Vypal2: A Versatile Peptide Ligase for Precision Tailoring of Proteins
Abstract
:1. Introduction
2. Results and Discussion
2.1. Demonstration of VML for Protein Labeling and the Use of the Labelled Protein in Cell Imaging
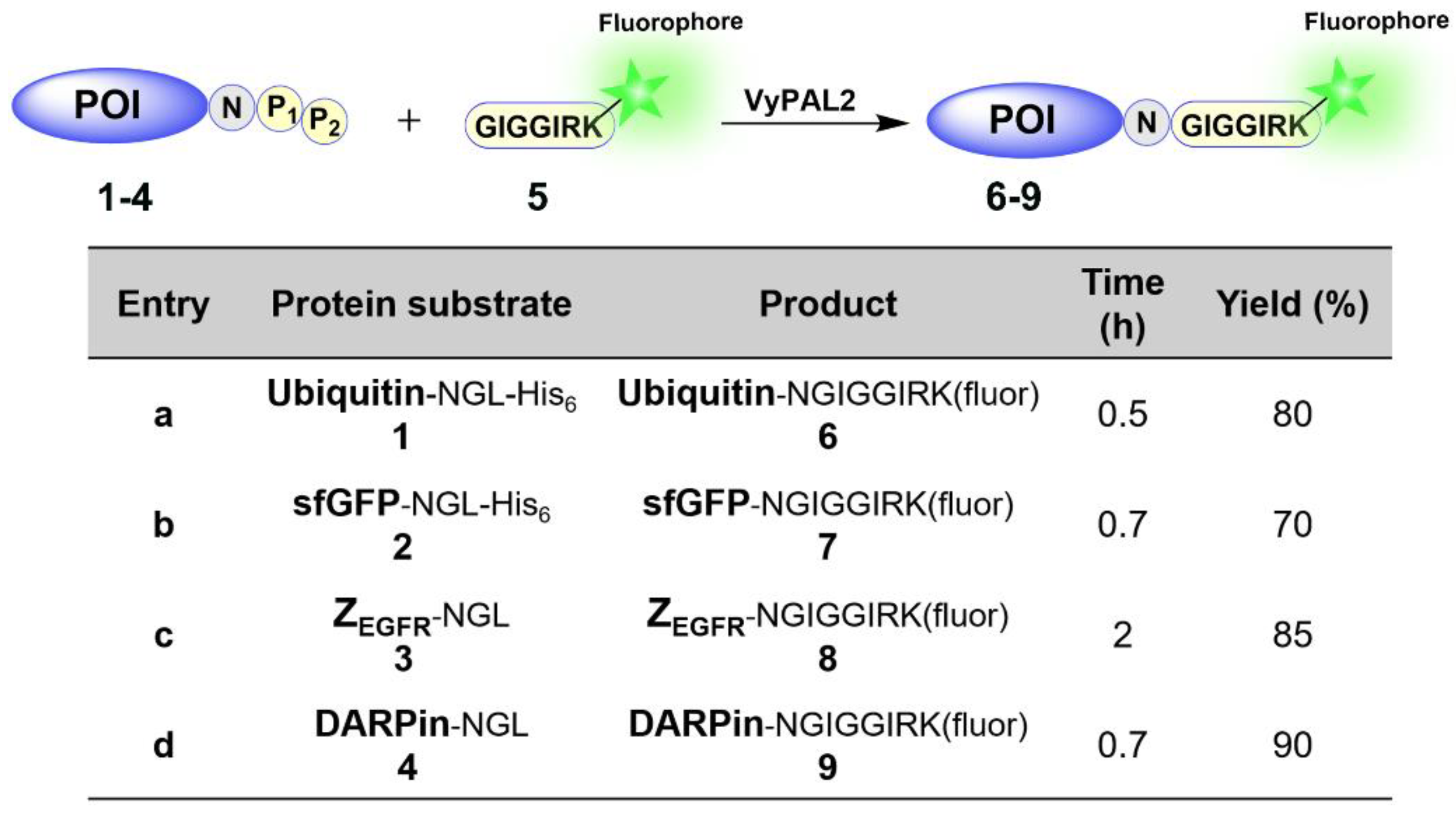
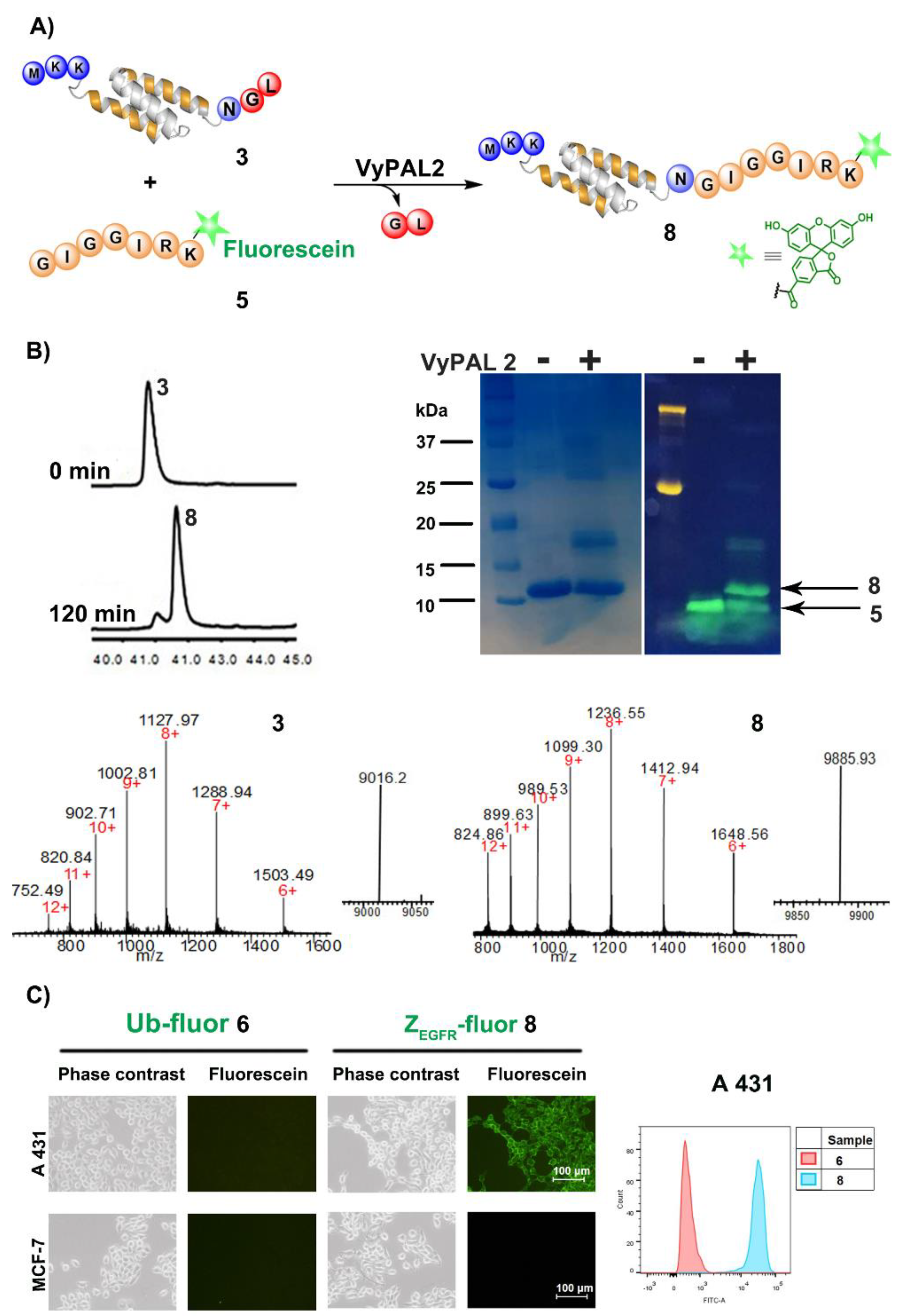
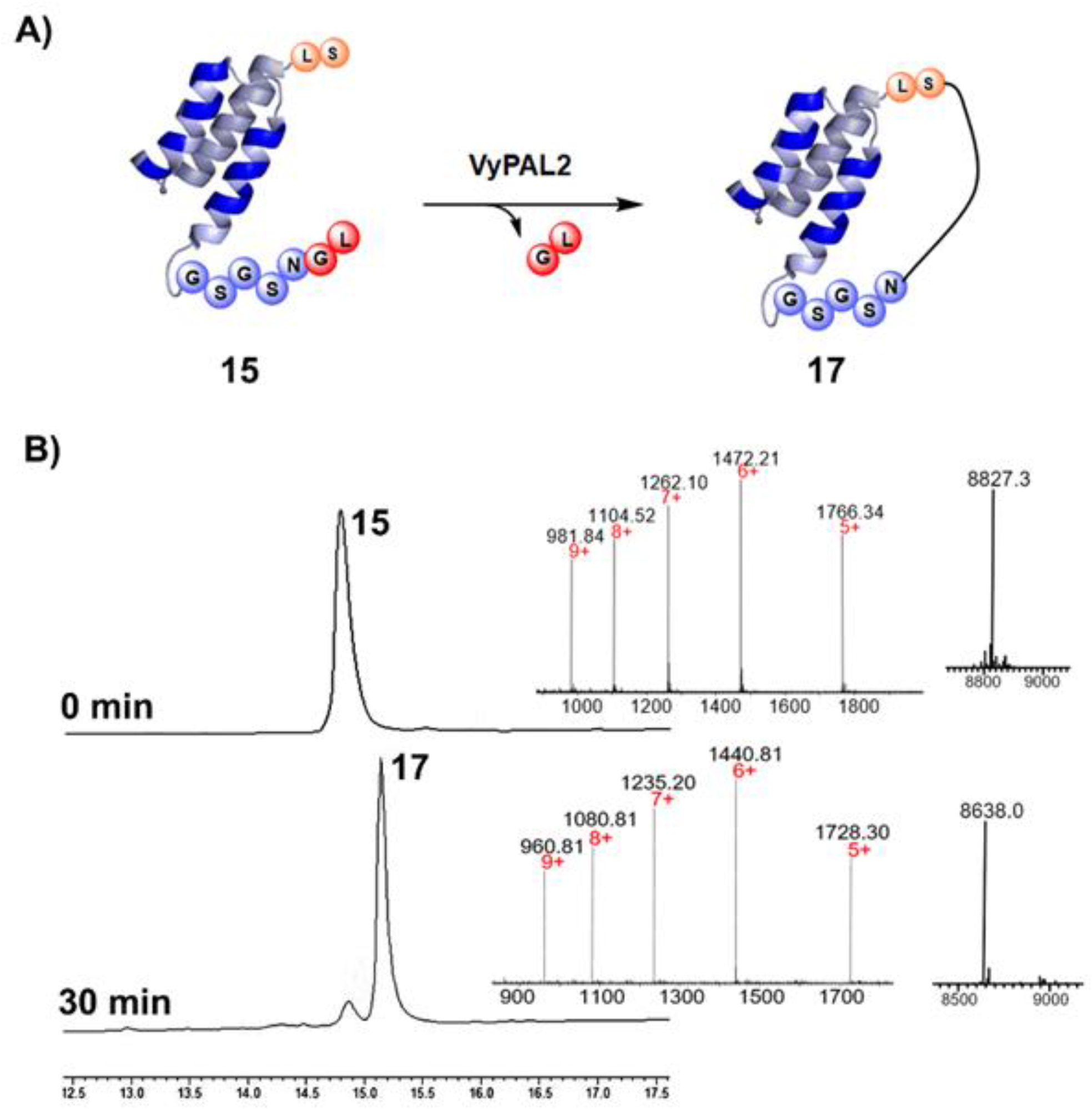
2.1.1. VyPAL2-Mediated Ligation for Protein C-Terminal Labeling
2.1.2. Binding of Fluorescence-Labeled Affibody 8 to A431 Cells
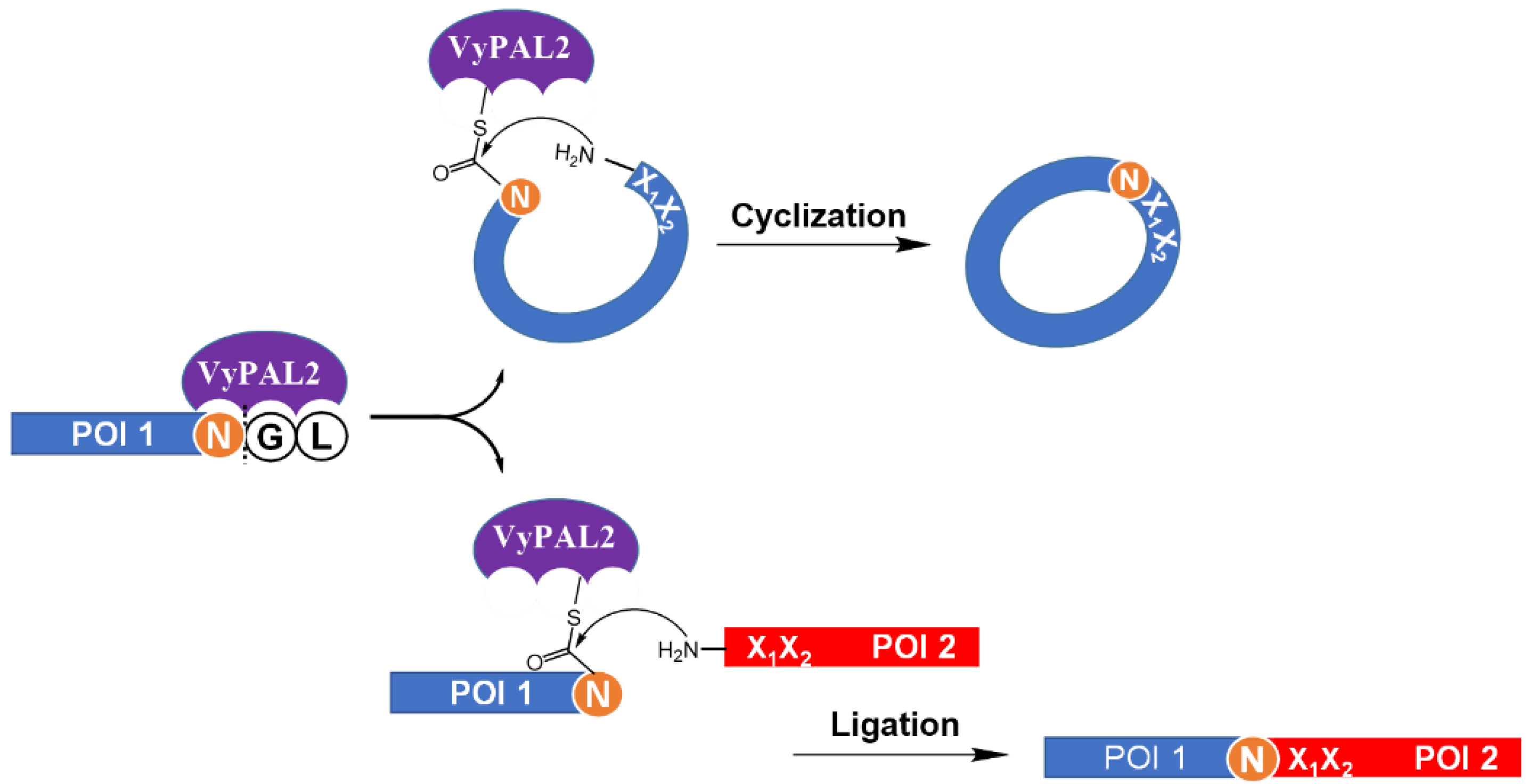
2.2. VyPAL-Mediated Inter-Protein Ligation
2.3. VML for Protein Cyclization
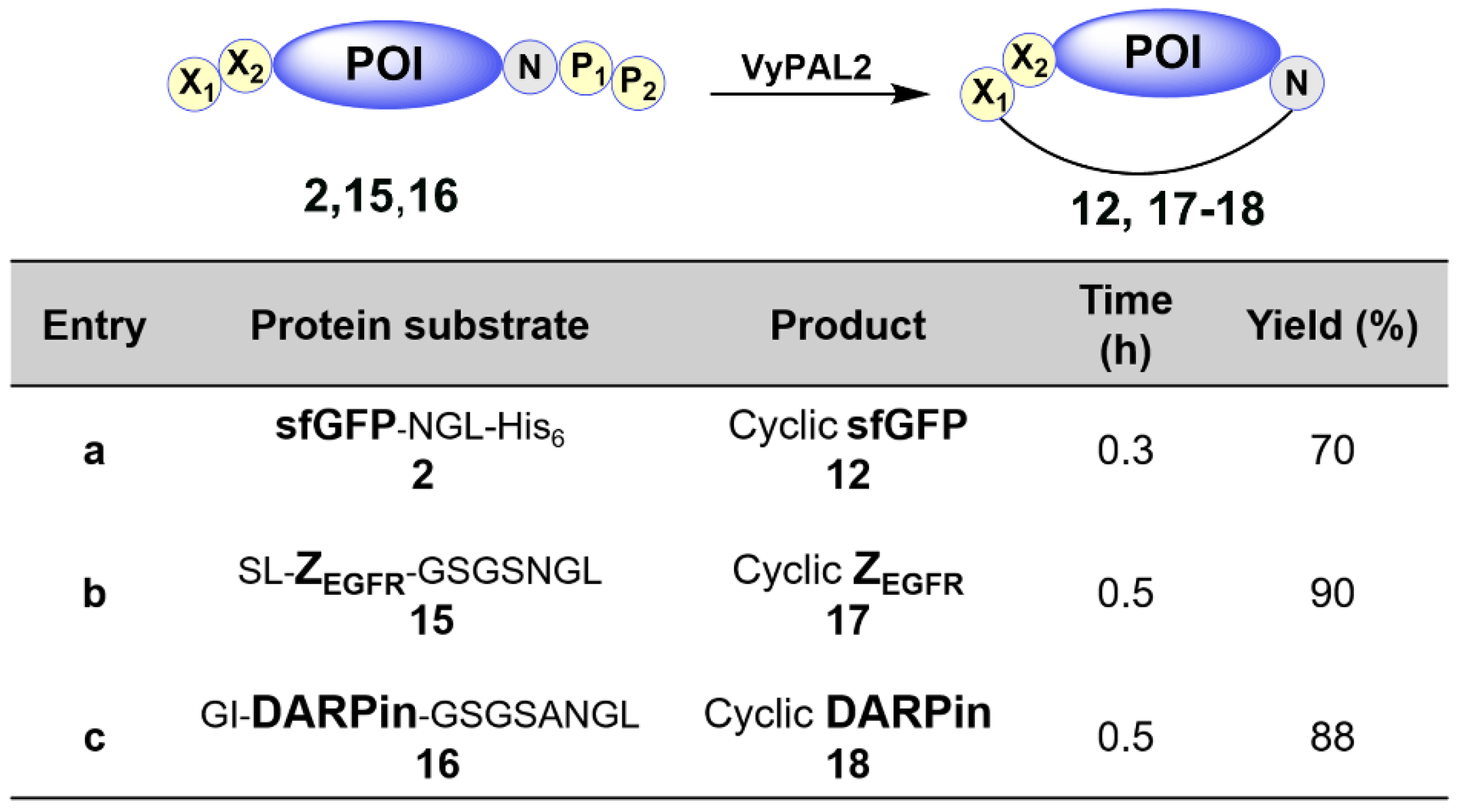
2.3.1. PAL-Mediated Affibody ZEGFR and DARPin Cyclization
2.3.2. Binding of Cyclic Affibody and Cyclic DARPin to Their Target Receptors
2.4. VyPAL2-Mediated Ligation on Cell Surface
2.4.1. VML for the Labelling of Affibody and DARPin Pre-Bound on Cell Surface
2.4.2. VML for Labeling a Cell-Surface Glycan Anchored Peptide Substrate Preinstalled via Oxime Conjugation
3. Materials and Methods
3.1. HPLC
3.2. Mass Spectrometry
3.3. Molecular Cloning and Protein Expression
3.4. Cell Culture and Imaging
3.5. Flow Cytometry (FACS) Analysis
3.6. Solid Phase Peptide Synthesis (SPPS)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nguyen, G.K.; Qiu, Y.; Cao, Y.; Hemu, X.; Liu, C.-F.; Tam, J.P. Butelase-mediated cyclization and ligation of peptides and proteins. Nat. Protoc. 2016, 11, 1977–1988. [Google Scholar] [CrossRef]
- Xu, S.; Zhao, Z.; Zhao, J. Recent advances in enzyme-mediated peptide ligation. Chin. Chem. Lett. 2018, 29, 1009–1016. [Google Scholar] [CrossRef]
- Pishesha, N.; Ingram, J.R.; Ploegh, H.L. Sortase A: A model for transpeptidation and its biological applications. Annu. Rev. Cell. Dev. Biol. 2018, 34, 163–188. [Google Scholar] [CrossRef] [PubMed]
- Weeks, A.M.; Wells, J.A. Subtiligase-catalyzed peptide ligation. Chem. Rev. 2019, 120, 3127–3160. [Google Scholar] [CrossRef]
- Jackson, M.A.; Nguyen, L.T.; Gilding, E.K.; Durek, T.; Craik, D.J. Make it or break it: Plant AEPs on stage in biotechnology. Biotechnol. Adv. 2020, 45, 107651. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Chan, N.-Y.; Liew, H.T.; Tan, S.J.; Chen, Y. Peptide asparaginyl ligases—Renegade peptide bond makers. Sci. China Chem. 2020, 63, 296–307. [Google Scholar] [CrossRef] [Green Version]
- Tang, T.M.S.; Luk, L.Y.P. Asparaginyl endopeptidases: Enzymology, applications and limitations. Org. Biomol. Chem. 2021, 19, 5048–5062. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.W.; Schneewind, O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 1999, 63, 174–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, S.-Y.; Budzik, J.M.; Schneewind, O. Sortases make pili from three ingredients. Proc. Natl. Acad. Sci. USA 2008, 105, 13703–13704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef]
- Donia, M.S.; Ravel, J.; Schmidt, E.W. A global assembly line for cyanobactins. Nat. Chem. Biol. 2008, 4, 341–343. [Google Scholar] [CrossRef]
- Chekan, J.R.; Estrada, P.; Covello, P.S.; Nair, S.K. Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants. Proc. Natl. Acad. Sci. USA 2017, 114, 6551–6556. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, G.K.; Wang, S.; Qiu, Y.; Hemu, X.; Lian, Y.; Tam, J.P. Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat. Chem. Biol. 2014, 10, 732–738. [Google Scholar] [CrossRef]
- Dall, E.; Brandstetter, H. Structure and function of legumain in health and disease. Biochimie 2016, 122, 126–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saska, I.; Gillon, A.D.; Hatsugai, N.; Dietzgen, R.G.; Hara-Nishimura, I.; Anderson, M.A.; Craik, D.J. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J. Biol. Chem. 2007, 282, 29721–29728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Hua, T.; Crowley, C.; Ru, H.; Ni, X.; Shaw, N.; Jiao, L.; Ding, W.; Qu, L.; Hung, L.-W. Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage. Cell Res. 2014, 24, 344–358. [Google Scholar] [CrossRef] [Green Version]
- Dall, E.; Fegg, J.C.; Briza, P.; Brandstetter, H. Structure and mechanism of an aspartimide-dependent peptide ligase in human legumain. Angew. Chem. Int. Ed. Engl. 2015, 54, 2917–2921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernath-Levin, K.; Nelson, C.; Elliott, A.G.; Jayasena, A.S.; Millar, A.H.; Craik, D.J.; Mylne, J.S. Peptide macrocyclization by a bifunctional endoprotease. Chem. Biol. 2015, 22, 571–582. [Google Scholar] [CrossRef] [Green Version]
- Harris, K.S.; Durek, T.; Kaas, Q.; Poth, A.G.; Gilding, E.K.; Conlan, B.F.; Saska, I.; Daly, N.L.; Van Der Weerden, N.L.; Craik, D.J. Efficient backbone cyclization of linear peptides by a recombinant asparaginyl endopeptidase. Nat. Commun. 2015, 6, 10199. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Wong, Y.H.; Nguyen, G.K.; Tam, J.P.; Lescar, J.; Wu, B. Engineering a catalytically efficient recombinant protein ligase. J. Am. Chem. Soc. 2017, 139, 5351–5358. [Google Scholar] [CrossRef]
- Zauner, F.B.; Dall, E.; Regl, C.; Grassi, L.; Huber, C.G.; Cabrele, C.; Brandstetter, H. Crystal structure of plant legumain reveals a unique two-chain state with pH-dependent activity regulation. Plant Cell 2018, 30, 686–699. [Google Scholar] [CrossRef]
- Jackson, M.; Gilding, E.; Shafee, T.; Harris, K.; Kaas, Q.; Poon, S.; Yap, K.; Jia, H.; Guarino, R.; Chan, L. Molecular basis for the production of cyclic peptides by plant asparaginyl endopeptidases. Nat. Commun. 2018, 9, 2411. [Google Scholar] [CrossRef]
- James, A.M.; Haywood, J.; Leroux, J.; Ignasiak, K.; Elliott, A.G.; Schmidberger, J.W.; Fisher, M.F.; Nonis, S.G.; Fenske, R.; Bond, C.S. The macrocyclizing protease butelase 1 remains autocatalytic and reveals the structural basis for ligase activity. Plant J. 2019, 98, 988–999. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Yap, K.; Chan, L.Y.; Rehm, F.B.; Looi, F.Y.; Poth, A.G.; Gilding, E.K.; Kaas, Q.; Durek, T.; Craik, D.J. A bifunctional asparaginyl endopeptidase efficiently catalyzes both cleavage and cyclization of cyclic trypsin inhibitors. Nat. Commun. 2020, 11, 1575. [Google Scholar] [CrossRef] [Green Version]
- Hemu, X.; El Sahili, A.; Hu, S.; Wong, K.; Chen, Y.; Wong, Y.H.; Zhang, X.; Serra, A.; Goh, B.C.; Darwis, D.A.; et al. Structural determinants for peptide-bond formation by asparaginyl ligases. Proc. Natl. Acad. Sci. USA. 2019, 116, 11737–11746. [Google Scholar] [PubMed] [Green Version]
- Cao, Y.; Nguyen, G.K.; Tam, J.P.; Liu, C.-F. Butelase-mediated synthesis of protein thioesters and its application for tandem chemoenzymatic ligation. Chem. Commun. 2015, 51, 17289–17292. [Google Scholar] [CrossRef]
- Nguyen, G.K.; Cao, Y.; Wang, W.; Liu, C.F.; Tam, J.P. Site-specific N-terminal labeling of peptides and proteins using butelase 1 and thiodepsipeptide. Angew. Chem. Int. Ed. Engl. 2015, 127, 15920–15924. [Google Scholar] [CrossRef]
- Nguyen, G.K.; Kam, A.; Loo, S.; Jansson, A.E.; Pan, L.X.; Tam, J.P. Butelase 1: A versatile ligase for peptide and protein macrocyclization. J. Am. Chem. Soc. 2015, 137, 15398–15401. [Google Scholar] [CrossRef]
- Nguyen, G.K.; Hemu, X.; Quek, J.P.; Tam, J.P. Butelase-mediated macrocyclization of d-amino-acid-containing peptides. Angew. Chem. Int. Ed. Engl. 2016, 128, 12994–12998. [Google Scholar] [CrossRef]
- Cao, Y.; Nguyen, G.K.; Chuah, S.; Tam, J.P.; Liu, C.-F. Butelase-mediated ligation as an efficient bioconjugation method for the synthesis of peptide dendrimers. Bioconjug. Chem. 2016, 27, 2592–2596. [Google Scholar] [CrossRef]
- Bi, X.; Yin, J.; Nguyen, G.K.; Rao, C.; Halim, N.B.A.; Hemu, X.; Tam, J.P.; Liu, C.F. Enzymatic engineering of live bacterial cell surfaces using butelase 1. Angew. Chem. Int. Ed. Engl. 2017, 56, 7822–7825. [Google Scholar] [CrossRef]
- Harmand, T.J.; Bousbaine, D.; Chan, A.; Zhang, X.; Liu, D.R.; Tam, J.P.; Ploegh, H.L. One-pot dual labeling of IgG 1 and preparation of C-to-C fusion proteins through a combination of sortase A and butelase 1. Bioconjug. Chem. 2018, 29, 3245–3249. [Google Scholar] [CrossRef]
- Bi, X.; Yin, J.; Hemu, X.; Rao, C.; Tam, J.P.; Liu, C.-F. Immobilization and Intracellular Delivery of Circular Proteins by Modifying a Genetically Incorporated Unnatural Amino Acid. Bioconjug. Chem. 2018, 29, 2170–2175. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Yin, J.; Zhang, D.; Zhang, X.; Balamkundu, S.; Lescar, J.; Dedon, P.C.; Tam, J.P.; Liu, C.-F. Tagging transferrin receptor with a disulfide FRET probe to gauge the redox state in endosomal compartments. Anal. Chem. 2020, 92, 12460–12466. [Google Scholar] [CrossRef] [PubMed]
- Hemu, X.; To, J.; Zhang, X.; Tam, J.P. Immobilized Peptide Asparaginyl Ligases Enhance Stability and Facilitate Macrocyclization and Site-specific Ligation. J. Org. Chem. 2020, 85, 1504–1512. [Google Scholar] [CrossRef] [PubMed]
- Hemu, X.; El Sahili, A.; Hu, S.; Zhang, X.; Serra, A.; Goh, B.C.; Darwis, D.A.; Chen, M.W.; Sze, S.K.; Liu, C.-F. Turning an asparaginyl endopeptidase into a peptide ligase. ACS Catal. 2020, 10, 8825–8834. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, D.; Hemu, X.; Hu, S.; To, J.; Zhang, X.; Lescar, J.; Tam, J.P.; Liu, C.-F. Engineering protein theranostics using bio-orthogonal asparaginyl peptide ligases. Theranostics 2021, 11, 5863–5875. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, Z.; Hu, S.; Balamkundu, S.; To, J.; Zhang, X.; Lescar, J.; Tam, J.P.; Liu, C.-F. pH-Controlled Protein Orthogonal Ligation Using Asparaginyl Peptide Ligases. J. Am. Chem. Soc. 2021, 143, 8704–8712. [Google Scholar] [CrossRef]
- Abrahmsen, L.; Tom, J.; Burnier, J.; Butcher, K.A.; Kossiakoff, A.; Wells, J.A. Engineering subtilisin and its substrates for efficient ligation of peptide bonds in aqueous solution. Biochemistry 1991, 30, 4151–4159. [Google Scholar] [CrossRef]
- Chang, T.K.; Jackson, D.Y.; Burnier, J.P.; Wells, J.A. Subtiligase: A tool for semisynthesis of proteins. Proc. Natl. Acad. Sci. USA 1994, 91, 12544–12548. [Google Scholar] [CrossRef] [Green Version]
- Henager, S.H.; Chu, N.; Chen, Z.; Bolduc, D.; Dempsey, D.R.; Hwang, Y.; Wells, J.; Cole, P.A. Enzyme-catalyzed expressed protein ligation. Nat. Methods 2016, 13, 925–927. [Google Scholar] [CrossRef] [Green Version]
- Tan, X.; Yang, R.; Liu, C.-F. Facilitating subtiligase-catalyzed peptide ligation reactions by using peptide thioester substrates. Org. Lett. 2018, 20, 6691–6694. [Google Scholar] [CrossRef]
- Weeks, A.M.; Wells, J.A. Engineering peptide ligase specificity by proteomic identification of ligation sites. Nat. Chem. Biol. 2018, 14, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Hart, S.A.; Schink, A.; Pollok, B.A. Sortase-mediated protein ligation: A new method for protein engineering. J. Am. Chem. Soc. 2004, 126, 2670–2671. [Google Scholar] [CrossRef] [PubMed]
- Popp, M.W.L.; Ploegh, H.L. Making and breaking peptide bonds: Protein engineering using sortase. Angew. Chem. Int. Ed. Engl. 2011, 50, 5024–5032. [Google Scholar] [CrossRef] [PubMed]
- Williamson, D.J.; Fascione, M.A.; Webb, M.E.; Turnbull, W.B. Efficient N-terminal labeling of proteins by use of sortase. Angew. Chem. Int. Ed. Engl. 2012, 51, 9377–9380. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.M.; Li, Y.T.; Pan, M.; Kong, X.Q.; Huang, Y.C.; Hong, Z.Y.; Liu, L. Irreversible site-specific hydrazinolysis of proteins by use of sortase. Angew. Chem. Int. Ed. Engl. 2014, 53, 2198–2202. [Google Scholar] [CrossRef]
- Steiner, D.; Forrer, P.; Plückthun, A. Efficient selection of DARPins with sub-nanomolar affinities using SRP phage display. J. Mol. Biol. 2008, 382, 1211–1227. [Google Scholar] [CrossRef] [Green Version]
- Friedman, M.; Orlova, A.; Johansson, E.; Eriksson, T.L.; Höidén-Guthenberg, I.; Tolmachev, V.; Nilsson, F.Y.; Ståhl, S. Directed evolution to low nanomolar affinity of a tumor-targeting epidermal growth factor receptor-binding affibody molecule. J. Mol. Biol. 2008, 376, 1388–1402. [Google Scholar] [CrossRef] [PubMed]
- Davidson, N.E.; Gelmann, E.P.; Lippman, M.E.; Dickson, R.B. Epidermal growth factor receptor gene expression in estrogen receptor-positive and negative human breast cancer cell lines. Mol. Endocrinol. 1987, 1, 216–223. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.T.; Rowlands, D.K.; Wong, C.H.; Lo, T.W.; Nguyen, G.K.; Li, H.Y.; Tam, J.P. Orally active peptidic bradykinin B1 receptor antagonists engineered from a cyclotide scaffold for inflammatory pain treatment. Angew. Chem. Int. Ed. Engl. 2012, 51, 5620–5624. [Google Scholar] [CrossRef] [PubMed]
- Popp, M.W.; Dougan, S.K.; Chuang, T.-Y.; Spooner, E.; Ploegh, H.L. Sortase-catalyzed transformations that improve the properties of cytokines. Proc. Natl. Acad. Sci. USA 2011, 108, 3169–3174. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Wang, Z.; Hu, S.; Lescar, J.; Tam, J.P.; Liu, C.-F. Vypal2: A Versatile Peptide Ligase for Precision Tailoring of Proteins. Int. J. Mol. Sci. 2022, 23, 458. https://doi.org/10.3390/ijms23010458
Zhang D, Wang Z, Hu S, Lescar J, Tam JP, Liu C-F. Vypal2: A Versatile Peptide Ligase for Precision Tailoring of Proteins. International Journal of Molecular Sciences. 2022; 23(1):458. https://doi.org/10.3390/ijms23010458
Chicago/Turabian StyleZhang, Dingpeng, Zhen Wang, Side Hu, Julien Lescar, James P. Tam, and Chuan-Fa Liu. 2022. "Vypal2: A Versatile Peptide Ligase for Precision Tailoring of Proteins" International Journal of Molecular Sciences 23, no. 1: 458. https://doi.org/10.3390/ijms23010458
APA StyleZhang, D., Wang, Z., Hu, S., Lescar, J., Tam, J. P., & Liu, C.-F. (2022). Vypal2: A Versatile Peptide Ligase for Precision Tailoring of Proteins. International Journal of Molecular Sciences, 23(1), 458. https://doi.org/10.3390/ijms23010458





