Genomics Associated Interventions for Heat Stress Tolerance in Cool Season Adapted Grain Legumes
Abstract
1. Introduction
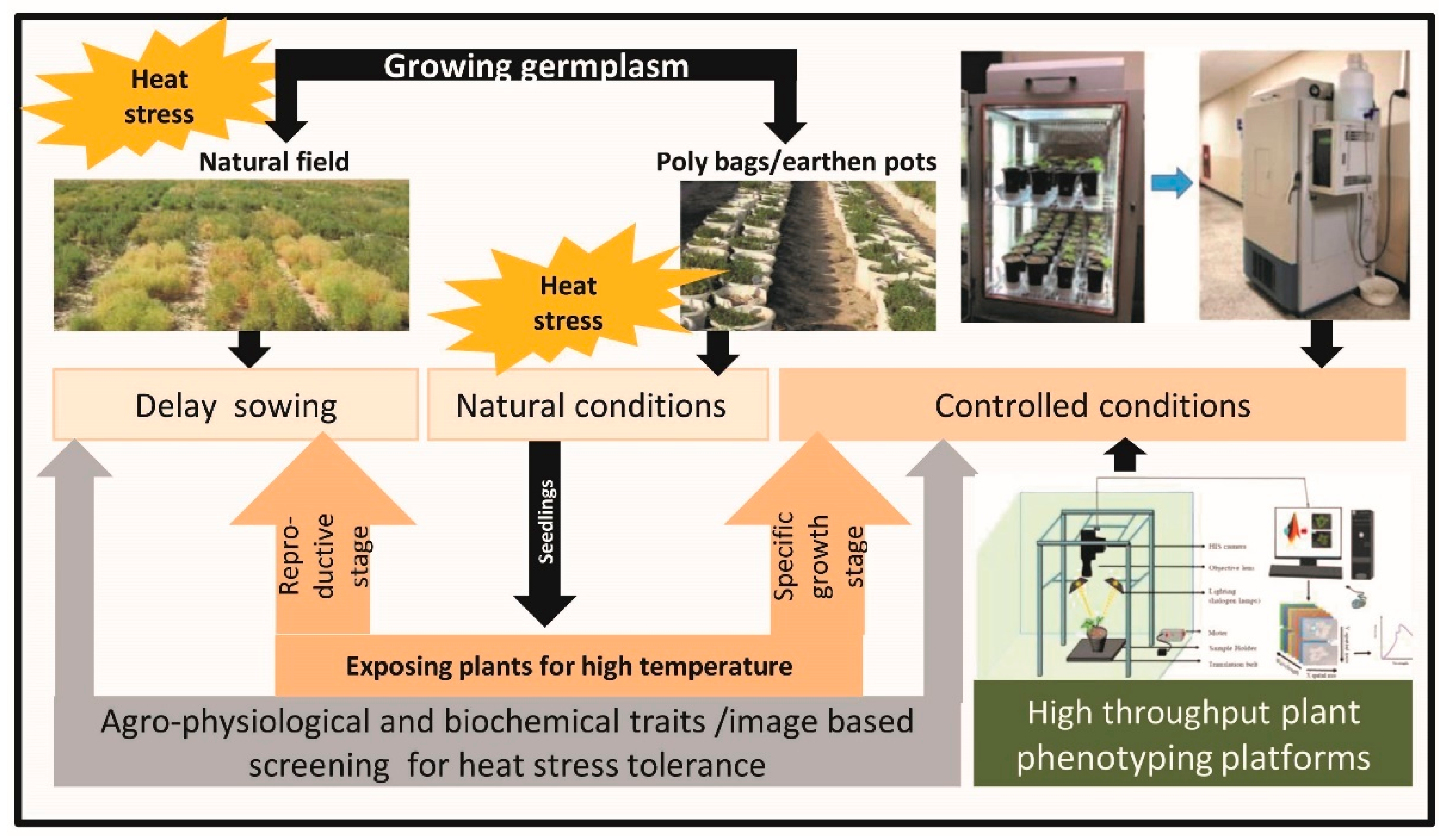
2. Advances in Screening Techniques for Heat Tolerance
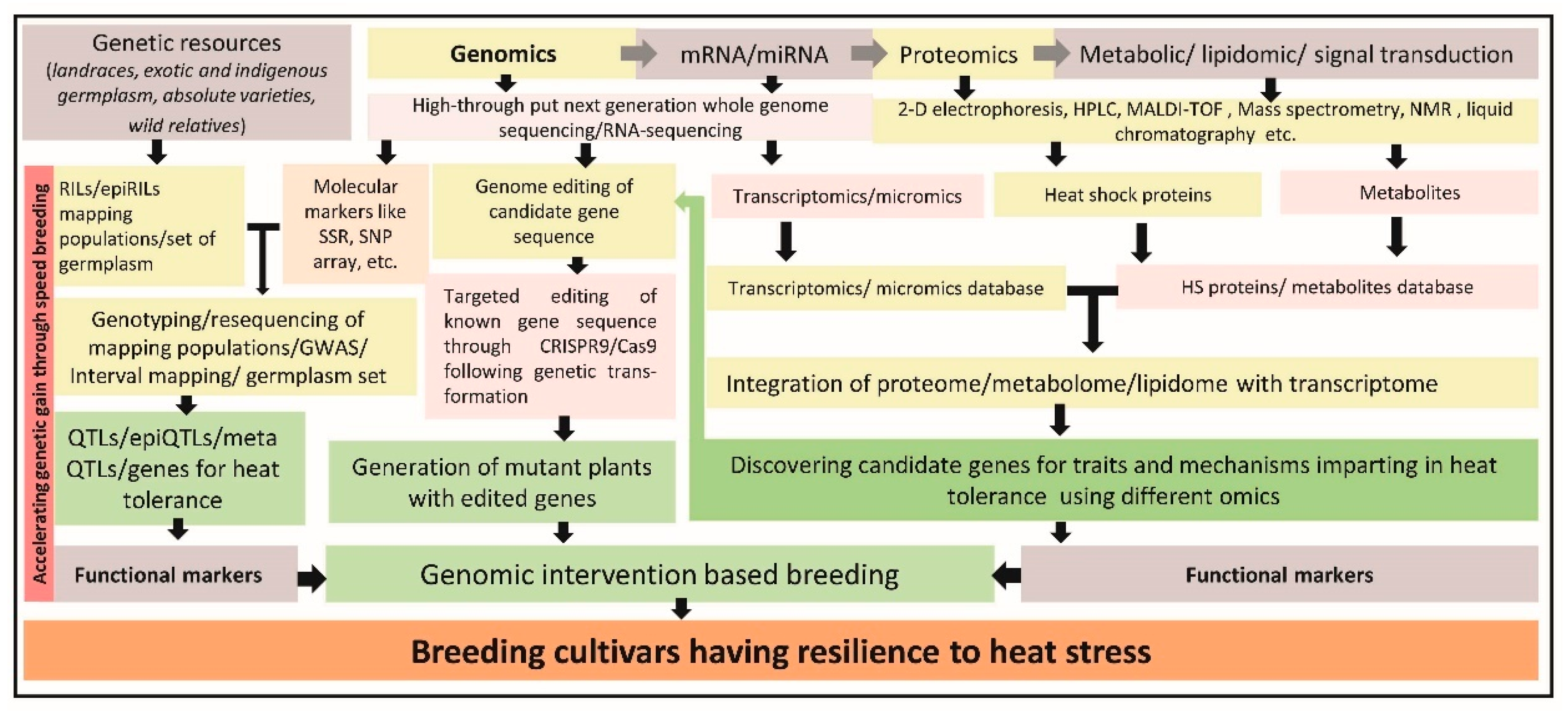
3. Genomics Interventions
3.1. QTL/Gene Mapped for Traits Imparting Heat Stress
3.2. Transcriptomics, Transcription Factors and Candidate Genes
3.2.1. microRNA
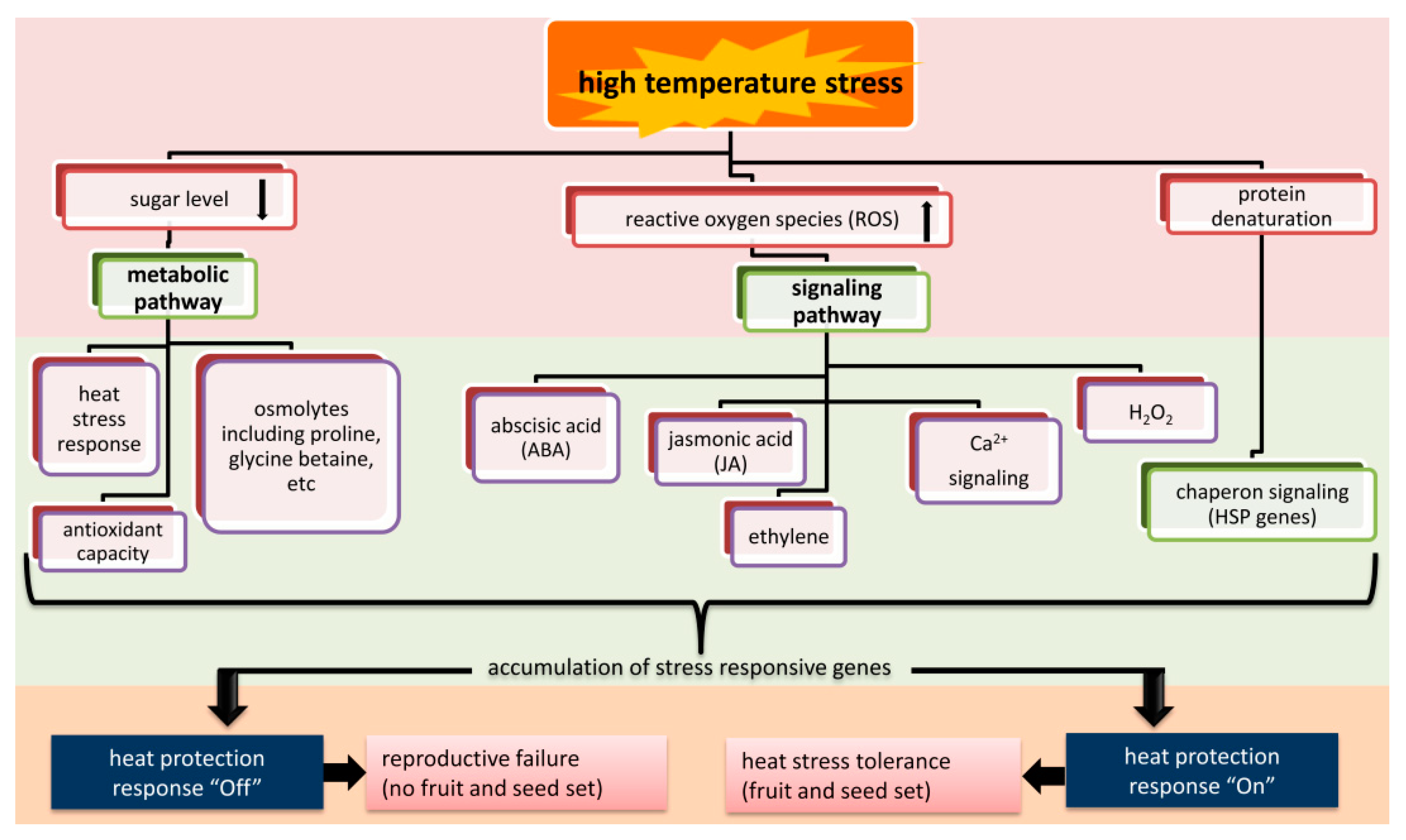
3.2.2. Signaling and Metabolic Pathways
4. Future Possibilities Genomic Intervention for Heat Stress Tolerance
4.1. Epigenetic Modifications
4.2. Mining Novel Allelic Variations for Heat Stress Tolerance
4.2.1. Genome Editing
4.2.2. Targeting-Induced Local Lesions in Genome (TILLING)
4.3. Genomics of Underground Traits
4.4. Nanoparticles Based Genomics and Proteomics
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Srivastava, R.P.; Vasishtha, H. Genotypic variation in pigeonpea for protein, dietary fibre, fatty acids and lectins. Indian J. Agric. Biochem. 2012, 25, 111–115. [Google Scholar]
- Kumar, J.; Sen Gupta, D.; Djalovic, I. Breeding, genetics, and genomics for tolerance against terminal heat in lentil: Current status and future directions. Legume Sci. 2020, 2, e38. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Zhu, Y.; Jones, A.; Rose, R.J.; Song, Y. Heat stress in legume seed setting: Effects, causes, and future prospects. Front. Plant Sci. 2019, 10, 938. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Srivastava, V.; Purayannur, S.; Kaladhar, V.C.; Cheruvu, P.J.; Verma, P.K. WRKY domain-encoding genes of a crop legume chickpea (Cicer arietinum L): Comparative analysis with Medicago truncatula WRKY family and characterization of group-III gene (s). DNA Res. 2016, 23, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, K.B.; Lu, Z.G.; Ren, S.X.; Jiang, H.R.; Cui, J.W.; Chen, G.; Teng, N.J.; Lam, H.M.; Jin, B. Differential physiological, transcriptomic and metabolomic responses of Arabidopsis leaves under prolonged warming and heat shock. BMC Plant Biol. 2020, 20, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ridge, P.E.; Pye, D.L. The effects of temperature and frost at flowering on the yield of peas grown in a Mediterranean environment. Field Crops Res. 1985, 12, 339–346. [Google Scholar] [CrossRef]
- Deva, C.R.; Urban, M.O.; Challinor, A.J.; Falloon, P.; Svitákova, L. Enhanced leaf cooling is a pathway to heat tolerance in common bean. Front. Plant Sci. 2020, 11, 19. [Google Scholar] [CrossRef]
- Sita, K.; Sehgal, A.; Hanumantha Rao, B.; Nair, R.M.; Prasad, P.V.; Kumar, S.; Gaur, P.M.; Farooq, M.; Siddique, K.H.; Varshney, R.K.; et al. Food legumes and rising temperatures: Effects, adaptive functional mechanisms specific to reproductive growth stage and strategies to improve heat tolerance. Front. Plant Sci. 2017, 8, 1658. [Google Scholar] [CrossRef]
- Gaur, P.M.; Samineni, S.; Krishnamurthy, L.; Kumar, S.; Ghanem, M.E.; Beebe, S.E.; Rao, I.M.; Chaturvedi, S.K.; Basu, P.S.; Nayyar, H.; et al. High temperature tolerance in grain legumes. Legume Perspect. 2015, 7, 23–24. [Google Scholar]
- Gaur, P.M.; Samineni, S.; Thudi, M.; Tripathi, S.; Sajja, S.B.; Jayalakshmi, V.; Mannur, D.M.; Vijayakumar, A.G.; Ganga Rao, N.V.; Ojiewo, C.; et al. Integrated breeding approaches for improving drought and heat adaptation in chickpea (Cicer arietinum L.). Plant Breed. 2019, 138, 389–400. [Google Scholar] [CrossRef]
- Ehlers, J.D.; Hall, A.E. Heat tolerance of contrasting cowpea lines in short and long days. Field Crops Res. 1998, 55, 11–21. [Google Scholar] [CrossRef]
- Porch, T.G.; Blair, M.W.; Lariguet, P.; Galeano, C.; Pankhurst, C.E.; Broughton, W.J. Generation of a mutant population for TILLING common bean genotype BAT 93. J. Am. Soc. Hortic. Sci. 2009, 134, 348–355. [Google Scholar] [CrossRef]
- Krishnamurthy, L.; Gaur, P.M.; Basu, P.S.; Chaturvedi, S.K.; Tripathi, S.; Vadez, V.; Rathore, A.; Varshney, R.K.; Gowda, C.L. Large genetic variation for heat tolerance in the reference collection of chickpea (Cicer arietinum L.) germplasm. Plant Genet. Resour. Charact. Util. 2011, 9, 59–69. [Google Scholar] [CrossRef]
- Porch, T.G.; Hall, A.E. Heat tolerance. In Genomics and Breeding for Climate-Resilient Crops; Kole, C., Ed.; Springer Publishing: Berlin, Germany, 2013; pp. 167–202. [Google Scholar]
- Kumar, A.P.; McKeown, P.C.; Boualem, A.; Ryder, P.; Brychkova, G.; Bendahmane, A.; Sarkar, A.; Chatterjee, M.; Spillane, C. TILLING by Sequencing (TbyS) for targeted genome mutagenesis in crops. Mol. Breed. 2017, 37, 14. [Google Scholar] [CrossRef]
- Govindaraj, M.; Pattanashetti, S.K.; Patne, N.; Kanatti, A.A.; Ciftci, Y.O. Breeding cultivars for heat stress tolerance in staple food crops. In Next Generation Plant Breeding; Çiftçi, Y.O., Ed.; Intech Open: London, UK, 2018; pp. 45–74. [Google Scholar]
- Kumar, J.; Sen Gupta, D.; Djalovic, I.; Kumar, S.; Siddique, K.H.M. Root-omics for drought tolerance in cool–season grain legumes. Physiol. Plant. 2021, 172, 629–644. [Google Scholar] [CrossRef]
- Andrews, M.; Hodge, S. Climate change, a challenge for cool season grain legume crop production. In Climate Change and Management of Cool Season Grain Legume Crops; Yadav, S., Redden, R., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 1–9. [Google Scholar]
- Farooq, M.; Nadeem, F.; Gogoi, N.; Ullah, A.; Alghamdi, S.S.; Nayyar, H.; Siddique, K.H.M. Heat stress in grain legumes during reproductive and grain-filling phases. Crop Pasture Sci. 2017, 68, 985–1005. [Google Scholar] [CrossRef]
- Kudapa, H.; Ramalingam, A.; Nayakoti, S.; Chen, X.; Zhuang, W.J.; Liang, X.; Kahl, G.; Edwards, D.; Varshney, R.K. Functional genomics to study stress responses in crop legumes: Progress and prospects. Funct. Plant Biol. 2013, 40, 1221–1233. [Google Scholar] [CrossRef]
- Rani, A.; Devi, P.; Jha, U.C.; Sharma, K.D.; Siddique, K.H.M.; Nayyar, H. Developing climate-resilient chickpea involving physiological and molecular approaches with a focus on temperature and drought stresses. Front. Plant Sci. 2020, 10, 1759. [Google Scholar] [CrossRef] [PubMed]
- Tafesse, E.G. Heat Stress Resistance in Pea (Pisum sativum L.) Based on Canopy and Leaf Traits. Doctoral Dissertation, University of Saskatchewan, Saskatoon, SK, Canada, 2018. [Google Scholar]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Lavania, D.; Siddiqui, M.H.; Al–Whaibi, M.H.; Singh, A.K.; Kumar, R.; Grover, A. Genetic approaches for breeding heat stress tolerance in faba bean (Vicia faba L.). Acta Physiol. Plant. 2015, 37, 1737. [Google Scholar] [CrossRef]
- Matthews, P.; Marcellos, H. Faba Bean. AgFact P4-2-7, 2nd ed.; NSW Department of Primary Industries: Tamworth, UK, 2003.
- Devasirvatham, V.; Gaur, P.M.; Mallikarjuna, N.; Raju, T.N.; Trethowan, R.M.; Tan, D.K. Reproductive biology of chickpea response to heat stress in the field is associated with the performance in controlled environments. Field Crops Res. 2013, 142, 9–19. [Google Scholar] [CrossRef]
- Sehgal, A.; Sita, K.; Kumar, J.; Kumar, S.; Singh, S.; Siddique, K.H.; Nayyar, H. Effects of drought, heat and their interaction on the growth, yield and photosynthetic function of lentil (Lens culinaris Medikus) genotypes varying in heat and drought sensitivity. Front. Plant Sci. 2017, 8, 1776. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.S.; Kaothien–Nakayama, P.; Iwano, M.; Takayama, S. A TILLING resource for functional genomics in Arabidopsis thaliana accession C24. Genes Genet. Syst. 2012, 87, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.R.; Paull, J.G.; Siddique, K.H.M.; Stoddard, F.L. Faba bean breeding for drought-affected environments: A physiological and agronomic perspective. Field Crops Res. 2010, 115, 279–286. [Google Scholar] [CrossRef]
- Burke, J.J.; Chen, J. Enhancement of reproductive heat tolerance in plants. PLoS ONE 2015, 10, e0122933. [Google Scholar] [CrossRef]
- Gao, G.; Tester, M.A.; Julkowska, M.M. The use of high-throughput phenotyping for assessment of heat stress-induced changes in Arabidopsis. Plant Phenomics 2020, 2020, 3723916. [Google Scholar] [CrossRef]
- Kiran, B.A.; Chimmad, V.P. Evaluation of chickpea genotypes for heat stress under partially controlled condition (elevated temperature) in polybags. Int. J. Chem. Stud. 2018, 6, 3008–3011. [Google Scholar]
- Zhang, C.; Craine, W.A.; McGee, R.J.; Vandemark, G.J.; Davis, J.B.; Brown, J.; Hulbert, S.H.; Sankaran, S. Image-based phenotyping of flowering intensity in cool-season crops. Sensors 2020, 20, 1450. [Google Scholar] [CrossRef]
- Confalonieri, R.; Paleari, L.; Foi, M.; Movedi, E.; Vesely, F.M.; Thoelke, W.; Agape, C.; Borlini, G.; Ferri, I.; Massara, F.; et al. PocketPlant3D: Analysing canopy structure using a smartphone. Biosyst. Eng. 2017, 164, 1–12. [Google Scholar] [CrossRef]
- Hui, F.; Zhu, J.; Hu, P.; Meng, L.; Zhu, B.; Guo, Y.; Li, B.; Ma, Y. Image-based dynamic quantification and high-accuracy 3D evaluation of canopy structure of plant populations. Ann. Bot. 2018, 121, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Di Gennaro, S.F.; Rizza, F.; Badeck, F.W.; Berton, A.; Delbono, S.; Gioli, B.; Toscano, P.; Zaldei, A.; Matese, A. UAV-based high-throughput phenotyping to discriminate barley vigour with visible and near-infrared vegetation indices. Int. J. Remote Sens. 2018, 39, 5330–5344. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Y.; Du, J.; Guo, X.; Wen, W.; Gu, S.; Wang, J.; Fan, J. Crop phenomics: Current status and perspectives. Front. Plant Sci. 2019, 10, 714. [Google Scholar] [CrossRef] [PubMed]
- Furbank, R.T.; Jimenez-Berni, J.A.; George-Jaeggli, B.; Potgieter, A.B.; Deery, D.M. Field crop phenomics: Enabling breeding for radiation use efficiency and biomass in cereal crops. New Phytol. 2019, 223, 1714–1727. [Google Scholar] [CrossRef]
- Bohra, A.; Jha, U.C.; Godwin, I.; Varshney, R.K. Genomic interventions for sustainable agriculture. Plant Biotechnol. J. 2020, 18, 2388–2405. [Google Scholar] [CrossRef]
- Roorkiwal, M.; Bharadwaj, C.; Barmukh, R.; Dixit, G.P.; Thudi, M.; Gaur, P.M.; Chaturvedi, S.K.; Fikre, A.; Hamwieh, A.; Kumar, S.; et al. Integrating genomics for chickpea improvement: Achievements and opportunities. Theor. Appl. Genet. 2020, 133, 1703–1720. [Google Scholar] [CrossRef]
- Kumar, J.; Gupta, D.S. Prospects of next generation sequencing in lentil breeding. Mol. Biol. Rep. 2020, 47, 9043–9053. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.S.; Kumar, J.; Gupta, S.; Dubey, S.; Gupta, P.; Singh, N.P.; Sablok, G. Identification, development, and application of cross-species intron-spanning markers in lentil (Lens culinaris Medik.). Crop J. 2018, 6, 299–305. [Google Scholar] [CrossRef]
- Kumar, J.; Sen Gupta, D.; Baum, M.; Varshney, R.K.; Kumar, S. Genomics-assisted lentil breeding: Current status and future strategies. Legume Sci. 2021, e71. [Google Scholar] [CrossRef]
- Gaur, R.; Azam, S.; Jeena, G.; Khan, A.W.; Choudhary, S.; Jain, M.; Yadav, G.; Tyagi, A.K.; Chattopadhyay, D.; Bhatia, S. High-throughput SNP discovery and genotyping for constructing a saturated linkage map of chickpea (Cicer arietinum L.). DNA Res. 2012, 19, 357–373. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Song, C.; Saxena, R.K.; Azam, S.; Yu, S.; Sharpe, A.G.; Cannon, S.; Baek, J.; Rosen, B.D.; Tarzan, B.; et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 2013, 31, 240–246. [Google Scholar] [CrossRef]
- Garg, R.; Patel, R.K.; Tyagi, A.K.; Jain, M. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 2011, 18, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Patel, R.K.; Jhanwar, S.; Priya, P.; Bhattacharjee, A.; Yadav, G.; Bhatia, S.; Chattopadhyay, D.; Tyagi, A.K.; Jain, M. Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiol. 2011, 156, 1661–1678. [Google Scholar] [CrossRef] [PubMed]
- Hiremath, P.J.; Farmer, A.; Cannon, S.B.; Woodward, J.; Kudapa, H.; Tuteja, R.; Kumar, A.; Bhanu Prakash, A.; Mulaosmanovic, B.; Gujaria, N.; et al. Large-scale transcriptome analysis in chickpea (Cicer arietinum L.) an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotechnol. J. 2011, 9, 922–931. [Google Scholar] [CrossRef] [PubMed]
- Hiremath, P.J.; Kumar, A.; Penmetsa, R.V.; Farmer, A.; Schlueter, J.A.; Chamarthi, S.K.; Whaley, A.M.; Carrasquilla-Garcia, N.; Gaur, P.M.; Upadhyaya, H.D.; et al. Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol. J. 2012, 10, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Jhanwar, S.; Priya, P.; Garg, R.; Parida, S.K.; Tyagi, A.K.; Jain, M. Transcriptome sequencing of wild chickpea as a rich resource for marker development. Plant Biotechnol. J. 2012, 10, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, G.; Jhanwar, S.; Priya, P.; Singh, V.K.; Saxena, M.S.; Parida, S.K.; Garg, R.; Tyagi, A.K.; Jain, M. Comparative analysis of kabuli chickpea transcriptome with desi and wild chickpea provides a rich resource for development of functional markers. PLoS ONE 2012, 7, e52443. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Cogan, N.O.; Pembleton, L.W.; Shinozuka, M.; Savin, K.W.; Materne, M.; Forster, J.W. Transcriptome sequencing of lentil based on second-generation technology permits large-scale unigene assembly and SSR marker discovery. BMC Genom. 2011, 12, 265. [Google Scholar] [CrossRef]
- Sharpe, A.G.; Ramsay, L.; Sanderson, L.A.; Fedoruk, M.J.; Clarke, W.E.; Li, R.; Kagale, S.; Vijayan, P.; Vandenberg, A.; Bett, K.E. Ancient orphan crop joins modern era: Gene-based SNP discovery and mapping in lentil. BMC Genom. 2013, 14, 1–3. [Google Scholar] [CrossRef]
- Verma, P.; Shah, N.; Bhatia, S. Development of an expressed gene catalogue and molecular markers from the de novo assembly of short sequence reads of the lentil (Lens culinaris M edik.) transcriptome. Plant Biotechnol. J. 2013, 11, 894–905. [Google Scholar] [CrossRef]
- Temel, H.Y.; Göl, D.; Akkale, H.B.; Kahriman, A.; Tanyolac, M.B. Single nucleotide polymorphism discovery through Illumina-based transcriptome sequencing and mapping in lentil. Turk. J. Agric. For. 2015, 39, 470–488. [Google Scholar] [CrossRef]
- Singh, D.; Singh, C.K.; Tomar, R.S.; Pal, M. Genetics and molecular mapping of heat tolerance for seedling survival and pod set in lentil. Crop Sci. 2017, 57, 3059–3067. [Google Scholar] [CrossRef]
- Pavan, S.; Bardaro, N.; Fanelli, V.; Marcotrigiano, A.R.; Mangini, G.; Taranto, F.; Catalano, D.; Montemurro, C.; De Giovanni, C.; Lotti, C.; et al. Genotyping by sequencing of cultivated lentil (Lens culinaris Medik.) highlights population structure in the Mediterranean gene pool associated with geographic patterns and phenotypic variables. Front. Genet. 2019, 10, 872. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Gupta, D.S.; Kesari, R.; Verma, R.; Murugesan, S.; Basu, P.S.; Soren, K.R.; Gupta, S.; Singh, N.P. Comprehensive RNAseq analysis for identification of genes expressed under heat stress in lentil. Physiol. Plant. 2021, 173, 1785–1807. [Google Scholar] [CrossRef] [PubMed]
- Gali, K.K.; Sackville, A.; Tafesse, E.G.; Lachagari, V.B.; McPhee, K.; Hybl, M.; Mikić, A.; Smýkal, P.; McGee, R.; Burstin, J.; et al. Genome-wide association mapping for agronomic and seed quality traits of field pea (Pisum sativum L.). Front. Plant Sci. 2019, 10, 1538. [Google Scholar] [CrossRef]
- Beji, S.; Fontaine, V.; Devaux, R.; Thomas, M.; Negro, S.S.; Bahrman, N.; Siol, M.; Aubert, G.; Burstin, J.; Hilbert, J.L.; et al. Genome-wide association study identifies favorable SNP alleles and candidate genes for frost tolerance in pea. BMC Genom. 2020, 21, 536. [Google Scholar] [CrossRef]
- Leonforte, A.; Sudheesh, S.; Cogan, N.O.; Salisbury, P.A.; Nicolas, M.E.; Materne, M.; Forster, J.W.; Kaur, S. SNP marker discovery, linkage map construction and identification of QTLs for enhanced salinity tolerance in field pea (Pisum sativum L.). BMC Plant Biol. 2013, 13, 161. [Google Scholar] [CrossRef]
- Gong, Y.M.; Xu, S.C.; Mao, W.H.; Hu, Q.Z.; Zhang, G.W.; Ding, J.; Li, Y.D. Developing new SSR markers from ESTs of pea (Pisum sativum L.). J. Zhejiang Univ. Sci. B 2010, 11, 702–707. [Google Scholar] [CrossRef]
- Deulvot, C.; Charrel, H.; Marty, A.; Jacquin, F.; Donnadieu, C.; Lejeune-Hénaut, I.; Aubert, G. Highly-multiplexed SNP genotyping for genetic mapping and germplasm diversity studies in pea. BMC Genom. 2010, 11, 468. [Google Scholar] [CrossRef]
- Cheng, P.; Holdsworth, W.; Ma, Y.; Coyne, C.J.; Mazourek, M.; Grusak, M.A.; Fuchs, S.; McGee, R.J. Association mapping of agronomic and quality traits in USDA pea single-plant collection. Mol. Breed. 2015, 35, 1–3. [Google Scholar] [CrossRef]
- Duarte, J.; Rivière, N.; Baranger, A.; Aubert, G.; Burstin, J.; Cornet, L.; Lavaud, C.; Lejeune-He′naut, I.; Martinant, J.P.; Pichon, J.P.; et al. Transcriptome sequencing for high throughput SNP development and genetic mapping in pea. BMC Genom. 2014, 15, 126. [Google Scholar] [CrossRef]
- Sindhu, A.; Ramsay, L.; Sanderson, L.A.; Stonehouse, R.; Li, R.; Condie, J.; Shunmugam, A.S.; Liu, Y.; Jha, A.B.; Diapari, M.; et al. Gene-based SNP discovery and genetic mapping in pea. Theor. Appl. Genet. 2014, 127, 2225–2241. [Google Scholar] [CrossRef]
- Franssen, S.U.; Shrestha, R.P.; Bräutigam, A.; Bornberg-Bauer, E.; Weber, A.P. Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genom. 2011, 12, 227. [Google Scholar] [CrossRef]
- Ferraro, K.; Jin, A.L.; Nguyen, T.D.; Reinecke, D.M.; Ozga, J.A.; Ro, D.K. Characterization of proanthocyanidin metabolism in pea (Pisum sativum) seeds. BMC Plant Biol. 2014, 14, 238. [Google Scholar] [CrossRef]
- Tayeh, N.; Aluome, C.; Falque, M.; Jacquin, F.; Klein, A.; Chauveau, A.; Bérard, A.; Houtin, H.; Rond, C.; Kreplak, J.; et al. Development of two major resources for pea genomics: The GenoPea 13.2 K SNP Array and a high-density, high-resolution consensus genetic map. Plant J. 2015, 84, 1257–1273. [Google Scholar] [CrossRef]
- Sudheesh, S.; Sawbridge, T.I.; Cogan, N.O.; Kennedy, P.; Forster, J.W.; Kaur, S. De novo assembly and characterisation of the field pea transcriptome using RNA-Seq. BMC Genom. 2015, 16, 611. [Google Scholar] [CrossRef] [PubMed]
- Alves–Carvalho, S.; Aubert, G.; Carrère, S.; Cruaud, C.; Brochot, A.L.; Jacquin, F.; Klein, A.; Martin, C.; Boucherot, K.; Kreplak, J.; et al. Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant J. 2015, 84, 1–19. [Google Scholar] [CrossRef]
- Zhukov, V.A.; Zhernakov, A.I.; Kulaeva, O.A.; Ershov, N.I.; Borisov, A.Y.; Tikhonovich, I.A. De novo assembly of the pea (Pisum sativum L.) nodule transcriptome. Int. J. Genom. 2015, 2015, 695947. [Google Scholar] [CrossRef]
- Kerr, S.C.; Gaiti, F.; Beveridge, C.A.; Tanurdzic, M. De novo transcriptome assembly reveals high transcriptional complexity in Pisum sativum axillary buds and shows rapid changes in expression of diurnally regulated genes. BMC Genom. 2017, 18, 221. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Gali, K.K.; Lachagari, R.V.; Chakravartty, N.; Bueckert, R.; Tarzan, B.; Warkentin, T.D. Identification of heat responsive genes in pea stipules and anthers through transcriptional profiling. bioRxiv 2021. [Google Scholar] [CrossRef]
- Yang, T.; Fang, L.; Zhang, X.; Hu, J.; Bao, S.; Hao, J.; Li, L.; He, Y.; Jiang, J.; Wang, F.; et al. High-throughput development of SSR markers from pea (Pisum sativum L.) based on next generation sequencing of a purified Chinese commercial variety. PLoS ONE 2015, 10, e0139775. [Google Scholar] [CrossRef]
- Mishra, R.K.; Gangadhar, B.H.; Nookaraju, A.; Kumar, S.; Park, S.W. Development of EST-derived SSR markers in pea (Pisum sativum) and their potential utility for genetic mapping and transferability. Plant Breed. 2012, 131, 118–124. [Google Scholar] [CrossRef]
- Kaur, S.; Pembleton, L.W.; Cogan, N.O.; Savin, K.W.; Leonforte, T.; Paull, J.; Materne, M.; Forster, J.W. Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genom. 2012, 13, 104. [Google Scholar] [CrossRef]
- Yang, T.; Jiang, J.; Burlyaeva, M.; Hu, J.; Coyne, C.J.; Kumar, S.; Redden, R.; Sun, X.; Wang, F.; Chang, J.; et al. Large-scale microsatellite development in grasspea (Lathyrus sativus L.), an orphan legume of the arid areas. BMC Plant Biol. 2014, 14, 65. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Yang, T.; Liu, R.; Hu, J.; Yao, Y.; Burlyaeva, M.; Wang, Y.; Ren, G.; Zhang, H.; Wang, D.; et al. An RNA sequencing transcriptome analysis of grasspea (Lathyrus sativus L.) and development of SSR and KASP markers. Front. Plant Sci. 2017, 8, 1873. [Google Scholar] [CrossRef] [PubMed]
- Almeida, N.F.; Krezdorn, N.; Rotter, B.; Winter, P.; Rubiales, D.; Vaz Patto, M.C. Lathyrus sativus transcriptome resistance response to Ascochyta lathyri investigated by deep SuperSAGE analysis. Front. Plant Sci. 2015, 6, 178. [Google Scholar] [CrossRef]
- Almeida, N.F.; Leitão, S.T.; Krezdorn, N.; Rotter, B.; Winter, P.; Rubiales, D.; Patto, M.C. Allelic diversity in the transcriptomes of contrasting rust-infected genotypes of Lathyrus sativus, a lasting resource for smart breeding. BMC Plant Biol. 2014, 14, 376. [Google Scholar] [CrossRef]
- Kaur, S.; Kimber, R.B.; Cogan, N.O.; Materne, M.; Forster, J.W.; Paull, J.G. SNP discovery and high-density genetic mapping in faba bean (Vicia faba L.) permits identification of QTLs for ascochyta blight resistance. Plant Sci. 2014, 217, 47–55. [Google Scholar] [CrossRef]
- Mokhtar, M.M.; Hussein, E.H.; El-Assal, S.E.; Atia, M.A. Vf ODB: A comprehensive database of ESTs, EST-SSRs, mtSSRs, microRNA-target markers and genetic maps in Vicia faba. AoB Plants 2020, 12, plaa064. [Google Scholar] [CrossRef]
- Khalifa, K.A.; Ibrahim, S.D.; El-Garhy, H.A.S.; Moustafa, M.M.M.; Maalouf, F.; Alsamman, A.M.; Hamwieh, A.; El Allali, A. Developing a new genic SSR primer database in faba bean (Vicia faba L.). J. Appl. Genet. 2021, 62, 373–387. [Google Scholar] [CrossRef]
- Ray, H.; Bock, C.; Georges, F. Faba bean: Transcriptome analysis from etiolated seedling and developing seed coat of key cultivars for synthesis of proanthocyanidins, phytate, raffinose family oligosaccharides, vicine, and convicine. Plant Genome 2015, 8, 1–11. [Google Scholar] [CrossRef]
- Ocaña, S.; Seoane, P.; Bautista, R.; Palomino, C.; Claros, G.M.; Torres, A.M.; Madrid, E. Large-scale transcriptome analysis in faba bean (Vicia faba L.) under Ascochyta fabae infection. PLoS ONE 2015, 10, e0135143. [Google Scholar]
- Yang, T.; Bao, S.Y.; Ford, R.; Jia, T.J.; Guan, J.P.; He, Y.H.; Sun, X.L.; Jiang, J.Y.; Hao, J.J.; Zhang, X.Y.; et al. High-throughput novel microsatellite marker of faba bean via next generation sequencing. BMC Genom. 2012, 13, 602. [Google Scholar] [CrossRef]
- Garzon, L.N.; Blair, M.W. Development and mapping of SSR markers linked to resistance-gene homologue clusters in common bean. Crop J. 2014, 2, 183–194. [Google Scholar] [CrossRef]
- Blair, M.W.; Hurtado, N.; Chavarro, C.M.; Muñoz–Torres, M.C.; Giraldo, M.C.; Pedraza, F.; Tomkins, J.; Wing, R. Gene-based SSR markers for common bean (Phaseolus vulgaris L.) derived from root and leaf tissue ESTs: An integration of the BMc series. BMC Plant Biol. 2011, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Jia, G.; Hyten, D.L.; Jenkins, J.; Hwang, E.Y.; Schroeder, S.G.; Osorno, J.M.; Schmutz, J.; Jackson, S.A.; McClean, P.E.; et al. SNP assay development for linkage map construction, anchoring whole-genome sequence, and other genetic and genomic applications in common bean. G3 Genes Genomes Genet. 2015, 5, 2285–2890. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, C.; Campa, A.; Garzón, A.S.; Miklas, P.; Ferreira, J.J. GWAS of pod morphological and color characters in common bean. BMC Plant Biol. 2021, 21, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Erdogmus, S.; Ates, D.; Nemli, S.; Yagmur, B.; Asciogul, T.K.; Ozkuru, E.; Karaca, N.; Yilmaz, H.; Esiyok, D.; Tanyolac, M.B. Genome-wide association studies of Ca and Mn in the seeds of the common bean (Phaseolus vulgaris L.). Genomics 2020, 112, 4536–4546. [Google Scholar] [CrossRef]
- Nkhata, W.; Shimelis, H.; Melis, R.; Chirwa, R.; Mzengeza, T.; Mathew, I.; Shayanowako, A. Genome-wide association analysis of bean fly resistance and agro-morphological traits in common bean. PLoS ONE 2021, 16, e0250729. [Google Scholar]
- Nemli, S.; Asciogul, T.K.; Ates, D.; Esiyok, D.; Tanyolac, M.B. SNP identification through genotyping by sequencing and genome-wide association study (GWAS) of pod traits in common bean. In Proceedings of the International Plant and Animal Genome Conference, San Diego, CA, USA, 10–12 January 2016; p. 19798. [Google Scholar]
- Ferreira, J.J.; Murube, E.; Campa, A. Introgressed genomic regions in a set of near-isogenic lines of common bean revealed by genotyping-by-sequencing. Plant Genome 2017, 10, plantgenome2016.08.0081. [Google Scholar] [CrossRef]
- Wu, L.; Chang, Y.; Wang, L.; Wu, J.; Wang, S. Genetic dissection of drought resistance based on root traits at the bud stage in common bean. Theor. Appl. Genet. 2021, 134, 1–15. [Google Scholar] [CrossRef]
- Ramírez, M.; Graham, M.A.; Blanco-López, L.; Silvente, S.; Medrano-Soto, A.; Blair, M.W.; Hernández, G.; Vance, C.P.; Lara, M. Sequencing and analysis of common bean ESTs. building a foundation for functional genomics. Plant Physiol. 2005, 137, 1211–1227. [Google Scholar] [CrossRef]
- Melotto, M.; Monteiro-Vitorello, C.B.; Bruschi, A.G.; Camargo, L.E. Comparative bioinformatic analysis of genes expressed in common bean (Phaseolus vulgaris L.) seedlings. Genome 2005, 48, 562–570. [Google Scholar] [CrossRef]
- McClean, P.E.; Mamidi, S.; McConnell, M.; Chikara, S.; Lee, R. Synteny mapping between common bean and soybean reveals extensive blocks of shared loci. BMC Genom. 2010, 11, 184. [Google Scholar] [CrossRef] [PubMed]
- Blair, M.W.; Fernandez, A.C.; Ishitani, M.; Moreta, D.; Seki, M.; Ayling, S.; Shinozaki, K. Construction and EST sequencing of full-length, drought stress cDNA libraries for common beans (Phaseolus vulgaris L.). BMC Plant Biol. 2011, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Thibivilliers, S.; Joshi, T.; Campbell, K.B.; Scheffler, B.; Xu, D.; Cooper, B.; Nguyen, H.T.; Stacey, G. Generation of Phaseolus vulgaris ESTs and investigation of their regulation upon Uromyces appendiculatus infection. BMC Plant Biol. 2009, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.A.; Rangel, P.N.; Brondani, C.; Martins, W.S.; Melo, L.C.; Carneiro, M.S.; Borba, T.C.; Brondani, R.P. The characterization of a new set of EST-derived simple sequence repeat (SSR) markers as a resource for the genetic analysis of Phaseolus vulgaris. BMC Genet. 2011, 12, 41. [Google Scholar] [CrossRef]
- Kalavacharla, V.; Liu, Z.; Meyers, B.C.; Thimmapuram, J.; Melmaiee, K. Identification and analysis of common bean (Phaseolus vulgaris L.) transcriptomes by massively parallel pyrosequencing. BMC Plant Biol. 2011, 11, 135. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.; Capella-Gutiérrez, S.; Rendón-Anaya, M.; Hernández-Oñate, M.; Minoche, A.E.; Erb, I.; Câmara, F.; Prieto-Barja, P.; Corvelo, A.; Sanseverino, W.; et al. Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome Biol. 2016, 17, 1–8. [Google Scholar] [CrossRef]
- Samantara, K.; Reyes, V.P.; Agrawal, N.; Mohapatra, S.R.; Jena, K.K. Advances and trends on the utilization of multi-parent advanced generation intercross (MAGIC) for crop improvement. Euphytica 2021, 217, 189. [Google Scholar] [CrossRef]
- Kitony, J.K.; Sunohara, H.; Tasaki, M.; Mori, J.-I.; Shimazu, A.; Reyes, V.P.; Yasui, H.; Yamagata, Y.; Yoshimura, A.; Yamasaki, M.; et al. Development of an Aus-derived nested association mapping (Aus-NAM) population in rice. Plants 2021, 10, 1255. [Google Scholar] [CrossRef]
- Spindel, J.; Iwata, H. Genomic selection in rice breeding. In Rice Genomics, Genetics and Breeding; Sasaki, T., Ashikari, M., Eds.; Springer: Singapore, 2018; pp. 473–496. [Google Scholar]
- Reyes, V.P.; Angeles-Shim, R.B.; Mendioro, M.S.; Manuel, M.C.C.; Lapis, R.S.; Shim, J.; Sunohara, H.; Nishiuchi, S.; Kikuta, M.; Makihara, D.; et al. Marker-assisted introgression and stacking of major qtls controlling grain number (Gn1a) and number of primary branching (WFP) to nerica cultivars. Plants 2021, 10, 844. [Google Scholar] [CrossRef]
- Jaganathan, D.; Thudi, M.; Kale, S.; Azam, S.; Roorkiwal, M.; Gaur, P.M.; Kishor, P.K.; Nguyen, H.; Sutton, T.; Varshney, R.K. Genotyping-by-sequencing based intra-specific genetic map refines a ‘‘QTL-hotspot” region for drought tolerance in chickpea. Mol. Genet. Genom. 2015, 290, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Kujur, A.; Upadhyaya, H.D.; Shree, T.; Bajaj, D.; Das, S.; Saxena, M.S.; Badoni, S.; Kumar, V.; Tripathi, S.; Gowda, C.L.; et al. Ultra-high density intra-specific genetic linkage maps accelerate identification of functionally relevant molecular tags governing important agronomic traits in chickpea. Sci. Rep. 2015, 5, 9468. [Google Scholar] [CrossRef]
- Ellis, T.H.N.; Turner, L.; Hellens, R.P.; Lee, D.; Harker, C.L.; Enard, C.; Domoney, C.; Davies, D.R.; Ambrose, M. Linkage maps in pea. Genetics 1992, 130, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Goyal, R.; Chahota, R.K.; Sharma, T.R.; Abdin, M.Z.; Bhatia, S. Construction of a genetic linkage map and identification of QTLs for seed weight and seed size traits in lentil (Lens culinaris Medik.). PLoS ONE 2015, 10, e0139666. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.J.; Samineni, S.; Thudi, M.; Sajja, S.B.; Rathore, A.; Das, R.R.; Khan, A.W.; Chaturvedi, S.K.; Lavanya, G.R.; Varshney, R.K.; et al. Molecular mapping of QTLs for heat tolerance in chickpea. Int. J. Mol. Sci. 2018, 19, 2166. [Google Scholar] [CrossRef] [PubMed]
- Jha, U.C.; Kole, P.C.; Singh, N.P. QTL mapping for heat stress tolerance in chickpea (Cicer arietinum L.). Legume Res. Int. J. 2021, 44, 382–387. [Google Scholar] [CrossRef]
- Singh, D.; Singh, C.K.; Taunk, J.; Jadon, V.; Pal, M.; Gaikwad, K. Genome wide transcriptome analysis reveals vital role of heat responsive genes in regulatory mechanisms of lentil (Lens culinaris Medikus). Sci. Rep. 2019, 9, 12976. [Google Scholar] [CrossRef]
- Tafesse, E.G.; Gali, K.K.; Lachagari, V.B.; Bueckert, R.; Warkentin, T.D. Genome-wide association mapping for heat stress responsive traits in field pea. Int. J. Mol. Sci. 2020, 21, 2043. [Google Scholar] [CrossRef]
- Bhat, J.A.; Ali, S.; Salgotra, R.K.; Mir, Z.A.; Dutta, S.; Jadon, V.; Tyagi, A.; Mushtaq, M.; Jain, N.; Singh, P.K.; et al. Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front. Genet. 2016, 7, 221. [Google Scholar] [CrossRef]
- George, A.W.; Cavanagh, C. Genome-wide association mapping in plants. Theor. Appl. Genet. 2015, 128, 1163–1174. [Google Scholar] [CrossRef] [PubMed]
- Verdeprado, H.; Kretzschmar, T.; Begum, H.; Raghavan, C.; Joyce, P.; Lakshmanan, P.; Cobb, J.N.; Collard, B.C. Association mapping in rice: Basic concepts and perspectives for molecular breeding. Plant Prod. Sci. 2018, 21, 159–176. [Google Scholar] [CrossRef]
- Thudi, M.; Upadhyaya, H.D.; Rathore, A.; Gaur, P.M.; Krishnamurthy, L.; Roorkiwal, M.; Nayak, S.N.; Chaturvedi, S.K.; Basu, P.S.; Gangarao, N.V.; et al. Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS ONE 2014, 9, e96758. [Google Scholar] [CrossRef] [PubMed]
- Vijayalakshmi, K.; Fritz, A.K.; Paulsen, G.M.; Bai, G.; Pandravada, S.; Gill, B.S. Modeling and mapping QTL for senescence-related traits in winter wheat under high temperature. Mol. Breed. 2010, 26, 163–175. [Google Scholar] [CrossRef]
- Paliwal, R.; Röder, M.S.; Kumar, U.; Srivastava, J.P.; Joshi, A.K. QTL mapping of terminal heat tolerance in hexaploid wheat (T. aestivum L.). Theor. Appl. Genet. 2012, 125, 561–575. [Google Scholar] [CrossRef]
- Talukder, S.K.; Babar, M.A.; Vijayalakshmi, K.; Poland, J.; Prasad, P.V.; Bowden, R.; Fritz, A. Mapping QTL for the traits associated with heat tolerance in wheat (Triticum aestivum L.). BMC Genet. 2014, 15, 97. [Google Scholar] [CrossRef] [PubMed]
- Shirdelmoghanloo, H.; Taylor, J.D.; Lohraseb, I.; Rabie, H.; Brien, C.; Timmins, A.; Martin, P.; Mather, D.E.; Emebiri, L.; Collins, N.C. A QTL on the short arm of wheat (Triticum aestivum L.) chromosome 3B affects the stability of grain weight in plants exposed to a brief heat shock early in grain filling. BMC Plant Biol. 2016, 16, 100. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.K.; Torp, A.M.; Rosenqvist, E.; Ottosen, C.O.; Andersen, S.B. QTLs and potential candidate genes for heat stress tolerance identified from the mapping populations specifically segregating for Fv/Fm in wheat. Front. Plant Sci. 2017, 8, 1668. [Google Scholar] [CrossRef]
- Maulana, F.; Ayalew, H.; Anderson, J.D.; Kumssa, T.T.; Huang, W.; Ma, X.F. Genome-wide association mapping of seedling heat tolerance in winter wheat. Front. Plant Sci. 2018, 9, 1272. [Google Scholar] [CrossRef]
- Chopra, R.; Burow, G.; Burke, J.J.; Gladman, N.; Xin, Z. Genome-wide association analysis of seedling traits in diverse Sorghum germplasm under thermal stress. BMC Plant Biol. 2017, 17, 12. [Google Scholar] [CrossRef]
- Ye, C.; Argayoso, M.A.; Redoña, E.D.; Sierra, S.N.; Laza, M.A.; Dilla, C.J.; Mo, Y.; Thomson, M.J.; Chin, J.; Delaviña, C.B.; et al. Mapping QTL for heat tolerance at flowering stage in rice using SNP markers. Plant Breed. 2012, 131, 33–41. [Google Scholar] [CrossRef]
- Ye, C.; Tenorio, F.A.; Argayoso, M.A.; Laza, M.A.; Koh, H.J.; Redoña, E.D.; Jagadish, K.S.; Gregorio, G.B. Identifying and confirming quantitative trait loci associated with heat tolerance at flowering stage in different rice populations. BMC Genet. 2015, 16, 41. [Google Scholar] [CrossRef] [PubMed]
- Kilasi, N.L.; Singh, J.; Vallejos, C.E.; Ye, C.; Jagadish, S.V.; Kusolwa, P.; Rathinasabapathi, B. Heat stress tolerance in rice (Oryza sativa L.): Identification of quantitative trait loci and candidate genes for seedling growth under heat stress. Front. Plant Sci. 2018, 9, 1578. [Google Scholar] [CrossRef] [PubMed]
- Frey, F.P.; Presterl, T.; Lecoq, P.; Orlik, A.; Stich, B. First steps to understand heat tolerance of temperate maize at adult stage: Identification of QTL across multiple environments with connected segregating populations. Theor. Appl. Genet. 2016, 129, 945–961. [Google Scholar] [CrossRef]
- Marfo, K.O.; Hall, A.E. Inheritance of heat tolerance during pod set in cowpea. Crop Sci. 1992, 32, 912–918. [Google Scholar] [CrossRef]
- Lucas, M.R.; Ehlers, J.D.; Huynh, B.L.; Diop, N.N.; Roberts, P.A.; Close, T.J. Markers for breeding heat-tolerant cowpea. Mol. Breed. 2013, 31, 529–536. [Google Scholar] [CrossRef]
- Pottorff, M.; Roberts, P.A.; Close, T.J.; Lonardi, S.; Wanamaker, S.; Ehlers, J.D. Identification of candidate genes and molecular markers for heat-induced brown discoloration of seed coats in cowpea [Vigna unguiculata (L.) Walp]. BMC Genom. 2014, 15, 1. [Google Scholar] [CrossRef]
- Kaga, A.; Han, O.K.; Wang, X.W.; Egawa, Y.; Tomooka, N.; Vaughan, D.A. Vigna angularis as a model for legume research. In “Conservation and Use of Wild Relatives of Crops”, Proceedings of the Joint Department of Agriculture, Sri Lanka and National Institute of Agrobiological Sciences, Japan Workshop, Okinawa, Japan, 3 February 2003; Department of Agriculture: Peradeniya, Sri Lanka, 2003; pp. 51–74. [Google Scholar]
- Vaughan, D.A.; Tomooka, N.; Kaga, A. Azuki Bean [Vigna angularis (Wild.) Ohwi & Ohashi]. In Genetic Resources Chromosome Engineering and Crop Improvement Series Grain Legumes; Singh, R.J., Jauhar, P.P., Eds.; Taylor & Francis Publishing: Boca Raton, FL, USA, 2005; pp. 347–359. [Google Scholar]
- Sudheesh, S.; Lombardi, M.; Leonforte, A.; Cogan, N.O.; Materne, M.; Forster, J.W.; Kaur, S. Consensus genetic map construction for field pea (Pisum sativum L.), trait dissection of biotic and abiotic stress tolerance and development of a diagnostic marker for the er1 powdery mildew resistance gene. Plant Mol. Biol. Rep. 2015, 33, 1391–1403. [Google Scholar]
- Desgroux, A.; L’anthoëne, V.; Roux-Duparque, M.; Rivière, J.P.; Aubert, G.; Tayeh, N.; Moussart, A.; Mangin, P.; Vetel, P.; Piriou, C.; et al. Genome-wide association mapping of partial resistance to Aphanomyces euteiches in pea. BMC Genom. 2016, 17, 124. [Google Scholar] [CrossRef]
- Diapari, M.; Sindhu, A.; Warkentin, T.D.; Bett, K.; Tar’an, B. Population structure and marker-trait association studies of iron, zinc and selenium concentrations in seed of field pea (Pisum sativum L.). Mol. Breed. 2015, 35, 30. [Google Scholar] [CrossRef]
- Ahmad, S.; Kaur, S.; Lamb-Palmer, N.D.; Lefsrud, M.; Singh, J. Genetic diversity and population structure of Pisum sativum accessions for marker-trait association of lipid content. Crop J. 2015, 3, 238–245. [Google Scholar] [CrossRef]
- Leonforte, A.; Forster, J.W.; Redden, R.J.; Nicolas, M.E.; Salisbury, P.A. Sources of high tolerance to salinity in pea (Pisum sativum L.). Euphytica 2013, 189, 203–216. [Google Scholar] [CrossRef]
- Klein, A.; Houtin, H.; Rond, C.; Marget, P.; Jacquin, F.; Boucherot, K.; Huart, M.; Rivière, N.; Boutet, G.; Lejeune-Hénaut, I.; et al. QTL analysis of frost damage in pea suggests different mechanisms involved in frost tolerance. Theor. Appl. Genet. 2014, 127, 1319–1930. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Diapari, M.; Bueckert, R.A.; Tar’an, B.; Warkentin, T.D. Population structure and association mapping of traits related to reproductive development in field pea. Euphytica 2017, 213, 1–20. [Google Scholar] [CrossRef]
- Wang, L.; Yu, X.; Wang, H.; Lu, Y.Z.; de Ruiter, M.; Prins, M.; He, Y.K. A novel class of heat-responsive small RNAs derived from the chloroplast genome of Chinese cabbage (Brassica rapa). BMC Genom. 2011, 12, 289. [Google Scholar] [CrossRef] [PubMed]
- Soares-Cavalcanti, N.M.; Belarmino, L.C.; Kido, E.A.; Pandolfi, V.; Marcelino-Guimarães, F.C.; Rodrigues, F.A.; Pereira, G.A.; Benko-Iseppon, A.M. Overall picture of expressed heat shock factors in Glycine max, Lotus japonicus and Medicago truncatula. Genet. Mol. Biol. 2012, 35, 247–259. [Google Scholar] [CrossRef][Green Version]
- González-Schain, N.; Dreni, L.; Lawas, L.M.; Galbiati, M.; Colombo, L.; Heuer, S.; Jagadish, K.S.; Kater, M.M. Genome-wide transcriptome analysis during anthesis reveals new insights into the molecular basis of heat stress responses in tolerant and sensitive rice varieties. Plant Cell Physiol. 2016, 57, 57–68. [Google Scholar] [CrossRef]
- Agarwal, G.; Garg, V.; Kudapa, H.; Doddamani, D.; Pazhamala, L.T.; Khan, A.W.; Thudi, M.; Lee, S.H.; Varshney, R.K. Genome-wide dissection of AP2/ERF and HSP90 gene families in five legumes and expression profiles in chickpea and pigeonpea. Plant Biotechnol. J. 2016, 14, 1563–1577. [Google Scholar] [CrossRef]
- Simões-Araújo, J.L.; Rodrigues, R.L.; Liliane, B.D.; Mondego, J.M.; Alves-Ferreira, M.; Rumjanek, N.G.; Margis-Pinheiro, M. Identification of differentially expressed genes by cDNA-AFLP technique during heat stress in cowpea nodules. FEBS Lett. 2012, 515, 44–50. [Google Scholar] [CrossRef]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Doring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef]
- Kumar, R.; Lavania, D.; Negi, M.; Siddiqui, M.H.; Al-Whaibi, M.; Grover, A. Identification and characterization of a small heat shock protein 17.9-CII gene from faba bean (Vicia faba L.). Acta Physiol. Plant. 2015, 37, 190. [Google Scholar] [CrossRef]
- Zhu, B.; Ye, C.; Lü, H.; Chen, X.; Chai, G.; Chen, J.; Wang, C. Identification and characterization of a novel heat shock transcription factor gene, GmHsfA1, in soybeans (Glycine max). J. Plant Res. 2006, 119, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.; Ye, C.J.; Lu, H.Y.; Xu, M.X.; Zhang, L.M.; Wang, C.; Luo, S.P.; Zhu, B.G. Cloning of GmHSFA1 gene and its over expression leading to enhancement of heat tolerance in transgenic soybean. Yi Chuan = Hered. 2006, 28, 1411–1420. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Caitar, V.S.; de Carvalho, M.C.; Darben, L.M.; Kuwahara, M.K.; Nepomuceno, A.L.; Dias, W.P.; Abdelnoor, R.V.; Marcelino-Guimarães, F.C. Genome-wide analysis of the Hsp20 gene family in soybean: Comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genom. 2013, 14, 577. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, R.I. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones and negative regulators. Genes Dev. 1998, 12, 3788–3796. [Google Scholar] [CrossRef] [PubMed]
- Baniwal, S.K.; Bharti, K.; Chan, K.Y.; Fauth, M.; Ganguli, A.; Kotak, S.; Mishra, S.K.; Nover, L.; Port, M.; Scharf, K.D.; et al. Heat stress response in plants: A complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 2004, 29, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Nover, L. Heat Shock Response; CRC Press Publishing: Boca Raton, FL, USA, 1991. [Google Scholar]
- Baniwal, S.K.; Chan, K.Y.; Scharf, K.D.; Nover, L. Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J. Biol. Chem. 2007, 282, 605–613. [Google Scholar] [CrossRef]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; Von Koskull-Döring, P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004, 39, 98–112. [Google Scholar] [CrossRef]
- Bharti, K.; Koskull-Döring, P.; Bharti, S.; Kumar, P.; Tintschl-Körbitzer, A.; Treuter, E.; Nover, L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 2004, 16, 1521–1535. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Bett, K.; Ramsay, L.; Chan, C.; Sharpe, A.; Cook, D.; Penmetsa, R.V. Lentil v1.0 and beyond. In Proceedings of the XXIV Plant and Animal Genome Conference, San Diego, CA, USA, 10–12 January 2016; pp. 9–13. [Google Scholar]
- Kreplak, J.; Madoui, M.A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Martin, L.; Fei, Z.; Giovannoni, J.; Rose, J.K. Catalyzing plant science research with RNA-seq. Front. Plant Sci. 2013, 4, 66. [Google Scholar] [CrossRef] [PubMed]
- Kudapa, H.; Agarwal, G.; Doddamani, D.; Kumar, V.; Khan, A.W.; Chitikineni, A.; Varshney, R.K. Comprehensive transcriptome analysis to identify heat stress responsive genes in chickpea (Cicer arietinum L.). In Proceedings of the International Plant and Animal Genome Conference Asia, Singapore, 19–21 May 2014. [Google Scholar]
- Yuan, C.; Li, C.; Zhao, X.; Yan, C.; Wang, J.; Mou, Y.; Sun, Q.; Shan, S. Genome-wide identification and characterization of HSP90-RAR1-SGT1-complex members from Arachis genomes and their responses to biotic and abiotic stresses. Front. Genet. 2021, 12, 689669. [Google Scholar] [CrossRef]
- Deokar, A.A.; Kondawar, V.; Kohli, D.; Aslam, M.; Jain, P.K.; Karuppayil, S.M.; Varshney, R.K.; Srinivasan, R. The CarERF genes in chickpea (Cicer arietinum L.) and the identification of CarERF116 as abiotic stress responsive transcription factor. Funct. Integr. Genom. 2015, 15, 27–46. [Google Scholar] [CrossRef]
- Zhang, J.; Li, J.; Liu, B.B.; Zhang, L.; Chen, J.; Lu, M.Z. Genome-wide analysis of the Populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genom. 2013, 14, 532. [Google Scholar] [CrossRef] [PubMed]
- Konda, A.K.; Farmer, R.; Soren, K.R.; Ps, S.; Setti, A. Structural modeling and molecular dynamics of a multi-stress responsive WRKY TF-DNA complex towards elucidating its role in stress signaling mechanisms in chickpea. J. Biomol. Struct. Dyn. 2018, 36, 2279–2291. [Google Scholar] [CrossRef] [PubMed]
- Chidambaranathan, P.; Jagannadham, P.T.; Satheesh, V.; Jain, P.K.; Srinivasan, R. Expression analysis of six chromatin remodeling complex genes (SWR1) in chickpea in different tissues during heat stress. Indian Jouranl Genet. 2016, 76, 1–11. [Google Scholar] [CrossRef]
- Parankusam, S.; Bhatnagar-Mathur, P.; Sharma, K.K. Heat responsive proteome changes reveal molecular mechanisms underlying heat tolerance in chickpea. Environ. Exp. Bot. 2017, 141, 132–144. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; Palakurthi, R.; Jha, R.; Valluri, V.; Bajaj, P.; Chitikineni, A.; Singh, N.P.; Varshney, R.K.; Thudi, M. Major QTLs and potential candidate genes for heat stress tolerance identified in chickpea (Cicer arietinum L.). Front. Plant Sci. 2021, 12, 655103. [Google Scholar] [CrossRef]
- Kudapa, H.; Garg, V.; Chitikineni, A.; Varshney, R.K. The RNA-Seq-based high resolution gene expression atlas of chickpea (Cicer arietinum L.) reveals dynamic spatio-temporal changes associated with growth and development. Plant Cell Environ. 2018, 41, 2209–2225. [Google Scholar] [CrossRef]
- Chidambaranathan, P.; Jagannadham, P.T.; Satheesh, V.; Kohli, D.; Basavarajappa, S.H.; Chellapilla, B.; Kumar, J.; Jain, P.K.; Srinivasan, R. Genome-wide analysis identifies chickpea (Cicer arietinum) heat stress transcription factors (Hsfs) responsive to heat stress at the pod development stage. J. Plant Res. 2018, 131, 525–542. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Thudi, M.; Roorkiwal, M.; He, W.; Upadhyaya, H.D.; Yang, W.; Bajaj, P.; Cubry, P.; Rathore, A.; Jian, J.; et al. Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat. Genet. 2019, 51, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Moin, M.; Bakshi, A.; Saha, A.; Dutta, M.; Madhav, S.M.; Kirti, P.B. Rice ribosomal protein large subunit genes and their spatio-temporal and stress regulation. Front. Plant Sci. 2016, 7, 1284. [Google Scholar] [CrossRef]
- Xing, H.; Fu, X.; Yang, C.; Tang, X.; Guo, L.; Li, C.; Xu, C.; Luo, K. Genome-wide investigation of pentatricopeptide repeat gene family in poplar and their expression analysis in response to biotic and abiotic stresses. Sci. Rep. 2018, 8, 2817. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Ragupathy, R.; Edwards, T.; Domaratzki, M.; Cloutier, S. MicroRNA-guided regulation of heat stress response in wheat. BMC Genom. 2019, 20, 488. [Google Scholar] [CrossRef] [PubMed]
- Panter, P.E.; Kent, O.; Dale, M.; Smith, S.J.; Skipsey, M.; Thorlby, G.; Cummins, I.; Ramsay, N.; Begum, R.A.; Sanhueza, D.; et al. MUR1-mediated cell-wall fucosylation is required for freezing tolerance in Arabidopsis thaliana. New Phytol. 2019, 224, 1518–1531. [Google Scholar] [CrossRef]
- Von Koskull-Doring, P.; Scharf, K.D.; Nover, L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007, 12, 452–457. [Google Scholar] [CrossRef]
- Kumar, J.; Choudhary, A.K.; Gupta, D.S.; Kumar, S. Towards exploitation of adaptive traits for climate-resilient smart pulses. Int. J. Mol. Sci. 2019, 20, 2971. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef]
- Dong, C.H.; Pei, H. Over-expression of miR397 improves plant tolerance to cold stress in Arabidopsis thaliana. J. Plant Biol. 2014, 57, 209–217. [Google Scholar] [CrossRef]
- Zhang, B. MicroRNA: A new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015, 66, 1749–1761. [Google Scholar] [CrossRef]
- Trumbo, J.L.; Zhang, B.; Stewart, C.N., Jr. Manipulating microRNAs for improved biomass and biofuels from plant feedstocks. Plant Biotechnol. J. 2015, 13, 337–354. [Google Scholar] [CrossRef]
- Hajyzadeh, M.; Turktas, M.; Khawar, K.M.; Unver, T. miR408 overexpression causes increased drought tolerance in chickpea. Gene 2015, 555, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Gupta, O.P.; Sharma, P.; Gupta, R.K.; Sharma, I. Current status on role of miRNAs during plant-fungus interaction. Physiol. Mol. Plant Pathol. 2014, 5, 1–7. [Google Scholar] [CrossRef]
- Ferdous, J.; Hussain, S.S.; Shi, B.J. Role of microRNAs in plant drought tolerance. Plant Biotechnol. J. 2015, 13, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Hackenberg, M.; Gustafson, P.; Langridge, P.; Shi, B.J. Differential expression of microRNAs and other small RNAs in barley between water and drought conditions. Plant Biotechnol. J. 2015, 13, 2–13. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2014, 16, 2001–2019. [Google Scholar] [CrossRef]
- Zhu, C.; Ding, Y.; Liu, H. MiR398 and plant stress responses. Physiol. Plant. 2011, 143, 1–9. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Lu, X.; Guan, Q.; Zhu, J. Downregulation of CSD2 by a heat-inducible miR398 is required for thermotolerance in Arabidopsis. Plant Signal. Behav. 2013, 8, e24952. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guan, Q.M.; Lu, X.Y.; Zeng, H.T.; Zhang, Y.Y.; Zhu, J.H. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013, 74, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Gurley, W.B. HSP101: A key component for the acquisition of thermotolerance in plants. Plant Cell 2000, 12, 457–460. [Google Scholar] [CrossRef]
- Yu, X.; Wang, H.; Lu, Y.; de Ruiter, M.; Cariaso, M.; Prins, M.; van Tunen, A.; He, Y. Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J. Exp. Bot. 2012, 63, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Szittya, G.; Moxon, S.; Santos, D.M.; Jing, R.; Fevereiro, M.P.; Moulton, V.; Dalmay, T. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genom. 2008, 9, 593. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Fu, Y.; Sunkar, R.; Barbazuk, W.B.; Zhu, J.K.; Yu, O. Novel and nodulation-regulated microRNAs in soybean roots. BMC Genom. 2008, 9, 160. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, P.; Cao, X.; Wang, X.; Zhang, A.; Li, X. Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochem. Biophys. Res. Commun. 2009, 378, 799–803. [Google Scholar] [CrossRef]
- Joshi, T.; Yan, Z.; Libault, M.; Jeong, D.H.; Park, S.; Green, P.J.; Sherrier, D.J.; Farmer, A.; May, G.; Meyers, B.C.; et al. Prediction of novel miRNAs and associated target genes in Glycine Max. BMC Bioinform. 2010, 11, 1–9. [Google Scholar] [CrossRef]
- Jagadeeswaran, G.; Zheng, Y.; Li, Y.F.; Shukla, L.I.; Matts, J.; Hoyt, P.; Macmil, S.L.; Wiley, G.B.; Roe, B.A.; Zhang, W.; et al. Cloning and characterization of small RNAs from Medicago truncatula reveals four novel legume-specific microRNA families. New Phytol. 2009, 184, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Lelandais-Brière, C.; Naya, L.; Sallet, E.; Calenge, F.; Frugier, F.; Hartmann, C.; Gouzy, J.; Crespi, M. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 2009, 21, 2780–2796. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, Y.; Mou, F.; Tian, Y.; Chen, L.; Zhang, S.; Jiang, Q.; Li, X. Genome-wide small rna analysis of soybean reveals auxin-responsive microRNAs that are differentially expressed in response to salt stress in root apex. Front. Plant Sci. 2016, 6, 1273. [Google Scholar] [CrossRef]
- Arenas-Huertero, C.; Pérez, B.; Rabanal, F.; Blanco-Melo, D.; De la Rosa, C.; Estrada-Navarrete, G.; Sanchez, F.; Covarrubias, A.A.; Reyes, J.L. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol. Biol. 2009, 70, 385–401. [Google Scholar] [CrossRef]
- Ramesh, V.; Admane, N.; Husain, S.M. Small RNAs landscape (sRNAome) of soybean [Glycine max (L.)]: Biogenesis, vital functions and potential applications. Plant Knowl. J. 2013, 2, 24–37. [Google Scholar]
- Li, H.; Dong, Y.; Yin, H.; Wang, N.; Yang, J.; Liu, X.; Wang, Y.; Wu, J.; Li, X. Characterization of the stress associated microRNAs in Glycine max by deep sequencing. BMC Plant Biol. 2011, 11, 170. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.; Ren, Y.; Xu, J.; Zhang, Z.; Wang, Y. Genome wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem. Biophys. Res. Commun. 2011, 417, 892–896. [Google Scholar] [CrossRef]
- Wang, T.; Chen, L.; Zhao, M.; Tian, Q.; Zhang, W.H. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genom. 2011, 12, 367. [Google Scholar] [CrossRef]
- Zhou, Z.S.; Zeng, H.Q.; Liu, Z.P.; Yang, Z.M. Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ. 2012, 35, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Samad, A.F.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.; Zainal, Z. MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef] [PubMed]
- Khandal, H.; Parween, S.; Roy, R.; Meena, M.K.; Chattopadhyay, D. MicroRNA profiling provides insights into post-transcriptional regulation of gene expression in chickpea root apex under salinity and water deficiency. Sci. Rep. 2017, 7, 4632. [Google Scholar] [CrossRef]
- Sun, X.; Lin, L.; Na, S. Regulation mechanism of microRNA in plant response to abiotic stress and breeding. Mol. Biol. Rep. 2019, 46, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Samad, A.F.A.; Ali, N.M.; Ismail, I.; Murad, A.M.A. Analysis of miRNAs targeting transcription factors in Persicaria minor induced by Fusarium oxysporum. AIP Conf. Proc. 2016, 1784, 020009. [Google Scholar]
- Sihag, P.; Sagwal, V.; Kumar, A.; Balyan, P.; Mir, R.R.; Dhankher, O.P.; Kumar, U. Discovery of miRNAs and development of heat-responsive miRNA-SSR markers for characterization of wheat germplasm for terminal heat tolerance breeding. Front. Genet. 2021, 12, 699420. [Google Scholar] [CrossRef] [PubMed]
- Larkindale, J.; Hall, J.D.; Knight, M.R.; Vierling, E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005, 138, 882–897. [Google Scholar] [CrossRef]
- Lee, U.; Wie, C.; Fernandez, B.O.; Feelisch, M.; Vierling, E. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 2008, 20, 786–802. [Google Scholar] [CrossRef] [PubMed]
- Sugio, A.; Dreos, R.; Aparicio, F.; Maule, A.J. The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 2009, 21, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Che, P.; Bussell, J.D.; Zhou, W.; Estavillo, G.M.; Pogson, B.J.; Smith, S.M. Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci. Signal. 2010, 3, ra69. [Google Scholar] [CrossRef]
- Kumar, S.V.; Wigge, P.A. H2A.Z-containing nucleosomes mediate the thermos sensory response in Arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef]
- Friedrich, T.; Faivre, L.; Bäurle, I.; Schubert, D. Chromatin-based mechanisms of temperature memory in plants. Plant Cell Environ. 2019, 42, 762–770. [Google Scholar] [CrossRef]
- Finka, A.; Cuendet, A.F.; Maathuis, F.J.; Saidi, Y.; Goloubinoff, P. Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 2012, 24, 3333–3348. [Google Scholar] [CrossRef]
- Sun, A.Z.; Guo, F.Q. Chloroplast retrograde regulation of heat stress responses in plants. Front. Plant Sci. 2016, 7, 398. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Quint, M.; Delker, C.; Franklin, K.A.; Wigge, P.A.; Halliday, K.J.; van Zanten, M. Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2016, 2, 15190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hong, Y.; Huang, L.; Li, D.; Song, F. Arabidopsis AtERF014 acts as a dual regulator that differentially modulates immunity against Pseudomonas syringae pv. tomato and Botrytis cinerea. Plant Physiol. 2016, 6, 30251. [Google Scholar]
- Zhang, L.; Zhang, F.; Melotto, M.; Yao, J.; He, S.Y. Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 2017, 68, 1371–1385. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Xu, N.; Huang, J.; Gao, F.; Zou, H.; Boudsocq, M.; Coaker, G.; Liu, J. A lectin receptor-like kinase mediates pattern-triggered salicylic acid signaling. Plant Physiol. 2017, 174, 2501–2514. [Google Scholar] [CrossRef]
- Awasthi, R.; Gaur, P.; Turner, N.C.; Vadez, V.; Siddique, K.H.M.; Nayyar, H. Effects of individual and combined heat and drought stress during seed filling on the oxidative metabolism and yield of chickpea (Cicer arietinum) genotypes differing in heat and drought tolerance. Crop Pasture Sci. 2017, 68, 823–841. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Kaushal, N.; Gupta, K.; Bhandhari, K.; Kumar, S.; Thakur, P.; Nayyar, H. Proline induces heat tolerance in chickpea (Cicer arietinum L.) plants by protecting vital enzymes of carbon and antioxidative metabolism. Physiol. Mol. Biol. Plants 2011, 17, 203–213. [Google Scholar] [CrossRef]
- Kumar, S.; Kaushal, N.; Nayyar, H.; Gaur, P. Abscisic acid induces heat tolerance in chickpea (Cicer arietinum L.) seedlings by facilitated accumulation of osmoprotectants. Acta Physiol. Plant. 2012, 34, 1651–1658. [Google Scholar] [CrossRef]
- Kaushal, N.; Awasthi, R.; Gupta, K.; Gaur, P.; Siddique, H.M.K.; Nayyar, H. Heat-stress-induced reproductive failures in chickpea (Cicer arietinum) are associated with impaired sucrose metabolism in leaves and anthers. Funct. Plant Biol. 2013, 40, 1334–1349. [Google Scholar] [CrossRef]
- Mutters, R.G.; Hall, A.E.; Patel, P.N. Photoperiod and light quality effects on cowpea floral development at high temperatures. Crop Sci. 1979, 29, 1501–1505. [Google Scholar] [CrossRef]
- De Leonardis, A.M.; Fragasso, M.; Beleggia, R.; Ficco, D.B.; De Vita, P.; Mastrangelo, A.M. Effects of heat stress on metabolite accumulation and composition, and nutritional properties of durum wheat grain. Int. J. Mol. Sci. 2015, 16, 30382–30404. [Google Scholar] [CrossRef] [PubMed]
- Obata, T.; Witt, S.; Lisec, J.; Palacios-Rojas, N.; Florez-Sarasa, I.; Yousfi, S.; Araus, J.L.; Cairns, J.E.; Fernie, A.R. Metabolite profiles of maize leaves in drought, heat, and combined stress field trials reveal the relationship between metabolism and grain yield. Plant Physiol. 2015, 169, 2665–2683. [Google Scholar] [CrossRef] [PubMed]
- Glaubitz, U.; Li, X.; Schaedel, S.; Erban, A.; Sulpice, R.; Kopka, J.; Hincha, D.K.; Zuther, E. Integrated analysis of rice transcriptomic and metabolomic responses to elevated night temperatures identifies sensitivity- and tolerance-related profiles. Plant Cell Environ. 2017, 40, 121–137. [Google Scholar] [CrossRef]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 152. [Google Scholar] [CrossRef]
- Vriet, C.; Hennig, L.; Laloi, C. Stress-induced chromatin changes in plants: Of memories, metabolites and crop improvement. Cell. Mol. Life Sci. 2015, 72, 1261–1273. [Google Scholar] [CrossRef]
- Sanyal, R.P.; Misra, H.S.; Saini, A. Heat-stress priming and alternative splicing-linked memory. J. Exp. Bot. 2018, 69, 2431–2434. [Google Scholar] [CrossRef]
- Sani, E.; Herzyk, P.; Perrella, G.; Colot, V.; Amtmann, A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013, 14, 1–24. [Google Scholar] [CrossRef]
- Mauch-Mani, B.; Baccelli, I.; Luna, E.; Flors, V. Defense priming: An adaptive part of induced resistance. Annu. Rev. Plant Biol. 2017, 68, 485–512. [Google Scholar] [CrossRef]
- Bäurle, I. Plant heat adaptation: Priming in response to heat stress. F1000 Res. 2016, 5, F1000 Faculty Rev-694. [Google Scholar] [CrossRef]
- Berry, S.; Dean, C. Environmental perception and epigenetic memory: Mechanistic insight through FLC. Plant J. 2015, 83, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Serrano, N.; Gao, G.; Atia, M.; Mokhtar, M.; Woo, Y.H.; Bazin, J.; Veluchamy, A.; Benhamed, M.; Crespi, M.; et al. Thermo priming triggers splicing memory in Arabidopsis. J. Exp. Bot. 2018, 69, 2659–2675. [Google Scholar] [CrossRef]
- Luco, R.F.; Allo, M.; Schor, I.E.; Kornblihtt, A.R.; Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 2011, 144, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Seki, M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014, 55, 1859–1863. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Sita, K.; Sehgal, A.; Bhandari, K.; Kumar, S.; Prasad, P.V.V.; Jha, U.; Kumar, J.; Siddique, K.H.M.; Nayyar, H. Heat Priming of Lentil (Lens culinaris Medik.) seeds and foliar treatment with γ-aminobutyric acid (GABA), confers protection to reproductive function and yield traits under high-temperature stress environments. Int. J. Mol. Sci. 2021, 22, 5825. [Google Scholar] [CrossRef]
- Ci, D.; Song, Y.; Tian, M.; Zhang, D. Methylation of miRNA genes in the response to temperature stress in Populus simonii. Front. Plant Sci. 2015, 6, 921. [Google Scholar] [CrossRef]
- Biswas, M.S.; Mano, J.I. Lipid peroxide-derived short-chain carbonyls mediate H2O2-induced and NaCl-induced programmed cell death in plants. Plant Physiol. 2015, 168, 885–898. [Google Scholar] [CrossRef]
- Jain, M.; Misra, G.; Patel, R.K.; Priya, P.; Jhanwar, S.; Khan, A.W.; Shah, N.; Singh, V.K.; Garg, R.; Jeena, G.; et al. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J. 2013, 74, 715–729. [Google Scholar] [CrossRef]
- Pellicer, J.; Hidalgo, O.; Dodsworth, S.; Leitch, I. Genome size diversity and its impact on the evolution of land plants. Genes 2018, 9, 88. [Google Scholar] [CrossRef]
- Emmrich, P.M.; Sarkar, A.; Njaci, I.; Kaithakottil, G.G.; Ellis, N.; Moore, C.; Edwards, A.; Heavens, D.; Waite, D.; Cheema, J.; et al. A draft genome of grass pea (Lathyrus sativus), a resilient diploid legume. bioRxiv 2020. [Google Scholar] [CrossRef]
- Sudheesh, S.; Verma, P.; Forster, J.W.; Cogan, N.O.; Kaur, S. Generation and characterisation of a reference transcriptome for lentil (Lens culinaris Medik.). Int. J. Mol. Sci. 2016, 17, 1887. [Google Scholar] [CrossRef]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef]
- Kang, Y.J.; Satyawan, D.; Shim, S.; Lee, T.; Lee, J.; Hwang, W.J.; Kim, S.K.; Lestari, P.; Laosatit, K.; Kim, K.H.; et al. Draft genome sequence of adzuki bean, Vigna angularis. Sci. Rep. 2015, 5, 8069. [Google Scholar] [CrossRef] [PubMed]
- Sivasakthi, K.; Thudi, M.; Tharanya, M.; Kale, S.M.; Kholová, J.; Halime, M.H.; Jaganathan, D.; Baddam, R.; Thirunalasundari, T.; Gaur, P.M.; et al. Plant vigour QTLs co-map with an earlier reported QTL hotspot for drought tolerance while water saving QTLs map in other regions of the chickpea genome. BMC Plant Biol. 2018, 18, 29. [Google Scholar] [CrossRef] [PubMed]
- Ricroch, A.; Clairand, P.; Harwood, W. Use of CRISPR systems in plant genome editing: Toward new opportunities in agriculture. Emerg. Top. Life Sci. 2017, 1, 169–182. [Google Scholar] [PubMed]
- Zaidi, S.S.; Mukhtar, M.S.; Mansoor, S. Genome editing: Targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 2018, 36, 898–906. [Google Scholar] [CrossRef]
- Guichard, A.; Haque, T.; Bobik, M.; Xu, X.R.; Klanseck, C.; Kushwah, R.B.; Berni, M.; Kaduskar, B.; Gantz, V.M.; Bier, E. Efficient allelic-drive in Drosophila. Nat. Commun. 2019, 10, 1640. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 2016, 7, 377. [Google Scholar] [CrossRef]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genom. 2018, 18, 31–41. [Google Scholar] [CrossRef]
- Janni, M.; Gullì, M.; Maestri, E.; Marmiroli, M.; Valliyodan, B.; Nguyen, H.T.; Marmiroli, N. Molecular and genetic bases of heat stress responses in crop plants and breeding for increased resilience and productivity. J. Exp. Bot. 2020, 71, 3780–3802. [Google Scholar] [CrossRef] [PubMed]
- Badhan, S.; Ball, A.S.; Mantri, N. First Report of CRISPR/Cas9 mediated DNA-free editing of 4CL and RVE7 genes in chickpea protoplasts. Int. J. Mol. Sci. 2021, 22, 396. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, P.; Konkin, D.; Polowick, P.; Hodgins, C.L.; Subedi, M.; Xiang, D.; Yu, B.; Patterson, N.; Rajagopalan, N.; Babic, V.; et al. CRISPR/Cas9 gene editing in legume crops: Opportunities and challenges. Legume Sci. 2021. [Google Scholar] [CrossRef]
- Uauy, C. Wheat genomics comes of age. Curr. Opin. Plant Biol. 2017, 36, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Uauy, C.; Till, B.; Liu, C.M. TILLING and associated technologies. J. Integr. Plant Biol. 2010, 52, 1027–1030. [Google Scholar] [CrossRef]
- Comastri, A.; Janni, M.; Simmonds, J.; Uauy, C.; Pignone, D.; Nguyen, H.T.; Marmiroli, N. Heat in wheat: Exploit reverse genetic techniques to discover new alleles within the Triticum durum sHsp26 family. Front. Plant Sci. 2018, 9, 1337. [Google Scholar] [CrossRef]
- Marko, D.; El-Shershaby, A.; Carriero, F.; Summerer, S.; Petrozza, A.; Iannacone, R.; Schleiff, E.; Fragkostefanakis, S. Identification and characterization of a thermos tolerant TILLING allele of heat shock binding protein 1 in tomato. Genes 2019, 10, 516. [Google Scholar] [CrossRef]
- Porch, T.G. Application of stress indices for heat tolerance screening of common bean. J. Agron. Crop Sci. 2006, 192, 390–394. [Google Scholar] [CrossRef]
- Xu, C.; Huang, B. Root proteomic responses to heat stress in two Agrostis grass species contrasting in heat tolerance. J. Exp. Bot. 2008, 59, 4183–4194. [Google Scholar] [CrossRef]
- Liu, X.; Huang, B. Root physiological factors involved in cool-season grass response to high soil temperature. Environ. Exp. Bot. 2005, 53, 233–245. [Google Scholar] [CrossRef]
- Valdés-López, O.; Batek, J.; Gomez-Hernandez, N.; Nguyen, C.T.; Isidra-Arellano, M.C.; Zhang, N.; Joshi, T.; Xu, D.; Hixson, K.K.; Weitz, K.K.; et al. Soybean roots grown under heat stress show global changes in their transcriptional and proteomic profiles. Front. Plant Sci. 2016, 7, 517. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; von Mikecz, A. Formation on nucleoplasmic protein aggregates impairs nuclear function in response to SiO2 nanoparticles. Exp. Cell Res. 2005, 305, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Buzea, C.; Pacheco, B.I.; Robbie, K. Nanomaterials and nanoparticles: Sources and toxicity. Biointerphases 2007, 2, 17–72. [Google Scholar] [CrossRef]
- Rico, C.M.; Duarte, S.; Garden, M.; Peralta-Videa, J.R.; Gardea-Torrcsdcy, J.L. Interaction of nanoparticles with edible plants and their possible implications in the food chain. J. Agric. Food Chem. 2011, 59, 3485–3498. [Google Scholar] [CrossRef]
- Hofmann, T.; Lowry, G.V.; Ghoshal, S.; Tufenkji, N.; Brambilla, D.; Dutcher, J.R.; Gilbertson, L.M.; Giraldo, J.P.; Kinsella, J.M.; Landry, M.P.; et al. Technology readiness and overcoming barriers to sustainably implement nanotechnology-enabled plant agriculture. Nat. Food 2020, 1, 416–425. [Google Scholar] [CrossRef]
- Al–Ahmadi, M.S. Cytogenetic and molecular assessment of some nanoparticles using Allium sativum assay. Afr. J. Biotechnol. 2019, 18, 783–796. [Google Scholar]
- Jha, S.; Pudake, R.N. Molecular mechanism of plant–nanoparticle interactions. In Plant Nanotechnology; Kole, C., Kumar, D., Khodakovskaya, M., Eds.; Springer Publishing: Cham, Switzerland, 2016; pp. 155–181. [Google Scholar]
| Crop | Reads/EST | Unigenes/Transcript | SSR | SNPs | References |
|---|---|---|---|---|---|
| Chickpea | - | 160,883 | 1022 | [44] | |
| - | 2619 | 81,845 | 76,084 | [45] | |
| - | 53,409 | 4816 | [46] | ||
| - | 34,760 | 4111 | 495 | [47] | |
| - | 103,215 | 26,252 | 26,082 | [48] | |
| - | - | - | 14,454 | [49] | |
| - | 37,265 | 4072 | 36,446 | [50] | |
| - | 43,389 | 5409 | 39,940 | [51] | |
| Lentil | 1,380,000 | 25,592 | - | - | [52] |
| 1,030,000 | 27,921 | - | - | [53] | |
| 119,855,798 | 20,009 | - | - | [54] | |
| 111,105,153 | 97,528 | - | - | [55] | |
| 58,621,121 | 77,346 | - | - | [56] | |
| 46,700,000 | - | - | - | [57] | |
| 26,165,023 | 96,824 | - | - | [2,58] | |
| - | - | - | - | ||
| Pea | 1005.1 million | - | 16,877 | [59] | |
| - | - | - | 10,739 | [60] | |
| - | - | 36,188 | [61] | ||
| 18,552 | 10,086 | 586 | - | [62] | |
| - | - | - | 520 | [63] | |
| - | - | - | 340 | [64] | |
| - | - | - | 956 | [61] | |
| 3,042,418 | - | 35,455 | [65] | ||
| - | 8822 | [66] | |||
| 2,209,735 | 195,661 | - | [67] | ||
| - | |||||
| 40,903 | 10,506 | - | [68] | ||
| - | 248,617 | [69] | |||
| 432 million | 27,145 | - | - | [70] | |
| one billion reads | 52,477 | - | - | [71] | |
| 69,706,469 | 48,628 | - | - | [72] | |
| ~55 million | 81,774 | - | - | [73] | |
| 88 million | 7946 | - | - | [74] | |
| - | 8899 | 3275 | - | [75] | |
| - | 10,800 | 2395 | - | [76] | |
| 720,324 | 70,682 | 2397 | - | [77] | |
| Grass pea | 493,364 | 651,827 | - | [78] | |
| 570 million | 27,431 | 3204 | 146,406 | [79] | |
| 46,994,629 + 72,566,465 | 134,914 | 200 | 4892 | [80] | |
| 399,648 | 14,386 | - | - | [81] | |
| Faba bean | - | - | 14,552 | [82] | |
| - | 37,378 | 9071 | - | [83] | |
| - | 25,502 + 12,319 | - | [84] | ||
| 1,247,881 | 343,325 | - | 560–2144 | [85] | |
| 87,269 | - | - | 39,060 | [86] | |
| - | - | 28,503 | - | [87] | |
| 304,680 | 60,440 | 802 | - | [77] | |
| Common bean | - | 629 | - | [88] | |
| 3123 | - | 184 | - | [89] | |
| - | - | 7015 | [90] | ||
| 418 million | - | - | 346,819 | [91] | |
| - | - | - | 19,204 | [92] | |
| - | - | - | 17,190 | [93] | |
| - | - | - | 43,018 | [94] | |
| - | - | - | 12,697 | [95] | |
| - | - | - | 230 | [96] | |
| 21,026 | 7969 | - | - | [97] | |
| - | 3126 | - | - | [98] | |
| - | - | 1800 | [99] | ||
| 7079 | 4219 | - | - | [100] | |
| 37,919 | 10,581 | - | - | [101] | |
| 9583 | - | 4764 | - | [102] | |
| - | 59,295 | - | - | [103] | |
| 900,000 | 30,491 | - | - | [104] |
| Crop | Traits | QTL Name/No. of MTAs | Population Size | PVE (%) | Reference |
|---|---|---|---|---|---|
| Chickpea | filled pods/plot | qfpod02_5 | 292 | 12.03 | [113] |
| total number of seeds/plot | qts02_5 | 292 | 10.00 | [113] | |
| grain yield per plot | Qgy02_5 | 292 | 16.56 | [113] | |
| % pod setting | q%podset06_5 | 292 | 13.30 | [39,113] | |
| chlorophyll content | - | 206 | 17.2 | [114] | |
| Lentil | seedling survival | qHt_ss | 142 | 12.1 | [115] |
| pod set | qHt_ps | 147 | 9.23 | [115] | |
| Field pea | chlorophyll concentration | 6 | 135 | 7–13 | [116] |
| photochemical reflectance index | 2 | 135 | 9 | [116] | |
| canopy temperature | 2 | 135 | 6 | [116] | |
| reproductive stem length | 6 | 135 | 4–6 | [116] | |
| internode length | 6 | 135 | 5–7 | [116] | |
| pod number | 9 | 135 | 7–10 | [116] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, J.; Mir, R.R.; Shafi, S.; Sen Gupta, D.; Djalovic, I.; Miladinovic, J.; Kumar, R.; Kumar, S.; Kumar, R. Genomics Associated Interventions for Heat Stress Tolerance in Cool Season Adapted Grain Legumes. Int. J. Mol. Sci. 2022, 23, 399. https://doi.org/10.3390/ijms23010399
Kumar J, Mir RR, Shafi S, Sen Gupta D, Djalovic I, Miladinovic J, Kumar R, Kumar S, Kumar R. Genomics Associated Interventions for Heat Stress Tolerance in Cool Season Adapted Grain Legumes. International Journal of Molecular Sciences. 2022; 23(1):399. https://doi.org/10.3390/ijms23010399
Chicago/Turabian StyleKumar, Jitendra, Reyazul Rouf Mir, Safoora Shafi, Debjyoti Sen Gupta, Ivica Djalovic, Jegor Miladinovic, Rahul Kumar, Sachin Kumar, and Rajeev Kumar. 2022. "Genomics Associated Interventions for Heat Stress Tolerance in Cool Season Adapted Grain Legumes" International Journal of Molecular Sciences 23, no. 1: 399. https://doi.org/10.3390/ijms23010399
APA StyleKumar, J., Mir, R. R., Shafi, S., Sen Gupta, D., Djalovic, I., Miladinovic, J., Kumar, R., Kumar, S., & Kumar, R. (2022). Genomics Associated Interventions for Heat Stress Tolerance in Cool Season Adapted Grain Legumes. International Journal of Molecular Sciences, 23(1), 399. https://doi.org/10.3390/ijms23010399








