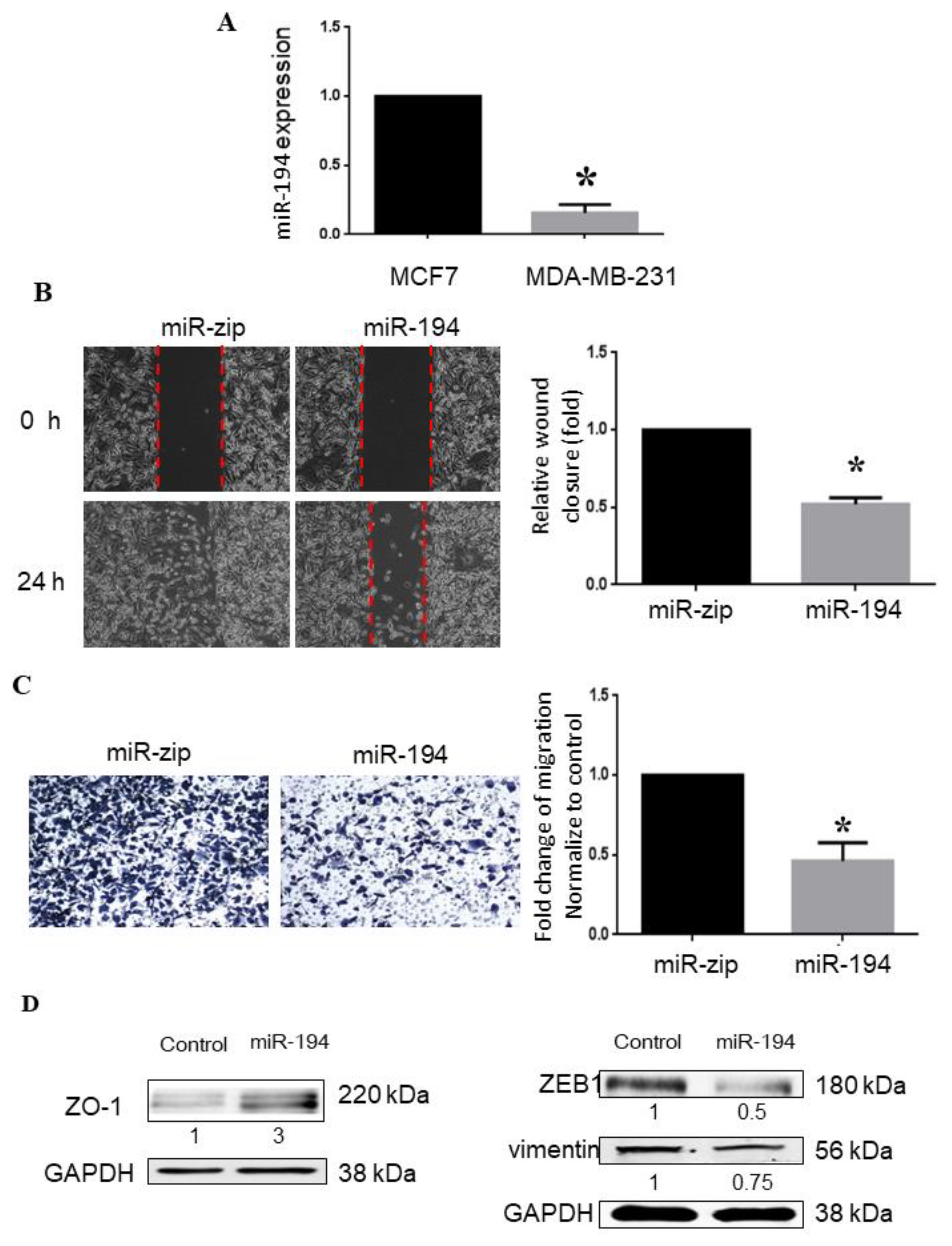
2.5. miR-194-5p Inhibits Metastasis of Breast Cancer
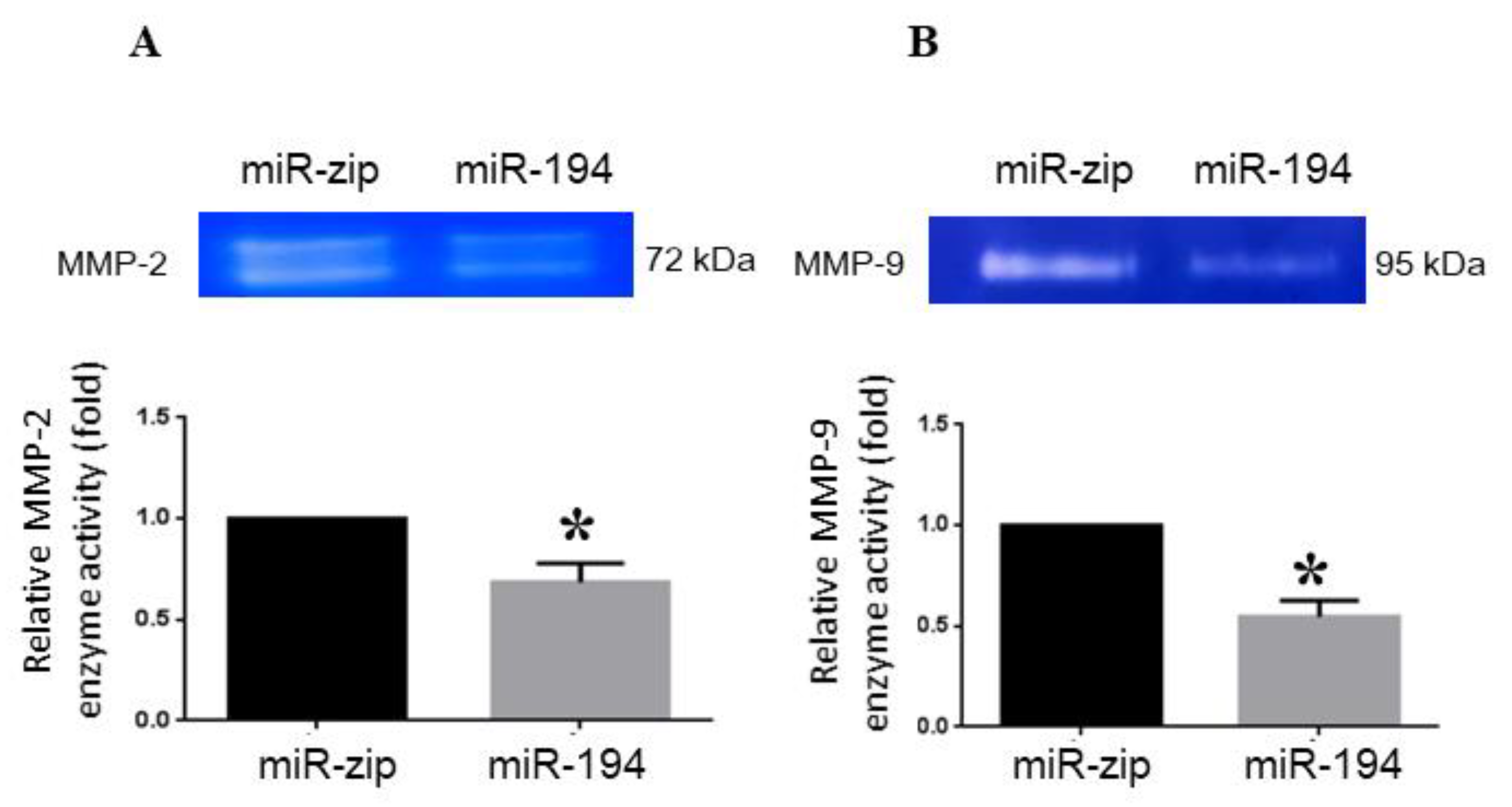
2.6. miRNA-194-5p Suppresses MMP2 and MMP9 Enzyme Activities
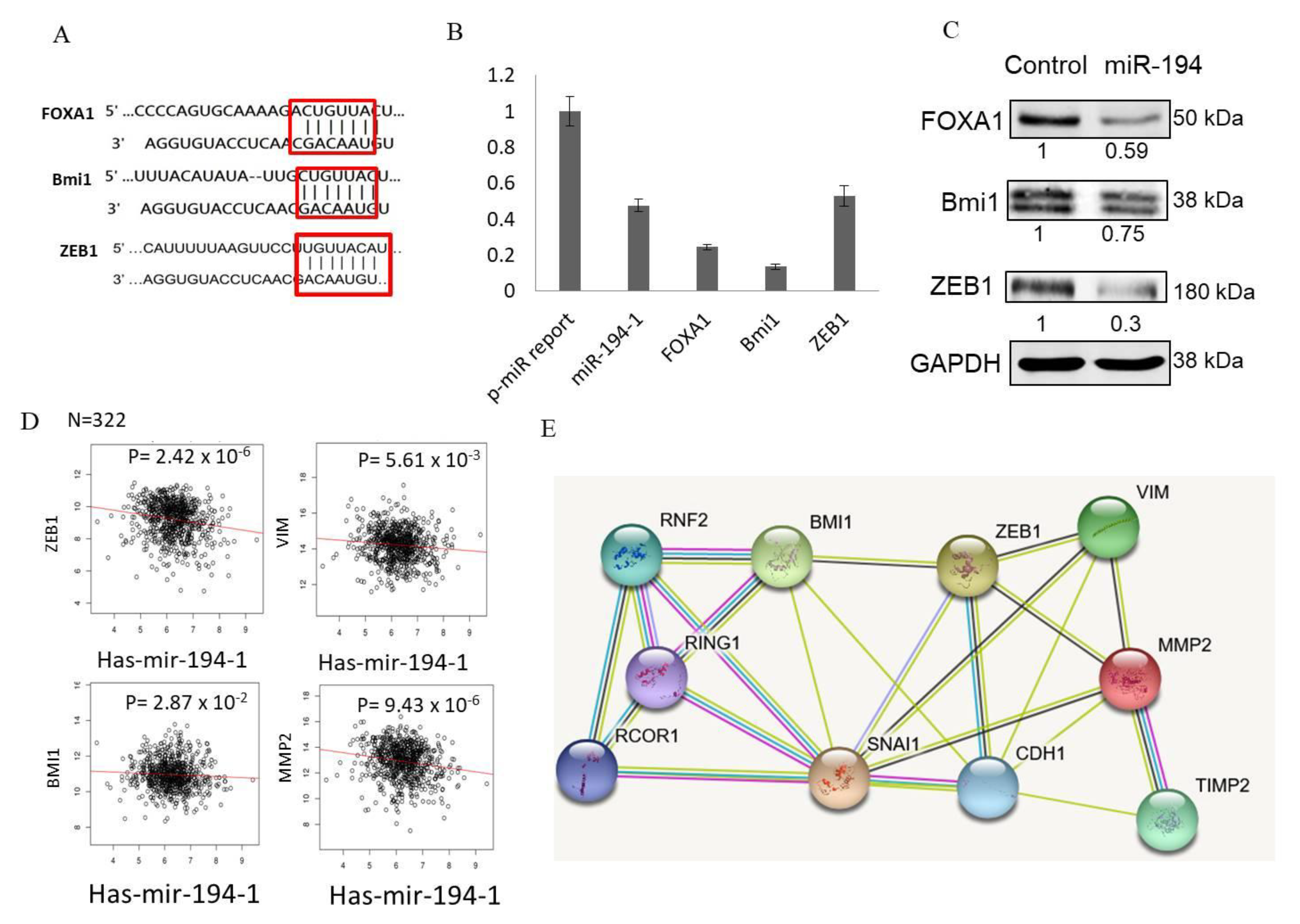
2.7. miR-194-5p Directly Targeted FOXA1, BMI1, and ZEB1
2.8. Expression of ZEB1, VIM, and MMP-2 Is Significantly Inversely Correlated with miR-194-5p Levels in TCGA-BRCA
3. Discussion
3.1. The Biological Functions of miR-194-5p in Breast Cancer Carcinogenesis
3.2. miR-194-5p Inhibits Invasion and Metastasis
4. Materials and Methods
4.1. Bioinformatic Analysis
4.2. MiRactDB
4.3. UALCAN Analysis
4.4. LinkedOmics Analysis
4.5. Analyses of Interactive Network and Modules
4.6. Cell Cultures
4.7. Isolation of RNA Samples and Quantification of MicroRNA Expression
4.8. Predicting miR-194-5p Target Genes and Constructs
4.9. Transfection of miR-194-5p into MDA-MB-231 Cells
4.10. Luciferase Reporter Gene Assays
4.11. Wound-Healing Assay
4.12. Trans-Well Migration Assays
4.13. Gelatin Zymography
4.14. Western Blot Analysis
4.15. Super-Enhancer (SE) Analysis
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Colorectal Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [Green Version]
- Weigelt, B.; Peterse, J.L.; van’t Veer, L.J. Breast cancer metastasis: Markers and models. Nat. Rev. Cancer 2005, 5, 591–602. [Google Scholar] [CrossRef]
- De Craene, B.; Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Zhang, C.; Lum, D.; Druso, J.E.; Blank, B.; Wilson, K.F.; Welm, A.; Antonyak, M.A.; Cerione, R.A. A class of extracellular vesicles from breast cancer cells activates VEGF receptors and tumour angiogenesis. Nat. Commun. 2017, 8, 14450. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [Green Version]
- Hudis, C.A.; Gianni, L. Triple-negative breast cancer: An unmet medical need. Oncologist 2011, 16 (Suppl. 1), 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Won, K.A.; Spruck, C. Triplenegative breast cancer therapy: Current and future perspectives (Review). Int. J. Oncol. 2020, 57, 1245–1261. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Yang, Y.; Zhao, L.; Yuan, J.; Niu, Y. Lin28A and androgen receptor expression in ER-/Her2+ breast cancer. Breast Cancer Res. Treat 2016, 156, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Nassar, F.J.; Nasr, R.; Talhouk, R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol. Ther. 2017, 172, 34–49. [Google Scholar] [CrossRef] [PubMed]
- Lages, E.; Ipas, H.; Guttin, A.; Nesr, H.; Berger, F.; Issartel, J.P. MicroRNAs: Molecular features and role in cancer. Front. Biosci. 2012, 17, 2508–2540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greene, S.B.; Herschkowitz, J.I.; Rosen, J.M. Small players with big roles: microRNAs as targets to inhibit breast cancer progression. Curr. Drug Targets 2010, 11, 1059–1073. [Google Scholar] [CrossRef]
- Zhu, X.; Li, D.; Yu, F.; Jia, C.; Xie, J.; Ma, Y.; Fan, S.; Cai, H.; Luo, Q.; Lv, Z.; et al. miR-194 inhibits the proliferation, invasion, migration, and enhances the chemosensitivity of non-small cell lung cancer cells by targeting forkhead box A1 protein. Oncotarget 2016, 7, 13139–13152. [Google Scholar] [CrossRef] [Green Version]
- Dell’Aversana, C.; Giorgio, C.; D’Amato, L.; Lania, G.; Matarese, F.; Saeed, S.; Di Costanzo, A.; Belsito Petrizzi, V.; Ingenito, C.; Martens, J.H.A.; et al. miR-194-5p/BCLAF1 deregulation in AML tumorigenesis. Leukemia 2017, 31, 2315–2325. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Shen, Z.L.; Gao, Z.D.; Zhao, G.; Wang, C.Y.; Yang, Y.; Zhang, J.Z.; Yan, Y.C.; Shen, C.; Jiang, K.W.; et al. MiR-194, commonly repressed in colorectal cancer, suppresses tumor growth by regulating the MAP4K4/c-Jun/MDM2 signaling pathway. Cell Cycle 2015, 14, 1046–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hironaka-Mitsuhashi, A.; Matsuzaki, J.; Takahashi, R.U.; Yoshida, M.; Nezu, Y.; Yamamoto, Y.; Shiino, S.; Kinoshita, T.; Ushijima, T.; Hiraoka, N.; et al. A tissue microRNA signature that predicts the prognosis of breast cancer in young women. PLoS ONE 2017, 12, e0187638. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Aigner, K.; Dampier, B.; Descovich, L.; Mikula, M.; Sultan, A.; Schreiber, M.; Mikulits, W.; Brabletz, T.; Strand, D.; Obrist, P.; et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 2007, 26, 6979–6988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spaderna, S.; Schmalhofer, O.; Wahlbuhl, M.; Dimmler, A.; Bauer, K.; Sultan, A.; Hlubek, F.; Jung, A.; Strand, D.; Eger, A.; et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008, 68, 537–544. [Google Scholar] [CrossRef] [Green Version]
- Maturi, V.; Enroth, S.; Heldin, C.H.; Moustakas, A. Genome-wide binding of transcription factor ZEB1 in triple-negative breast cancer cells. J. Cell Physiol. 2018, 233, 7113–7127. [Google Scholar] [CrossRef] [Green Version]
- Mock, K.; Preca, B.T.; Brummer, T.; Brabletz, S.; Stemmler, M.P.; Brabletz, T. The EMT-activator ZEB1 induces bone metastasis associated genes including BMP-inhibitors. Oncotarget 2015, 6, 14399–14412. [Google Scholar] [CrossRef] [Green Version]
- Meidhof, S.; Brabletz, S.; Lehmann, W.; Preca, B.T.; Mock, K.; Ruh, M.; Schuler, J.; Berthold, M.; Weber, A.; Burk, U.; et al. ZEB1-associated drug resistance in cancer cells is reversed by the class I HDAC inhibitor mocetinostat. EMBO Mol. Med. 2015, 7, 831–847. [Google Scholar] [CrossRef]
- Li, Z.; Ying, X.; Chen, H.; Ye, P.; Shen, Y.; Pan, W.; Zhang, L. MicroRNA-194 inhibits the epithelial-mesenchymal transition in gastric cancer cells by targeting FoxM1. Dig. Dis. Sci. 2014, 59, 2145–2152. [Google Scholar] [CrossRef]
- Li, R.; Qu, H.; Wang, S.; Chater, J.M.; Wang, X.; Cui, Y.; Yu, L.; Zhou, R.; Jia, Q.; Traband, R.; et al. CancerMIRNome: An interactive analysis and visualization database for miRNome profiles of human cancer. bioRxiv 2021. [Google Scholar] [CrossRef] [PubMed]
- Sereno, M.; Hasko, J.; Molnar, K.; Medina, S.J.; Reisz, Z.; Malho, R.; Videira, M.; Tiszlavicz, L.; Booth, S.A.; Wilhelm, I.; et al. Downregulation of circulating miR 802-5p and miR 194-5p and upregulation of brain MEF2C along breast cancer brain metastasization. Mol. Oncol. 2020, 14, 520–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eslami Amirabadi, H.; Tuerlings, M.; Hollestelle, A.; SahebAli, S.; Luttge, R.; van Donkelaar, C.C.; Martens, J.W.M.; den Toonder, J.M.J. Characterizing the invasion of different breast cancer cell lines with distinct E-cadherin status in 3D using a microfluidic system. Biomed. Microdevices 2019, 21, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ma, L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012, 31, 653–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaravinos, A. The Regulatory Role of MicroRNAs in EMT and Cancer. J. Oncol. 2015, 2015, 865816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, H.; Liu, C.; Zou, X.; Wu, W.; Zhang, C.; Yuan, D. MiRNA-194 Regulates Palmitic Acid-Induced Toll-Like Receptor 4 Inflammatory Responses in THP-1 Cells. Nutrients 2015, 7, 3483–3496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Kang, Y.; Liao, W.M.; Yu, L. MiR-194 regulates chondrogenic differentiation of human adipose-derived stem cells by targeting Sox5. PLoS ONE 2012, 7, e31861. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, H.; Zhang, R.; Zhang, S.; Shu, Q.; Zhang, D.; Xu, G. Altered expression of microRNAs in the neuronal differentiation of human Wharton’s Jelly mesenchymal stem cells. Neurosci. Lett. 2015, 600, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Barad, O.; Meiri, E.; Avniel, A.; Aharonov, R.; Barzilai, A.; Bentwich, I.; Einav, U.; Gilad, S.; Hurban, P.; Karov, Y.; et al. MicroRNA expression detected by oligonucleotide microarrays: System establishment and expression profiling in human tissues. Genome Res. 2004, 14, 2486–2494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hino, K.; Fukao, T.; Watanabe, M. Regulatory interaction of HNF1-alpha to microRNA-194 gene during intestinal epithelial cell differentiation. In Nucleic Acids Symposium Series; Oxford University Press: Oxford, UK, 2007; pp. 415–416. [Google Scholar]
- Meng, Z.; Fu, X.; Chen, X.; Zeng, S.; Tian, Y.; Jove, R.; Xu, R.; Huang, W. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology 2010, 52, 2148–2157. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Zhao, F.; Wang, Z.; Liu, Z.; Chiang, Y.; Xu, Y.; Gao, P.; Xu, H. Inverse association between miR-194 expression and tumor invasion in gastric cancer. Ann. Surg. Oncol. 2012, 19 (Suppl. 3), S509–S517. [Google Scholar] [CrossRef]
- Cai, H.K.; Chen, X.; Tang, Y.H.; Deng, Y.C. MicroRNA-194 modulates epithelial-mesenchymal transition in human colorectal cancer metastasis. Oncol. Targets 2017, 10, 1269–1278. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Xiao, Z.; Zhang, S. Knockdown of miR-194-5p inhibits cell proliferation, migration and invasion in breast cancer by regulating the Wnt/beta-catenin signaling pathway. Int. J. Mol. Med. 2018, 42, 3355–3363. [Google Scholar]
- Dong, P.; Kaneuchi, M.; Watari, H.; Hamada, J.; Sudo, S.; Ju, J.; Sakuragi, N. MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol. Cancer 2011, 10, 99. [Google Scholar] [CrossRef] [Green Version]
- Tan, H.; Kim, P.; Sun, P.; Zhou, X. miRactDB characterizes miRNA-gene relation switch between normal and cancer tissues across pan-cancer. Brief. Bioinform. 2020, 22, bbaa089. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, S.C.; Huang, S.F.; Chiang, J.H.; Chen, Y.F.; Huang, C.C.; Tsai, M.H.; Tsai, F.J.; Kao, M.C.; Yang, J.S. The differential regulation of microRNAs is associated with oral cancer. Oncol. Rep. 2017, 38, 1613–1620. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.C.; Tsai, M.H.; Chiu, C.F.; Lu, C.C.; Kuo, S.C.; Chang, N.W.; Yang, J.S. AMPK-dependent signaling modulates the suppression of invasion and migration by fenofibrate in CAL 27 oral cancer cells through NF-kappaB pathway. Environ. Toxicol. 2016, 31, 866–876. [Google Scholar] [CrossRef]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yen, Y.-T.; Yang, J.-C.; Chang, J.-B.; Tsai, S.-C. Down-Regulation of miR-194-5p for Predicting Metastasis in Breast Cancer Cells. Int. J. Mol. Sci. 2022, 23, 325. https://doi.org/10.3390/ijms23010325
Yen Y-T, Yang J-C, Chang J-B, Tsai S-C. Down-Regulation of miR-194-5p for Predicting Metastasis in Breast Cancer Cells. International Journal of Molecular Sciences. 2022; 23(1):325. https://doi.org/10.3390/ijms23010325
Chicago/Turabian StyleYen, Yu-Ting, Jou-Chun Yang, Jiun-Bo Chang, and Shih-Chang Tsai. 2022. "Down-Regulation of miR-194-5p for Predicting Metastasis in Breast Cancer Cells" International Journal of Molecular Sciences 23, no. 1: 325. https://doi.org/10.3390/ijms23010325





