Exosomal MicroRNAs as Potential Biomarkers of Hepatic Injury and Kidney Disease in Glycogen Storage Disease Type Ia Patients
Abstract
:1. Introduction
2. Results and Discussion
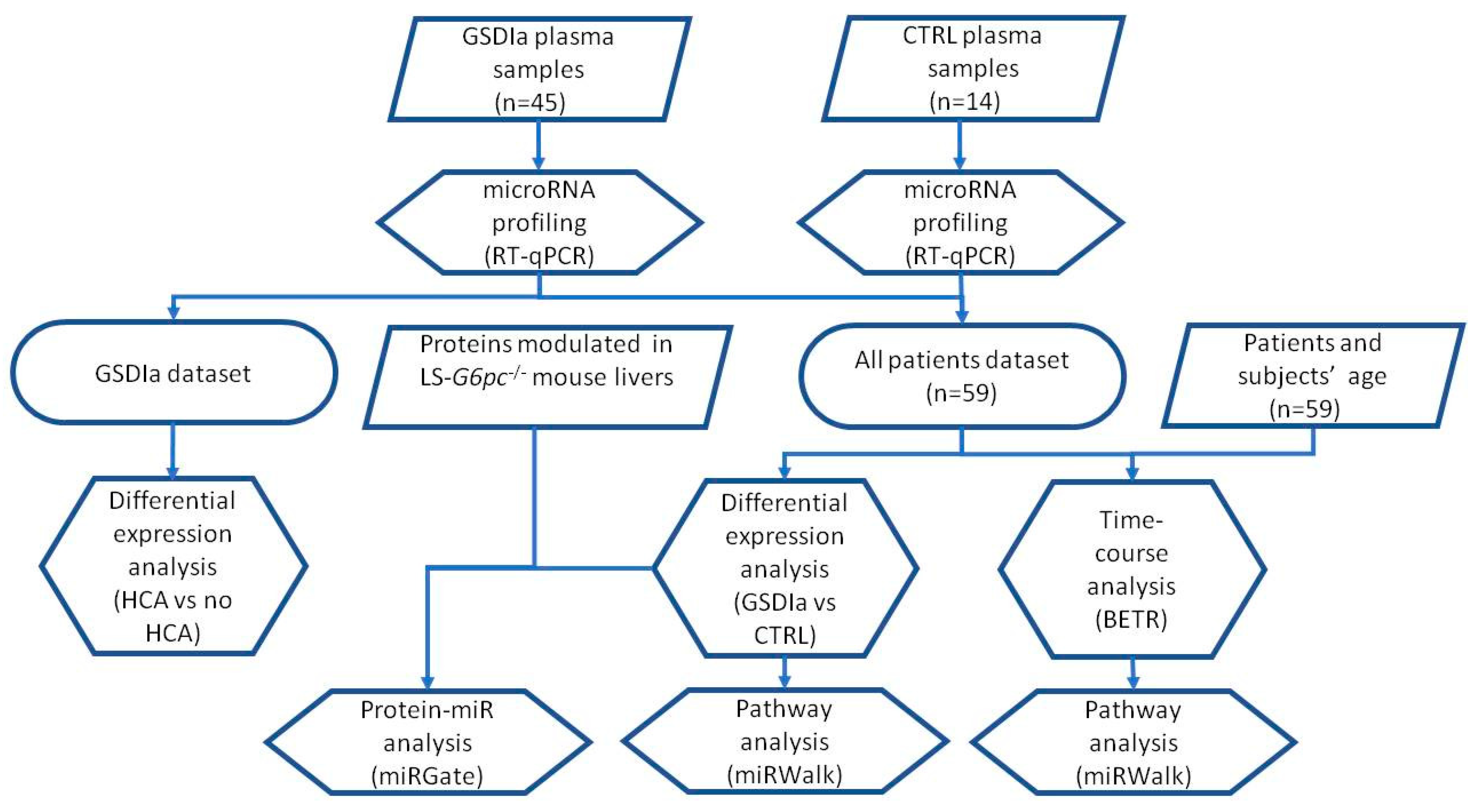
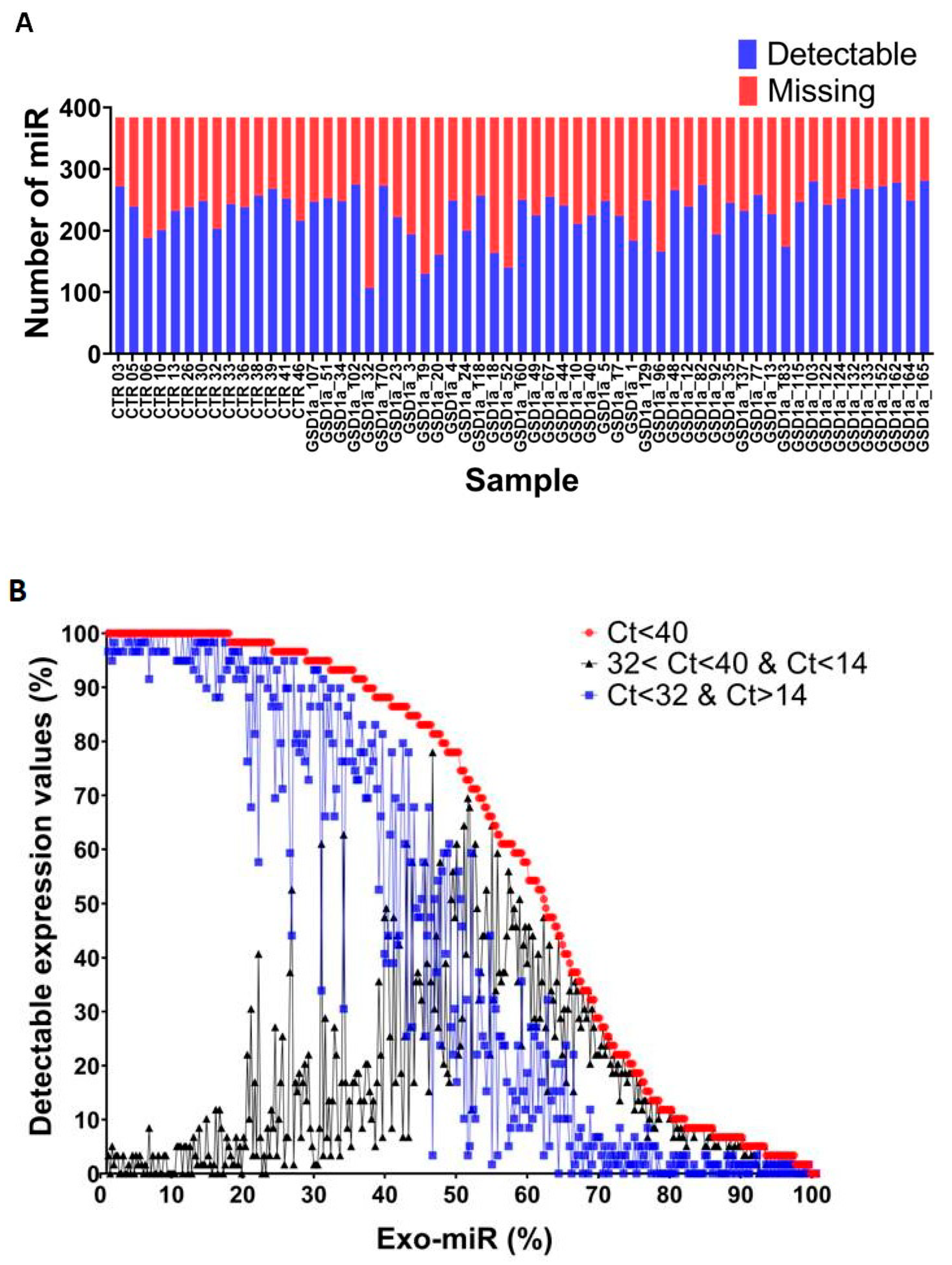
2.1. Exo-miRs Expression Profiling of GSDIa and CTRL Plasma Exosomes
2.2. Deregulation of Exo-miRs in GSDIa Patients
2.3. Age-Dependent Modulation of Exo-miRs in GSDIa Patients
2.4. Enrichment of Specific GO Biological Processes and KEGG Pathways in the Exo-miR Expression Profile of GSDIa Patients
2.5. Overlapping between MicroRNA Targets and Proteins Modulated in LS-G6pc−/− Mouse Livers
3. Materials and Methods
3.1. Patients Blood Sample Collection
3.2. Exosome Isolation, MicroRNA Purification and Quantitative Real-Time PCR (qRT-PCR)
3.3. Bioinformatic Procedures and Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chou, J.Y.; Matern, D.; Mansfield, B.C.; Chen, Y.-T. Type I Glycogen Storage Diseases: Disorders of the Glucose-6-Phosphatase Complex. Curr. Mol. Med. 2002, 2, 121–143. [Google Scholar] [CrossRef]
- Chen, Y.T.; Cornblath, M.; Sidbury, J.B. Cornstarch Therapy in Type I Glycogen-Storage Disease. N. Engl. J. Med. 1984, 310, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Dambska, M.; Labrador, E.B.; Kuo, C.L.; Weinstein, D.A. Prevention of Complications in Glycogen Storage Disease Type Ia with Optimization of Metabolic Control. Pediatric Diabetes 2017, 18, 327–331. [Google Scholar] [CrossRef]
- Okechuku, G.O.; Shoemaker, L.R.; Dambska, M.; Brown, L.M.; Mathew, J.; Weinstein, D.A. Tight Metabolic Control plus ACE Inhibitor Therapy Improves GSD I Nephropathy. J. Inherit. Metab. Dis. 2017, 40, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Hoogerland, J.A.; Peeks, F.; Hijmans, B.S.; Wolters, J.C.; Kooijman, S.; Bos, T.; Bleeker, A.; van Dijk, T.H.; Wolters, H.; Gerding, A.; et al. Impaired Very-Low-Density Lipoprotein Catabolism Links Hypoglycemia to Hypertriglyceridemia in Glycogen Storage Disease Type Ia. J. Inherit. Metab. Dis. 2021, 44, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Franco, L.M.; Krishnamurthy, V.; Bali, D.; Weinstein, D.A.; Arn, P.; Clary, B.; Boney, A.; Sullivan, J.; Frush, D.P.; Chen, Y.-T.; et al. Hepatocellular Carcinoma in Glycogen Storage Disease Type Ia: A Case Series. J. Inherit. Metab. Dis. 2005, 28, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Rake, J.P.; Visser, G.; Labrune, P.; Leonard, J.V.; Ullrich, K.; Smit, G.P.A. Glycogen Storage Disease Type I: Diagnosis, Management, Clinical Course and Outcome. Results of the European Study on Glycogen Storage Disease Type I (ESGSD I). Eur. J. Pediatr. 2002, 161 (Suppl. S1), S20–S34. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, X.; Wang, J.; Hao, Y.; Liu, F.; Wang, X.; Yang, L.; Lu, Z. The Roles of Exosomes as Future Therapeutic Agents and Diagnostic Tools for Glioma. Front. Oncol. 2021, 11, 733529. [Google Scholar] [CrossRef]
- Su, L.; Zhao, X.; Li, R.; Zhang, Z.; Liu, J.; Du, J.; Wei, H. Identification of Altered Exosomal MicroRNAs and MRNAs in Alzheimer’s Disease. Ageing Res. Rev. 2021, 73, 101497. [Google Scholar] [CrossRef]
- Yi, Y.; Wu, M.; Zeng, H.; Hu, W.; Zhao, C.; Xiong, M.; Lv, W.; Deng, P.; Zhang, Q.; Wu, Y. Tumor-Derived Exosomal Non-Coding RNAs: The Emerging Mechanisms and Potential Clinical Applications in Breast Cancer. Front. Oncol. 2021, 11, 738945. [Google Scholar] [CrossRef]
- Resaz, R.; Vanni, C.; Segalerba, D.; Sementa, A.R.; Mastracci, L.; Grillo, F.; Murgia, D.; Bosco, M.C.; Chou, J.Y.; Barbieri, O.; et al. Development of Hepatocellular Adenomas and Carcinomas in Mice with Liver-Specific G6Pase-α Deficiency. Dis. Model. Mech. 2014, 7, 1083–1091. [Google Scholar] [CrossRef] [Green Version]
- Resaz, R.; Cangelosi, D.; Morini, M.; Segalerba, D.; Mastracci, L.; Grillo, F.; Bosco, M.C.; Bottino, C.; Colombo, I.; Eva, A. Circulating Exosomal MicroRNAs as Potential Biomarkers of Hepatic Injury and Inflammation in a Murine Model of Glycogen Storage Disease Type 1a. Dis. Model. Mech. 2020, 13, dmm043364. [Google Scholar] [CrossRef]
- Zanardi, N.; Morini, M.; Tangaro, M.A.; Zambelli, F.; Bosco, M.C.; Varesio, L.; Eva, A.; Cangelosi, D. PIPE-T: A New Galaxy Tool for the Analysis of RT-QPCR Expression Data. Sci. Rep. 2019, 9, 17550. [Google Scholar] [CrossRef] [Green Version]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A Novel and Universal Method for MicroRNA RT-QPCR Data Normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef] [Green Version]
- Ma, N.; Li, F.; Li, D.; Hui, Y.; Wang, X.; Qiao, Y.; Zhang, Y.; Xiang, Y.; Zhou, J.; Zhou, L.; et al. Igf2-Derived Intronic MiR-483 Promotes Mouse Hepatocellular Carcinoma Cell Proliferation. Mol. Cell. Biochem. 2012, 361, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Umezu, T.; Tsuneyama, K.; Kanekura, K.; Hayakawa, M.; Tanahashi, T.; Kawano, M.; Taguchi, Y.-H.; Toyoda, H.; Tamori, A.; Kuroda, M.; et al. Comprehensive Analysis of Liver and Blood MiRNA in Precancerous Conditions. Sci. Rep. 2020, 10, 21766. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Wong, Y.S.; Goh, B.K.P.; Chan, C.Y.; Cheow, P.C.; Chow, P.K.H.; Lim, T.K.H.; Goh, G.B.B.; Krishnamoorthy, T.L.; Kumar, R.; et al. Circulating MicroRNAs as Potential Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. Sci. Rep. 2019, 9, 10464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Wang, Q.; Sun, J.; Liu, H.; Wang, H. Long Non-Coding RNA TPTEP1 Exerts Inhibitory Effects on Hepatocellular Carcinoma by Impairing MicroRNA-454-3p-Mediated DLG5 Downregulation. Dig. Liver Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiao, Y.; Fu, Z.; Luo, Z.; Su, J.; Li, Y. High MiR-454-3p Expression Predicts Poor Prognosis in Hepatocellular Carcinoma. Cancer Manag. Res. 2019, 11, 2795–2802. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Zhang, J.; Apaer, S.; Yao, G.; Li, T. MicroRNA-19a-3p and MicroRNA-376c-3p Promote Hepatocellular Carcinoma Progression Through SOX6-Mediated Wnt/β-Catenin Signaling Pathway. Int. J. Gen. Med. 2021, 14, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chang, W.; Chang, W.; Chang, X.; Zhai, S.; Pan, G.; Dang, S. MicroRNA-376c-3p Facilitates Human Hepatocellular Carcinoma Progression via Repressing AT-Rich Interaction Domain 2. J. Cancer 2018, 9, 4187–4196. [Google Scholar] [CrossRef]
- Wang, S.; Wang, T.; Gu, P. MicroRNA-145-5p Inhibits Migration, Invasion, and Metastasis in Hepatocellular Carcinoma by Inhibiting ARF6. Cancer Manag. Res. 2021, 13, 3473–3484. [Google Scholar] [CrossRef]
- Hussen, B.M.; Honarmand Tamizkar, K.; Hidayat, H.J.; Taheri, M.; Ghafouri-Fard, S. The Role of Circular RNAs in the Development of Hepatocellular Carcinoma. Pathol. Res. Pract. 2021, 223, 153495. [Google Scholar] [CrossRef]
- Cao, L.; Xie, B.; Yang, X.; Liang, H.; Jiang, X.; Zhang, D.; Xue, P.; Chen, D.; Shao, Z. MiR-324-5p Suppresses Hepatocellular Carcinoma Cell Invasion by Counteracting ECM Degradation through Post-Transcriptionally Downregulating ETS1 and SP1. PLoS ONE 2015, 10, e0133074. [Google Scholar] [CrossRef] [Green Version]
- Rong, Z.; Wang, Z.; Wang, X.; Qin, C.; Geng, W. Molecular Interplay between Linc01134 and YY1 Dictates Hepatocellular Carcinoma Progression. J. Exp. Clin. Cancer Res. 2020, 39, 61. [Google Scholar] [CrossRef] [Green Version]
- Komoll, R.-M.; Hu, Q.; Olarewaju, O.; von Döhlen, L.; Yuan, Q.; Xie, Y.; Tsay, H.-C.; Daon, J.; Qin, R.; Manns, M.P.; et al. MicroRNA-342-3p Is a Potent Tumour Suppressor in Hepatocellular Carcinoma. J. Hepatol. 2021, 74, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Mens, M.M.J.; Abozaid, Y.J.; Bos, D.; Darwish Murad, S.; de Knegt, R.J.; Ikram, M.A.; Pan, Q.; Ghanbari, M. Circulatory MicroRNAs as Potential Biomarkers for Fatty Liver Disease: The Rotterdam Study. Aliment. Pharmacol. Ther. 2021, 53, 432–442. [Google Scholar] [CrossRef]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. MiR-122 Regulation of Lipid Metabolism Revealed by In Vivo Antisense Targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [Green Version]
- de Frias, F.T.; Rocha, K.C.E.; de Mendonça, M.; Murata, G.M.; Araujo, H.N.; de Sousa, L.G.O.; de Sousa, É.; Hirabara, S.M.; de Leite, N.C.; Carneiro, E.M.; et al. Fenofibrate Reverses Changes Induced by High-Fat Diet on Metabolism in Mice Muscle and Visceral Adipocytes. J. Cell Physiol. 2018, 233, 3515–3528. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Yin, Z.; Zhong, H.; Liang, T.; Guo, L. Analysis of the Expression, Function, and Evolution of MiR-27 Isoforms and Their Responses in Metabolic Processes. Genomics 2019, 111, 1249–1257. [Google Scholar] [CrossRef]
- Liu, L.; Chen, H.; Yun, J.; Song, L.; Ma, X.; Luo, S.; Song, Y. MiRNA-483-5p Targets HDCA4 to Regulate Renal Tubular Damage in Diabetic Nephropathy. Horm. Metab. Res. 2021, 53, 562–569. [Google Scholar] [CrossRef]
- Regmi, A.; Liu, G.; Zhong, X.; Hu, S.; Ma, R.; Gou, L.; Zafar, M.I.; Chen, L. Evaluation of Serum MicroRNAs in Patients with Diabetic Kidney Disease: A Nested Case-Controlled Study and Bioinformatics Analysis. Med. Sci. Monit. 2019, 25, 1699–1708. [Google Scholar] [CrossRef]
- Assmann, T.S.; Recamonde-Mendoza, M.; de Souza, B.M.; Bauer, A.C.; Crispim, D. MicroRNAs and Diabetic Kidney Disease: Systematic Review and Bioinformatic Analysis. Mol. Cell Endocrinol. 2018, 477, 90–102. [Google Scholar] [CrossRef]
- Jiang, Z.-H.; Tang, Y.-Z.; Song, H.-N.; Yang, M.; Li, B.; Ni, C.-L. MiRNA-342 Suppresses Renal Interstitial Fibrosis in Diabetic Nephropathy by Targeting SOX6. Int. J. Mol. Med. 2020, 45, 45–52. [Google Scholar] [CrossRef] [Green Version]
- Bai, L.; Lin, Y.; Xie, J.; Zhang, Y.; Wang, H.; Zheng, D. MiR-27b-3p Inhibits the Progression of Renal Fibrosis via Suppressing STAT1. Hum. Cell 2021, 34, 383–393. [Google Scholar] [CrossRef]
- Prabu, P.; Rome, S.; Sathishkumar, C.; Gastebois, C.; Meugnier, E.; Mohan, V.; Balasubramanyam, M. MicroRNAs from Urinary Extracellular Vesicles Are Non-Invasive Early Biomarkers of Diabetic Nephropathy in Type 2 Diabetes Patients with the “Asian Indian Phenotype”. Diabetes Metab. 2019, 45, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Zhang, H.; Zhao, C.; Luo, Y.; Chen, Y. Silencing of MiR-150-5p Ameliorates Diabetic Nephropathy by Targeting SIRT1/P53/AMPK Pathway. Front. Physiol. 2021, 12, 624989. [Google Scholar] [CrossRef]
- Melis, D.; Cozzolino, M.; Minopoli, G.; Balivo, F.; Parini, R.; Rigoldi, M.; Paci, S.; Dionisi-Vici, C.; Burlina, A.; Andria, G.; et al. Progression of Renal Damage in Glycogen Storage Disease Type I Is Associated to Hyperlipidemia: A Multicenter Prospective Italian Study. J. Pediatr. 2015, 166, 1079–1082. [Google Scholar] [CrossRef]
- Melis, D.; Parenti, G.; Gatti, R.; Casa, R.D.; Parini, R.; Riva, E.; Burlina, A.B.; Dionisi Vici, C.; Di Rocco, M.; Furlan, F.; et al. Efficacy of ACE-Inhibitor Therapy on Renal Disease in Glycogen Storage Disease Type 1: A Multicentre Retrospective Study. Clin. Endocrinol. (Oxf.) 2005, 63, 19–25. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, N.; Zhao, S.; Chen, X.; Li, F.; Tao, X. MiR-221-3p and MiR-15b-5p Promote Cell Proliferation and Invasion by Targeting Axin2 in Liver Cancer. Oncol. Lett 2019, 18, 6491–6500. [Google Scholar] [CrossRef]
- Chen, Z.; Xiang, B.; Qi, L.; Zhu, S.; Li, L. MiR-221-3p Promotes Hepatocellular Carcinogenesis by Downregulating O6-Methylguanine-DNA Methyltransferase. Cancer Biol. Ther. 2020, 21, 915–926. [Google Scholar] [CrossRef]
- de Conti, A.; Ortega, J.F.; Tryndyak, V.; Dreval, K.; Moreno, F.S.; Rusyn, I.; Beland, F.A.; Pogribny, I.P. MicroRNA Deregulation in Nonalcoholic Steatohepatitis-Associated Liver Carcinogenesis. Oncotarget 2017, 8, 88517–88528. [Google Scholar] [CrossRef] [Green Version]
- Shi, K.-Q.; Lin, Z.; Chen, X.-J.; Song, M.; Wang, Y.-Q.; Cai, Y.-J.; Yang, N.-B.; Zheng, M.-H.; Dong, J.-Z.; Zhang, L.; et al. Hepatocellular Carcinoma Associated MicroRNA Expression Signature: Integrated Bioinformatics Analysis, Experimental Validation and Clinical Significance. Oncotarget 2015, 6, 25093–25108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.-B.; Chen, X.-B.; Xu, L.-L.; Zhang, M.; Feng, L.; Yi, P.-S.; Tang, J.-W.; Xu, M.-Q. MiR-203 Inhibits Augmented Proliferation and Metastasis of Hepatocellular Carcinoma Residual in the Promoted Regenerating Liver. Cancer Sci. 2017, 108, 338–346. [Google Scholar] [CrossRef] [Green Version]
- Furuta, M.; Kozaki, K.; Tanaka, S.; Arii, S.; Imoto, I.; Inazawa, J. MiR-124 and MiR-203 Are Epigenetically Silenced Tumor-Suppressive MicroRNAs in Hepatocellular Carcinoma. Carcinogenesis 2010, 31, 766–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.-J.; Yuan, M.-X.; Ji, C.-Y.; Zhang, Y.-B.; Peng, Y.-M.; Zhang, T.; Gao, H.-Q.; Sheng, X.-Y.; Liu, Z.-Y.; Xie, W.-X.; et al. Long Non-Coding RNA CRNDE Regulates Angiogenesis in Hepatoblastoma by Targeting the MiR-203/VEGFA Axis. Pathobiology 2020, 87, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Feng, R.; Li, X.; Li, D.; Zhai, W. TGF-β- and Lipopolysaccharide-Induced Upregulation of Circular RNA PWWP2A Promotes Hepatic Fibrosis via Sponging MiR-203 and MiR-223. Aging (Albany N. Y.) 2019, 11, 9569–9580. [Google Scholar] [CrossRef]
- Aryee, M.J.; Gutiérrez-Pabello, J.A.; Kramnik, I.; Maiti, T.; Quackenbush, J. An Improved Empirical Bayes Approach to Estimating Differential Gene Expression in Microarray Time-Course Data: BETR (Bayesian Estimation of Temporal Regulation). BMC Bioinform. 2009, 10, 409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, B.; Ding, F.; Huang, C.-Y.; Xiao, H.; Fei, F.-Y.; Li, J. Role of MiR-16-5p in the Proliferation and Metastasis of Hepatocellular Carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 137–145. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Wang, L.; Yao, B.; Sun, L.; Liu, R.; Chen, T.; Niu, Y.; Tu, K.; Liu, Q. Long Non-Coding RNA AGAP2-AS1, Functioning as a Competitive Endogenous RNA, Upregulates ANXA11 Expression by Sponging MiR-16-5p and Promotes Proliferation and Metastasis in Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 194. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.; Ge, G.; Pan, T.; Wen, D.; Chen, L.; Yu, X.; Zhou, X.; Gan, J. A Serum MicroRNA Panel as Potential Biomarkers for Hepatocellular Carcinoma Related with Hepatitis B Virus. PLoS ONE 2014, 9, e107986. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.-W.; Qian, Z.-L.; Li, C.; Cui, S.-C. Identification of MiRNA and MRNA Expression Profiles by PCR Microarray in Hepatitis B Virus-associated Hepatocellular Carcinoma. Mol. Med. Rep. 2018, 18, 5123–5132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MiR-126-3p Suppresses Tumor Metastasis and Angiogenesis of Hepatocellular Carcinoma by Targeting LRP6 and PIK3R2—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/25240815/ (accessed on 4 November 2021).
- Lou, W.; Chen, J.; Ding, B.; Chen, D.; Zheng, H.; Jiang, D.; Xu, L.; Bao, C.; Cao, G.; Fan, W. Identification of Invasion-Metastasis-Associated MicroRNAs in Hepatocellular Carcinoma Based on Bioinformatic Analysis and Experimental Validation. J. Transl. Med. 2018, 16, 266. [Google Scholar] [CrossRef] [PubMed]
- Aguado-Fraile, E.; Ramos, E.; Conde, E.; Rodríguez, M.; Martín-Gómez, L.; Lietor, A.; Candela, Á.; Ponte, B.; Liaño, F.; García-Bermejo, M.L. A Pilot Study Identifying a Set of MicroRNAs As Precise Diagnostic Biomarkers of Acute Kidney Injury. PLoS ONE 2015, 10, e0127175. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wu, H.; Wang, G.; Feng, Y. Identification of Potential Candidate Genes for Hypertensive Nephropathy Based on Gene Expression Profile. BMC Nephrol. 2016, 17, 149. [Google Scholar] [CrossRef] [Green Version]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. MiRWalk: An Online Resource for Prediction of MicroRNA Binding Sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef] [PubMed]
- Gautam, S.; Zhang, L.; Arnaoutova, I.; Lee, C.; Mansfield, B.C.; Chou, J.Y. The Signaling Pathways Implicated in Impairment of Hepatic Autophagy in Glycogen Storage Disease Type Ia. Hum. Mol. Genet. 2020, 29, 834–844. [Google Scholar] [CrossRef] [Green Version]
- Rossi, A.; Ruoppolo, M.; Formisano, P.; Villani, G.; Albano, L.; Gallo, G.; Crisci, D.; Moccia, A.; Parenti, G.; Strisciuglio, P.; et al. Insulin-Resistance in Glycogen Storage Disease Type Ia: Linking Carbohydrates and Mitochondria? J. Inherit. Metab. Dis. 2018, 41, 985–995. [Google Scholar] [CrossRef]
- Cangelosi, D.; Resaz, R.; Petretto, A.; Segalerba, D.; Ognibene, M.; Raggi, F.; Mastracci, L.; Grillo, F.; Bosco, M.C.; Varesio, L.; et al. A Proteomic Analysis of GSD-1a in Mouse Livers: Evidence for Metabolic Reprogramming, Inflammation, and Macrophage Polarization. J. Proteome Res. 2019, 18, 2965–2978. [Google Scholar] [CrossRef] [PubMed]
- Andrés-León, E.; Gómez-López, G.; Pisano, D.G. Prediction of MiRNA-MRNA Interactions Using MiRGate. Methods Mol. Biol 2017, 1580, 225–237. [Google Scholar] [CrossRef]
- Qiu, L.; Wang, T.; Ge, Q.; Xu, H.; Wu, Y.; Tang, Q.; Chen, K. Circular RNA Signature in Hepatocellular Carcinoma. J. Cancer 2019, 10, 3361–3372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simile, M.M.; Peitta, G.; Tomasi, M.L.; Brozzetti, S.; Feo, C.F.; Porcu, A.; Cigliano, A.; Calvisi, D.F.; Feo, F.; Pascale, R.M. MicroRNA-203 Impacts on the Growth, Aggressiveness and Prognosis of Hepatocellular Carcinoma by Targeting MAT2A and MAT2B Genes. Oncotarget 2019, 10, 2835–2854. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Controls | Age (Years) | Gender | Tumor |
|---|---|---|---|
| CTR 01 | 10 | M | no |
| CTR 02 | 2 | M | no |
| CTR 03 | 6 | M | no |
| CTR 04 | 4 | F | no |
| CTR 05 | 12 | F | no |
| CTR 06 | 16 | F | no |
| CTR 07 | 14 | F | no |
| CTR 08 | 18 | M | no |
| CTR 09 | 21 | M | no |
| CTR 10 | 38 | F | no |
| CTR 11 | 16 | F | no |
| CTR 12 | 61 | F | no |
| CTR 13 | 52 | F | no |
| CTR 14 | 48 | M | no |
| Patients | |||
| GSD 01 | 6 | M | no |
| GSD 02 | 7 | F | no |
| GSD 03 | 7 | F | no |
| GSD 04 | 8 | M | no |
| GSD 05 | 10 | F | no |
| GSD 06 | 11 | M | no |
| GSD 07 | 12 | F | no |
| GSD 08 | 14 | F | no |
| GSD 09 | 16 | F | no |
| GSD 10 | 16 | M | no |
| GSD 11 | 19 | M | no |
| GSD 12 | 19 | F | no |
| GSD 13 | 19 | M | no |
| GSD 14 | 20 | F | no |
| GSD 15 | 20 | F | yes |
| GSD 16 | 21 | F | no |
| GSD 17 | 22 | F | no |
| GSD 18 | 22 | M | no |
| GSD 19 | 22 | M | yes |
| GSD 20 | 22 | M | yes |
| GSD 21 | 22 | F | no |
| GSD 22 | 24 | F | no |
| GSD 23 | 26 | F | yes |
| GSD 24 | 26 | M | no |
| GSD 25 | 26 | F | no |
| GSD 26 | 28 | M | no |
| GSD 27 | 28 | F | yes |
| GSD 28 | 29 | M | no |
| GSD 29 | 30 | M | yes |
| GSD 30 | 31 | M | yes |
| GSD 31 | 31 | F | yes |
| GSD 32 | 31 | M | yes |
| GSD 33 | 32 | M | yes |
| GSD 34 | 34 | F | yes |
| GSD 35 | 35 | F | no |
| GSD 36 | 36 | F | yes |
| GSD 37 | 36 | M | no |
| GSD 38 | 37 | M | no |
| GSD 39 | 37 | F | no |
| GSD 40 | 39 | M | yes |
| GSD 41 | 40 | F | yes |
| GSD 42 | 45 | M | yes |
| GSD 43 | 49 | F | yes |
| GSD 44 | 53 | M | yes |
| GSD 45 | 63 | F | yes |
| Exo-miR a | GSD HCA vs. GSD NO HCA b | p-Value c | GSD vs. CTRL c | p-Value d |
|---|---|---|---|---|
| miR-221-3p | 2.46 | 0.01 | ||
| miR-195-5p | 2.15 | 0.01 | ||
| miR-19a-3p | −0.64 | 0.006 | 1.15 | 0.01 |
| miR-203-3p | −1.39 | 0.03 | ||
| miR-483-5p | 2.17 | 0.0003 | ||
| miR-454-3p | 1.47 | 0.007 | ||
| miR-122-5p | 1.33 | 0.01 | ||
| miR-342-3p | 1.46 | 0.01 | ||
| miR-376c-3p | 1.20 | 0.01 | ||
| miR-145-5p | −1.54 | 0.01 | ||
| miR-103-3p | −1.11 | 0.03 | ||
| miR-27b-3p | −1.01 | 0.04 | ||
| miR-324-5p | −1.08 | 0.04 | ||
| miR-150-5p | 1.34 | 0.04 |
| Inflammatory and Immune Response | miR 27-b-3p | miR-103a-3p | miR-324-5p | miR-19a-3p | miR-145-5p | miR-203a-3p | miR-195-3p | miR-454-3p | miR-122-5p | miR-150-5p | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CD163 | acute-phase response (GO:0006953) | + | |||||||||
| MBL2 | innate immune response (GO:0045087) | + | + | + | |||||||
| C3 | complement and coagulation cascades (map04610) | + | |||||||||
| C5 | complement and coagulation cascades (map04610) | + | |||||||||
| Glucose and Lipid Metabolism | |||||||||||
| ACACA | fatty acid biosynthetic process (GO:0006633) | + | + | + | + | ||||||
| ACACB | fatty acid biosynthetic process (GO:0006633) | + | + | + | + | ||||||
| FDPS | cholesterol biosynthetic process (GO:0006695) | + | |||||||||
| GOT2 | 2-oxocarboxylic acid metabolism (map01210) | + | |||||||||
| GPT | 2-oxocarboxylic acid metabolism (map01210) | + | + | ||||||||
| HMGCS1 | cholesterol biosynthetic process (GO:0006695) | + | + | + | |||||||
| HSD17B7 | cholesterol biosynthetic process (GO:0006695) | + | |||||||||
| IDH4 | 2-oxocarboxylic acid metabolism (map01210) | + | |||||||||
| LDHA | 2-oxocarboxylic acid metabolism (map01210) | + | |||||||||
| MVD | cholesterol biosynthetic process (GO:0006695) | + | |||||||||
| TM7SF2 | cholesterol biosynthetic process (GO:0006695) | + | + | ||||||||
| Response to Hypoxia | |||||||||||
| LDHA, | pyruvate metabolism (map00620) | + | + | ||||||||
| PKLR | glycolytic process (GO:0006096) | + | + | ||||||||
| GAPDH | glycolytic process (GO:0006096) | + | + | ||||||||
| DCN | homeostatic process (GO:0042592) | + | |||||||||
| FBP1 | glycolytic process (GO:0006096) | + | |||||||||
| GBE1 | glycogen biosynthetic process (GO:0005978) | + | + | + | |||||||
| GLRX | homeostatic process (GO:0042592) | + | |||||||||
| NEDD4L | homeostatic process (GO:0042592) | + | + | + | |||||||
| PGK1 | glycolytic process (GO:0006096) | + | + | + | + | + | + | + | + | ||
| PLIN2 | homeostatic process (GO:0042592) | + | + | + | |||||||
| UGP2 | glycogen biosynthetic process (GO:0005978) | + | + | ||||||||
| ALDOA | glycolytic process (GO:0006096) | + | |||||||||
| ALDOB | glycolytic process (GO:0006096) | + | + | ||||||||
| GALK1 | glycolytic process (GO:0006096) | + | |||||||||
| MIF | homeostatic process (GO:0042592) | + | + | ||||||||
| PCK1 | homeostatic process (GO:0042592) | + | + | + | |||||||
| S100A4 | epithelial to mesenchymal transition (GO:0001837) | + | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Resaz, R.; Cangelosi, D.; Segalerba, D.; Morini, M.; Uva, P.; Bosco, M.C.; Banderali, G.; Estrella, A.; Wanner, C.; Weinstein, D.A.; et al. Exosomal MicroRNAs as Potential Biomarkers of Hepatic Injury and Kidney Disease in Glycogen Storage Disease Type Ia Patients. Int. J. Mol. Sci. 2022, 23, 328. https://doi.org/10.3390/ijms23010328
Resaz R, Cangelosi D, Segalerba D, Morini M, Uva P, Bosco MC, Banderali G, Estrella A, Wanner C, Weinstein DA, et al. Exosomal MicroRNAs as Potential Biomarkers of Hepatic Injury and Kidney Disease in Glycogen Storage Disease Type Ia Patients. International Journal of Molecular Sciences. 2022; 23(1):328. https://doi.org/10.3390/ijms23010328
Chicago/Turabian StyleResaz, Roberta, Davide Cangelosi, Daniela Segalerba, Martina Morini, Paolo Uva, Maria Carla Bosco, Giuseppe Banderali, Ana Estrella, Corbinian Wanner, David A. Weinstein, and et al. 2022. "Exosomal MicroRNAs as Potential Biomarkers of Hepatic Injury and Kidney Disease in Glycogen Storage Disease Type Ia Patients" International Journal of Molecular Sciences 23, no. 1: 328. https://doi.org/10.3390/ijms23010328
APA StyleResaz, R., Cangelosi, D., Segalerba, D., Morini, M., Uva, P., Bosco, M. C., Banderali, G., Estrella, A., Wanner, C., Weinstein, D. A., Sechi, A., Paci, S., Melis, D., Di Rocco, M., Lee, Y. M., & Eva, A. (2022). Exosomal MicroRNAs as Potential Biomarkers of Hepatic Injury and Kidney Disease in Glycogen Storage Disease Type Ia Patients. International Journal of Molecular Sciences, 23(1), 328. https://doi.org/10.3390/ijms23010328







