Searching for the Best Transthyretin Aggregation Protocol to Study Amyloid Fibril Disruption
Abstract
:1. Introduction
2. Results
2.1. Characterization of TTRwt Fibril Formation Protocols
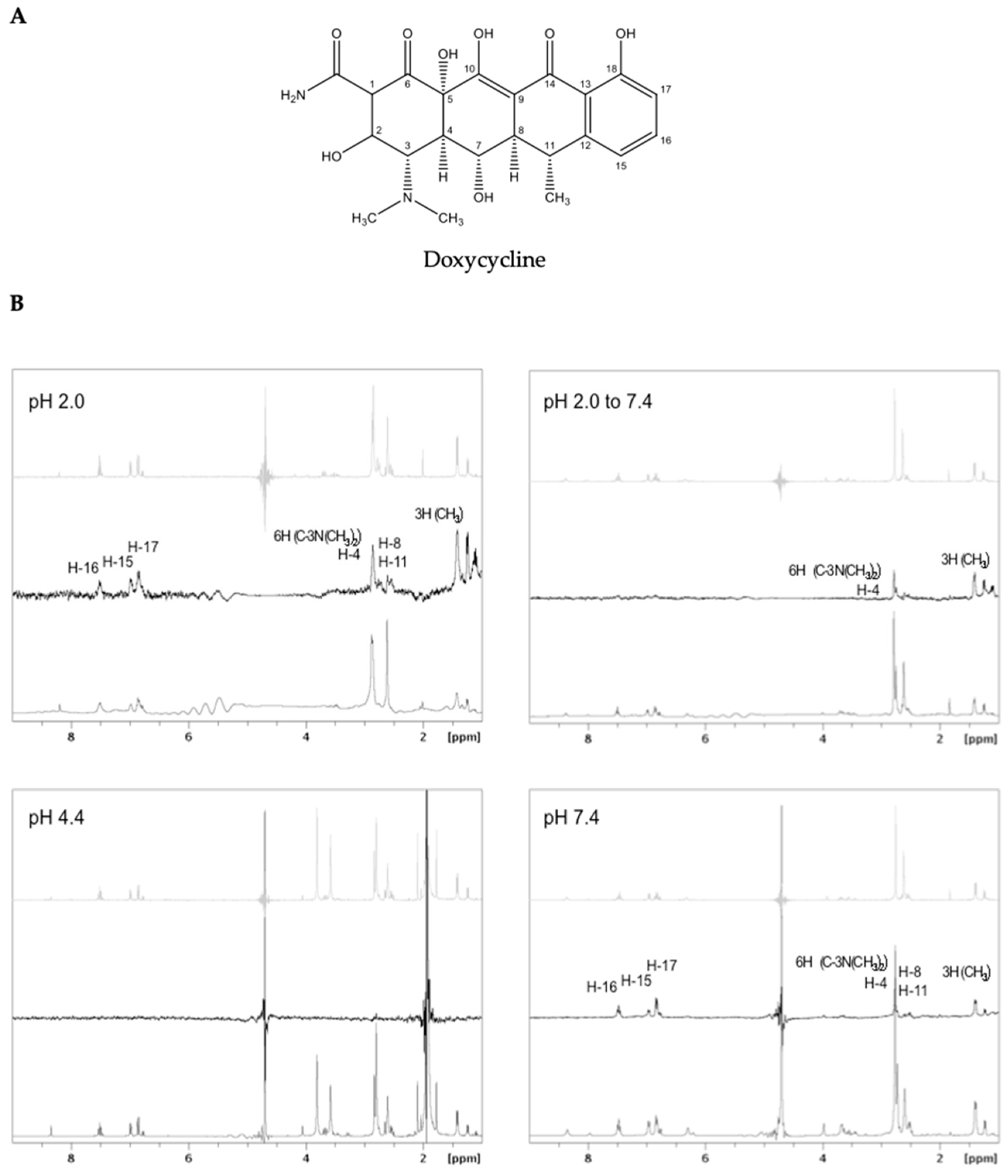
2.2. Interaction of Doxycycline with Preformed TTRwt Fibrils
2.3. Effect of Doxycycline on TTRwt Fibril Disaggregation
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Samples
4.3. Amyloid Fibril Formation Protocols
4.4. Thioflavin-T Assay
4.5. Circular Dichroism Spectroscopy (CD)
4.6. Turbidity Assays
4.7. Saturation Transfer Difference Nuclear Magnetic Resonance (STD NMR)
4.8. Dynamic Light Scattering (DLS)
4.9. Transmission Electron Microscopy (TEM)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Almeida, Z.L.; Brito, R.M.M. Structure and aggregation mechanisms in amyloids. Molecules 2020, 25, 1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefani, M.; Dobson, C.M. Protein aggregation and aggregate toxicity: New insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. 2003, 81, 678–699. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Iadanza, M.G.; Jackson, M.P.; Hewitt, E.W.; Ranson, N.A.; Radford, S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 755–773. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vassar, P.S.; Culling, C.F. Fluorescent stains, with special reference to amyloid and connective tissues. Arch. Pathol. 1959, 68, 487–498. [Google Scholar] [PubMed]
- Tanskanen, M.; Peuralinna, T.; Polvikoski, T.; Notkola, I.-L.; Sulkava, R.; Hardy, J.; Singleton, A.; Kiuru-Enari, S.; Paetau, A.; Tienari, P.J.; et al. Senile systemic amyloidosis affects 25% of the very aged and associates with genetic variation in alpha2-macroglobulin and tau: A population-based autopsy study. Ann. Med. 2008, 40, 232–239. [Google Scholar] [CrossRef]
- Ueda, M.; Horibata, Y.; Shono, M.; Misumi, Y.; Oshima, T.; Su, Y.; Tasaki, M.; Shinriki, S.; Kawahara, S.; Jono, H.; et al. Clinicopathological features of senile systemic amyloidosis: An ante- and post-mortem study. Mod. Pathol. 2011, 24, 1533–1544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, P.P.; Figueira, A.S.; Bravo, F.R. Amyloid fibril protein related to prealbumin in familial amyloidotic polyneuropathy. Proc. Natl. Acad. Sci. USA 1978, 75, 4499–4503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobson, D.R.; Pastore, R.D.; Yaghoubian, R.; Kane, I.; Gallo, G.; Buck, F.S.; Buxbaum, J.N. Variant-sequence transthyretin (isoleucine 122) in late-onset cardiac amyloidosis in black Americans. N. Engl. J. Med. 1997, 336, 466–473. [Google Scholar] [CrossRef]
- Benson, M.D. Leptomeningeal amyloid and variant transthyretins. Am. J. Pathol. 1996, 148, 351–354. [Google Scholar]
- Nakamura, M.; Yamashita, T.; Ueda, M.; Obayashi, K.; Sato, T.; Ikeda, T.; Washimi, Y.; Hirai, T.; Kuwahara, Y.; Yamamoto, M.T.; et al. Neuroradiologic and clinicopathologic features of oculoleptomeningeal type amyloidosis. Neurology 2005, 65, 1051–1056. [Google Scholar] [CrossRef]
- Benson, M.D.; Kincaid, J.C. The molecular biology and clinical features of amyloid neuropathy. Muscle Nerve 2007, 36, 411–423. [Google Scholar] [CrossRef]
- Mutations in Hereditary Amyloidosis. Available online: http://amyloidosismutations.com/mut-attr.php (accessed on 16 April 2021).
- Blaner, W.S.; Bonifácio, M.J.; Feldman, H.D.; Piantedosi, R.; Saraiva, M.J.M. Studies on the synthesis and secretion of transthyretin by the human hepatoma cell line Hep G2. FEBS Lett. 1991, 287, 193–196. [Google Scholar] [CrossRef] [Green Version]
- Dickson, P.W.; Schreiber, G. High levels of messenger RNA for transthyretin (prealbumin) in human choroid plexus. Neurosci. Lett. 1986, 66, 311–315. [Google Scholar] [CrossRef]
- Ong, D.E.; Davis, J.T.; O’Day, W.T.; Bok, D. Synthesis and secretion of retinol-binding protein and transthyretin by cultured retinal pigment epithelium. Biochemistry 1994, 33, 1835–1842. [Google Scholar] [CrossRef]
- Gião, T.; Saavedra, J.; Cotrina, E.; Quintana, J.; Llop, J.; Arsequell, G.; Cardoso, I. Undiscovered roles for transthyretin: From a transporter protein to a new therapeutic target for Alzheimer’s disease. Int. J. Mol. Sci. 2020, 21, 2075. [Google Scholar] [CrossRef] [Green Version]
- Morris, A.M.; Watzky, M.A.; Finke, R.G. Protein aggregation kinetics, mechanism, and curve-fitting: A review of the literature. Biochim. Biophys. Acta 2009, 1794, 375–397. [Google Scholar] [CrossRef]
- Faria, T.Q.; Almeida, Z.L.; Cruz, P.F.; Jesus, C.S.H.; Castanheira, P.; Brito, R.M.M. A look into amyloid formation by transthyretin: Aggregation pathway and a novel kinetic model. Phys. Chem. Chem. Phys. 2015, 17, 7255–7263. [Google Scholar] [CrossRef]
- Lai, Z.; Colón, W.; Kelly, J.W. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry 1996, 35, 6470–6482. [Google Scholar] [CrossRef]
- Quintas, A.; Saraiva, M.J.; Brito, R.M. The tetrameric protein transthyretin dissociates to a non-native monomer in solution. A novel model for amyloidogenesis. J. Biol. Chem. 1999, 274, 32943–32949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintas, A.; Vaz, D.C.; Cardoso, I.; Saraiva, M.J.; Brito, R.M. Tetramer dissociation and monomer partial unfolding precedes protofibril formation in amyloidogenic transthyretin variants. J. Biol. Chem. 2001, 276, 27207–27213. [Google Scholar] [CrossRef] [Green Version]
- Frangolho, A.; Correia, B.E.; Vaz, D.C.; Almeida, Z.L.; Brito, R.M.M. Oligomerization Profile of Human Transthyretin Variants with Distinct Amyloidogenicity. Molecules 2020, 25, 5698. [Google Scholar] [CrossRef]
- Saraiva, M.J.; Magalhaes, J.; Ferreira, N.; Almeida, M.R. Transthyretin Deposition in Familial Amyloidotic Polyneuropathy. Curr. Med. Chem. 2012, 19, 2304–2311. [Google Scholar] [CrossRef]
- Azevedo, E.P.C.; Pereira, H.M.; Garratt, R.C.; Kelly, J.W.; Foguel, D.; Palhano, F.L. Dissecting the structure, thermodynamic stability, and aggregation properties of the A25T transthyretin (A25T-TTR) variant involved in leptomeningeal amyloidosis: Identifying protein partners that co-aggregate during A25T-TTR fibrillogenesis in cerebrospinal fluid. Biochemistry 2011, 50, 11070–11083. [Google Scholar]
- Bulawa, C.E.; Connelly, S.; DeVit, M.; Wang, L.; Weigel, C.; Fleming, J.A.; Packman, J.; Powers, E.T.; Wiseman, R.L.; Foss, T.R.; et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc. Natl. Acad. Sci. USA 2012, 109, 9629–9634. [Google Scholar] [CrossRef] [Green Version]
- Damas, A.M.; Saraiva, M.J. Review: TTR amyloidosis-structural features leading to protein aggregation and their implications on therapeutic strategies. J. Struct. Biol. 2000, 130, 290–299. [Google Scholar] [CrossRef]
- Ueda, M.; Ando, Y. Recent advances in transthyretin amyloidosis therapy. Transl. Neurodegener. 2014, 3, 19. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.L.; Butler, J.; Heidecker, B. Emerging therapies in transthyretin amyloidosis—A new wave of hope after years of stagnancy? Eur. J. Heart Fail. 2020, 22, 39–53. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, E.J.; Guo, S.; Benson, M.D.; Booten, S.; Freier, S.; Hughes, S.G.; Kim, T.W.; Jesse Kwoh, T.; Matson, J.; Norris, D.; et al. Suppressing transthyretin production in mice, monkeys and humans using 2nd-Generation antisense oligonucleotides. Amyloid 2016, 23, 148–157. [Google Scholar] [CrossRef]
- Benson, M.D.; Waddington-Cruz, M.; Berk, J.L.; Polydefkis, M.; Dyck, P.J.; Wang, A.K.; Planté-Bordeneuve, V.; Barroso, F.A.; Merlini, G.; Obici, L.; et al. Inotersen treatment for patients with Hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018, 379, 22–31. [Google Scholar] [CrossRef]
- Brannagan, T.H.; Wang, A.K.; Coelho, T.; Waddington-Cruz, M.; Polydefkis, M.J.; Dyck, P.J.; Plante-Bordeneuve, V.; Berk, J.L.; Barroso, F.; Merlini, G.; et al. Early data on long-term efficacy and safety of inotersen in patients with hereditary transthyretin amyloidosis: A 2-year update from the open-label extension of the NEURO-TTR trial. Eur. J. Neurol. 2020, 27, 1374–1381. [Google Scholar] [CrossRef] [PubMed]
- Suhr, O.B.; Coelho, T.; Buades, J.; Pouget, J.; Conceicao, I.; Berk, J.; Schmidt, H.; Waddington-Cruz, M.; Campistol, J.M.; Bettencourt, B.R.; et al. Efficacy and safety of patisiran for familial amyloidotic polyneuropathy: A phase II multi-dose study. Orphanet. J. Rare Dis. 2015, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Adams, D.; Suhr, O.B.; Dyck, P.J.; Litchy, W.J.; Leahy, R.G.; Chen, J.; Gollob, J.; Coelho, T. Trial design and rationale for APOLLO, a Phase 3, placebo-controlled study of patisiran in patients with hereditary ATTR amyloidosis with polyneuropathy. BMC Neurol. 2017, 17, 181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Goel, V.; Robbie, G.J. Pharmacokinetics of Patisiran, the First Approved RNA Interference Therapy in Patients with Hereditary Transthyretin-Mediated Amyloidosis. J. Clin. Pharmacol. 2019, 60, 573–585. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, N.; Saraiva, M.J.; Almeida, M.R. Epigallocatechin-3-gallate as a potential therapeutic drug for TTR-related amyloidosis: “in vivo” evidence from FAP mice models. PLoS ONE 2012, 7, e29933. [Google Scholar] [CrossRef]
- Kristen, A.V.; Lehrke, S.; Buss, S.; Mereles, D.; Steen, H.; Ehlermann, P.; Hardt, S.; Giannitsis, E.; Schreiner, R.; Haberkorn, U.; et al. Green tea halts progression of cardiac transthyretin amyloidosis: An observational report. Clin. Res. Cardiol. 2012, 101, 805–813. [Google Scholar] [CrossRef] [Green Version]
- Merlini, G.; Ascari, E.; Amboldi, N.; Bellotti, V.; Arbustini, E.; Perfetti, V.; Ferrari, M.; Zorzoli, I.; Marinone, M.G.; Garini, P. Interaction of the anthracycline 4′-iodo-4′-deoxydoxorubicin with amyloid fibrils: Inhibition of amyloidogenesis. Proc. Natl. Acad. Sci. USA 1995, 92, 2959–2963. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, I.; Merlini, G.; Saraiva, M.J. 4′-iodo-4′-Deoxydoxorubicin and tetracyclines disrupt transthyretin amyloid fibrils in vitro producing noncytotoxic species: Screening for TTR fibril disrupters. FASEB J. 2003, 17, 803–809. [Google Scholar] [CrossRef]
- Cardoso, I.; Brito, M.; Saraiva, M.J. Extracellular matrix markers for disease progression and follow-up of therapies in familial amyloid polyneuropathy V30M TTR-related. Dis. Markers 2008, 25, 37–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardoso, I.; Saraiva, M.J. Doxycycline disrupts transthyretin amyloid: Evidence from studies in a FAP transgenic mice model. FASEB J. 2006, 20, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Obici, L.; Cortese, A.; Lozza, A.; Lucchetti, J.; Gobbi, M.; Palladini, G.; Perlini, S.; Saraiva, M.J.; Merlini, G. Doxycycline plus tauroursodeoxycholic acid for transthyretin amyloidosis: A phase II study. Amyloid 2012, 19 (Suppl. S1), 34–36. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, N.; Saraiva, M.J.; Almeida, M.R. Natural polyphenols inhibit different steps of the process of transthyretin (TTR) amyloid fibril formation. FEBS Lett. 2011, 585, 2424–2430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Xing, Y.F.; Huang, B.; Zhang, Y.Z.; Zeng, C.M. Tea catechins induce the conversion of preformed lysozyme amyloid fibrils to amorphous aggregates. J. Agric. Food Chem. 2009, 57, 11391–11396. [Google Scholar] [CrossRef] [PubMed]
- Forloni, G.; Colombo, L.; Girola, L.; Tagliavini, F.; Salmona, M. Anti-amyloidogenic activity of tetracyclines: Studies in vitro. FEBS Lett. 2001, 487, 404–407. [Google Scholar] [CrossRef] [Green Version]
- Skoulakis, S.; Goodfellow, J.M. The pH-dependent stability of wild-type and mutant transthyretin oligomers. Biophys. J. 2003, 84, 2795–2804. [Google Scholar] [CrossRef] [Green Version]
- Shnyrov, V.L.; Villar, E.; Zhadan, G.G.; Sanchez-Ruiz, J.M.; Quintas, A.; Saraiva, M.J.; Brito, R.M. Comparative calorimetric study of non-amyloidogenic and amyloidogenic variants of the homotetrameric protein transthyretin. Biophys. Chem. 2000, 88, 61–67. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Smith, C.S.; Petrassi, H.M.; Hammarström, P.; White, J.T.; Sacchettini, J.C.; Kelly, J.W. An engineered transthyretin monomer that is nonamyloidogenic, unless it is partially denatured. Biochemistry 2001, 40, 11442–11452. [Google Scholar] [CrossRef]
- Colon, W.; Kelly, J.W. Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry 1992, 31, 8654–8660. [Google Scholar] [CrossRef]
- Lundberg, E.; Olofsson, A.; Westermark, G.T.; Sauer-Eriksson, A.E. Stability and fibril formation properties of human and fish transthyretin, and of the Escherichia coli transthyretin-related protein. FEBS J. 2009, 276, 1999–2011. [Google Scholar] [CrossRef]
- Karlstedt, E.; Jimenez-Zepeda, V.; Howlett, J.G.; White, J.A.; Fine, N.M. Clinical experience with the use of doxycycline and ursodeoxycholic acid for the treatment of transthyretin cardiac amyloidosis. J. Card. Fail. 2019, 25, 147–153. [Google Scholar] [CrossRef]
- Teilum, K.; Kunze, M.B.A.; Erlendsson, S.; Kragelund, B.B. (S)Pinning down protein interactions by NMR. Protein Sci. 2017, 26, 436–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindgren, M.; Sörgjerd, K.; Hammarström, P. Detection and characterization of aggregates, prefibrillar amyloidogenic oligomers, and protofibrils using fluorescence spectroscopy. Biophys. J. 2005, 88, 4200–4212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brito, R.; Damas, A.; Saraiva, M. Amyloid Formation by Transthyretin: From Protein Stability to Protein Aggregation. Curr. Med. Chem. Endocr. Metab. Agents 2003, 3, 349–360. [Google Scholar] [CrossRef] [Green Version]
- Dolado, I.; Nieto, J.; Saraiva, M.J.M.; Arsequell, G.; Valencia, G.; Planas, A. Kinetic assay for high-throughput screening of in vitro transthyretin amyloid fibrillogenesis inhibitors. J. Comb. Chem. 2005, 7, 246–252. [Google Scholar] [CrossRef]
- Arsequell, G. Planas, Methods to evaluate the inhibition of TTR fibrillogenesis induced by small ligands. Curr. Med. Chem. 2012, 19, 2343–2355. [Google Scholar] [CrossRef]
- Simões, C.J.V.; Almeida, Z.L.; Costa, D.; Jesus, C.S.H.; Cardoso, A.L.; Almeida, M.R.; Saraiva, M.J.; Pinho e Melo, T.M.V.D.; Brito, R.M.M. A novel bis-furan scaffold for transthyretin stabilization and amyloid inhibition. Eur. J. Med. Chem. 2016, 121, 823–840. [Google Scholar] [CrossRef]
- Viegas, A.; Manso, J.; Nobrega, F.L.; Cabrita, E.J. Saturation-transfer difference (STD) NMR: A simple and fast method for ligand screening and characterization of protein binding. J. Chem. Educ. 2011, 88, 990–994. [Google Scholar] [CrossRef]
- Teixeira, C.; Costelha, S.; Martins, H.S.; Teixeira, A.; Saraiva, M.J. Doxycycline-tauroursodeoxycholic acid treatment: Effects in the heart of a transthyretin V30M transgenic mouse model. Amyloid 2017, 24, 80. [Google Scholar] [CrossRef]
- Cardoso, I.; Pereira, P.J.; Damas, A.M.; Saraiva, M.J. Aprotinin binding to amyloid fibrils. Eur. J. Biochem. 2000, 267, 2307–2311. [Google Scholar] [CrossRef] [Green Version]
- Micsonai, A.; Wien, F.; Bulyáki, E.; Kun, J.; Moussong, E.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef] [PubMed]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Sousa, M.M.; Fernandes, R.; Palha, J.A.; Taboada, A.; Vieira, P.; Saraiva, M.J. Evidence for early cytotoxic aggregates in transgenic mice for human transthyretin Leu55Pro. Am. J. Pathol. 2002, 161, 1935–1948. [Google Scholar] [CrossRef] [Green Version]



| pH | Temperature | Incubation Period | Protein Concentration | Stirring |
|---|---|---|---|---|
| 2 | 25 °C | At least 1 week | 80 μM and then diluted to 3.6 μM | No |
| 4.4 | 37 °C | 3 days | 3.6 μM | No |
| 7.4 | 60 °C | 6 days | 3.6 μM | No |
| Aggregation Protocol | Morphology of the Aggregates and Fibrils | Length/Diameter of Aggregated Species | Thioflavin-T Assay | Effect of pH Adjustment to pH 7.4 | |
|---|---|---|---|---|---|
| pH 2.0 | Long, unbranched, mature fibrils | 50 to 170 nm / 5 to 9 nm | Positive | Alterations in the size and secondary structure of fibrils | Fibrils remain ThT positive |
| pH 4.4 | Mixture of spheroid structures, amorphous aggregates, and short unbranched fibrils | Spheroid aggregates (4 to 6 nm); fibrils (25 nm / 4–5 nm) | Positive | Slight effect on size, secondary structure, and morphology of aggregates to more amorphous and less fibrillar species | Fibrils remain ThT positive |
| pH 7.4 | Unbranched fibrils | 10 to 50 nm/4 to 6 nm | Positive | Not necessary | |
| Aggregation Protocol | pH 2.0 | pH 2.0 → pH 7.4 | pH 4.4 | pH 4.4 → pH 7.4 | pH 7.4 (60 °C) | pH 7.4 (37 °C) |
|---|---|---|---|---|---|---|
| α-Helix | 0.14 | 0.03 | 0.07 | 0.08 | 0.11 | 0.09 |
| β-Sheet | 0.43 | 0.32 | 0.32 | 0.24 | 0.4 | 0.39 |
| Turn | 0.09 | 0.13 | 0.16 | 0.17 | 0.11 | 0.13 |
| Unordered | 0.34 | 0.52 | 0.45 | 0.51 | 0.38 | 0.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferreira, E.; Almeida, Z.L.; Cruz, P.F.; Silva e Sousa, M.; Veríssimo, P.; Brito, R.M.M. Searching for the Best Transthyretin Aggregation Protocol to Study Amyloid Fibril Disruption. Int. J. Mol. Sci. 2022, 23, 391. https://doi.org/10.3390/ijms23010391
Ferreira E, Almeida ZL, Cruz PF, Silva e Sousa M, Veríssimo P, Brito RMM. Searching for the Best Transthyretin Aggregation Protocol to Study Amyloid Fibril Disruption. International Journal of Molecular Sciences. 2022; 23(1):391. https://doi.org/10.3390/ijms23010391
Chicago/Turabian StyleFerreira, Elisabete, Zaida L. Almeida, Pedro F. Cruz, Marta Silva e Sousa, Paula Veríssimo, and Rui M. M. Brito. 2022. "Searching for the Best Transthyretin Aggregation Protocol to Study Amyloid Fibril Disruption" International Journal of Molecular Sciences 23, no. 1: 391. https://doi.org/10.3390/ijms23010391
APA StyleFerreira, E., Almeida, Z. L., Cruz, P. F., Silva e Sousa, M., Veríssimo, P., & Brito, R. M. M. (2022). Searching for the Best Transthyretin Aggregation Protocol to Study Amyloid Fibril Disruption. International Journal of Molecular Sciences, 23(1), 391. https://doi.org/10.3390/ijms23010391






