RNA Exosome Component EXOSC4 Amplified in Multiple Cancer Types Is Required for the Cancer Cell Survival
Abstract
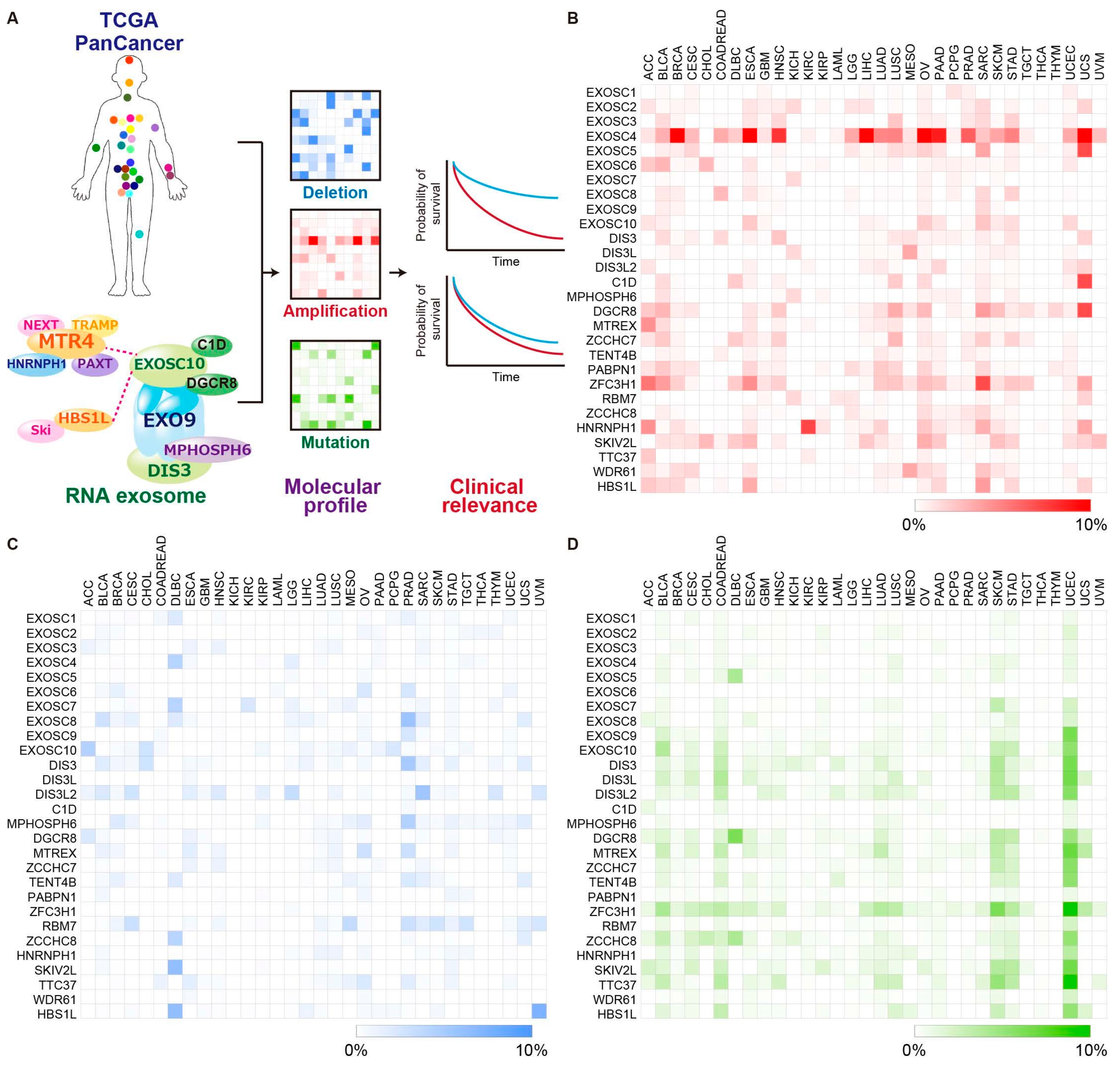
:1. Introduction
2. Results
2.1. EXOSC4 Is Amplified and Upregulated in Pancreatic Cancer Tissue
2.2. EXOSC4 Knockdown Causes a Reduction in the Growth of Pancreatic Cancer Cells
2.3. EXOSC4 Downregulates BIK and SESN2 mRNAs in Pancreatic Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Copy Number Alteration and Mutation Analysis
4.2. Gene Expression Analysis
4.3. Cell Culture
4.4. Antibodies and Reagents
4.5. Immunohistochemical Analysis
4.6. Immunofluorescence
4.7. RNA Interference
4.8. The qRT-PCR Analysis
4.9. Immunoblotting
4.10. Cell Proliferation Assay
4.11. Apoptosis
4.12. Data Analysis
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Łabno, A.; Tomecki, R.; Dziembowski, A. Cytoplasmic RNA decay pathways—Enzymes and mechanisms. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 3125–3147. [Google Scholar] [CrossRef] [PubMed]
- Kilchert, C.; Wittmann, S.; Vasiljeva, L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 2016, 17, 227–239. [Google Scholar] [CrossRef]
- Vanacova, S.; Stef, R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007, 8, 651–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Januszyk, K.; Lima, C.D. The eukaryotic RNA exosome. Curr. Opin. Struct. Biol. 2014, 24, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Schmid, M.; Jensen, T.H. Controlling nuclear RNA levels. Nat. Rev. Genet. 2018, 19, 518–529. [Google Scholar] [CrossRef]
- Schneider, C.; Tollervey, D. Threading the barrel of the RNA exosome. Trends Biochem. Sci. 2013, 38, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Laffleur, B.; Basu, U.; Lim, J. RNA Exosome and Non-coding RNA-Coupled Mechanisms in AID-Mediated Genomic Alterations. J. Mol. Biol. 2017, 429, 3230–3241. [Google Scholar] [CrossRef]
- Zinder, J.C.; Lima, C.D. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 2017, 31, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Macias, S.; Cordiner, R.A.; Gautier, P.; Plass, M.; Cáceres, J.F. DGCR8 Acts as an Adaptor for the Exosome Complex to Degrade Double-Stranded Structured RNAs. Mol. Cell 2015, 60, 873–885. [Google Scholar] [CrossRef] [Green Version]
- Meola, N.; Jensen, T.H. Targeting the nuclear RNA exosome: Poly(A) binding proteins enter the stage. RNA Biol. 2017, 14, 820–826. [Google Scholar] [CrossRef] [Green Version]
- Wyers, F.; Rougemaille, M.; Badis, G.; Rousselle, J.C.; Dufour, M.E.; Boulay, J.; Régnault, B.; Devaux, F.; Namane, A.; Séraphin, B.; et al. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 2005, 121, 725–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lubas, M.; Christensen, M.S.; Kristiansen, M.S.; Domanski, M.; Falkenby, L.G.; Lykke-Andersen, S.; Andersen, J.S.; Dziembowski, A.; Jensen, T.H. Interaction Profiling Identifies the Human Nuclear Exosome Targeting Complex. Mol. Cell 2011, 43, 624–637. [Google Scholar] [CrossRef] [PubMed]
- LaCava, J.; Houseley, J.; Saveanu, C.; Petfalski, E.; Thompson, E.; Jacquier, A.; Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005, 121, 713–724. [Google Scholar] [CrossRef]
- San Paolo, S.; Vanacova, S.; Schenk, L.; Scherrer, T.; Blank, D.; Keller, W.; Gerber, A.P. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet. 2009, 5, e1000555. [Google Scholar] [CrossRef] [Green Version]
- Meola, N.; Domanski, M.; Karadoulama, E.; Chen, Y.; Gentil, C.; Pultz, D.; Vitting-Seerup, K.; Lykke-Andersen, S.; Andersen, J.S.; Sandelin, A.; et al. Identification of a Nuclear Exosome Decay Pathway for Processed Transcripts. Mol. Cell 2016, 64, 520–533. [Google Scholar] [CrossRef] [Green Version]
- Tanu, T.; Taniue, K.; Imamura, K.; Onoguchi-Mizutani, R.; Han, H.; Jensen, T.H.; Akimitsu, N. hnRNPH1-MTR4 complex-mediated regulation of NEAT1v2 stability is critical for IL8 expression. RNA Biol. 2021, 18, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Chlebowski, A.; Lubas, M.; Jensen, T.H.; Dziembowski, A. RNA decay machines: The exosome. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Morton, D.J.; Kuiper, E.G.; Jones, S.K.; Leung, S.W.; Corbett, A.H.; Fasken, M.B. The RNA exosome and RNA exosome-linked disease. RNA 2018, 24, 127–142. [Google Scholar] [CrossRef] [Green Version]
- Kalisiak, K.; Kuliński, T.M.; Tomecki, R.; Cysewski, D.; Pietras, Z.; Chlebowski, A.; Kowalska, K.; Dziembowski, A. A short splicing isoform of HBS1L links the cytoplasmic exosome and SKI complexes in humans. Nucleic Acids Res. 2017, 45, 2068–2080. [Google Scholar] [CrossRef] [Green Version]
- Kowalinski, E.; Kögel, A.; Ebert, J.; Reichelt, P.; Stegmann, E.; Habermann, B.; Conti, E. Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex. Mol. Cell 2016, 63, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Schmid, M.; Jensen, T.H. The Nuclear RNA Exosome and Its Cofactors. In Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2019; Volume 1203, pp. 113–132. [Google Scholar]
- Wan, J.; Yourshaw, M.; Mamsa, H.; Rudnik-Schöneborn, S.; Menezes, M.P.; Hong, J.E.; Leong, D.W.; Senderek, J.; Salman, M.S.; Chitayat, D.; et al. Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat. Genet. 2012, 44, 704–708. [Google Scholar] [CrossRef] [Green Version]
- Weißbach, S.; Langer, C.; Puppe, B.; Nedeva, T.; Bach, E.; Kull, M.; Bargou, R.; Einsele, H.; Rosenwald, A.; Knop, S.; et al. The molecular spectrum and clinical impact of DIS3 mutations in multiple myeloma. Br. J. Haematol. 2015, 169, 57–70. [Google Scholar] [CrossRef]
- Di Donato, N.; Neuhann, T.; Kahlert, A.K.; Klink, B.; Hackmann, K.; Neuhann, I.; Novotna, B.; Schallner, J.; Krause, C.; Glass, I.A.; et al. Mutations in EXOSC2 are associated with a novel syndrome characterised by retinitis pigmentosa, progressive hearing loss, premature ageing, short stature, mild intellectual disability and distinctive gestalt. J. Med. Genet. 2016, 53, 419–425. [Google Scholar] [CrossRef]
- Schottmann, G.; Picker-Minh, S.; Schwarz, J.M.; Gill, E.; Rodenburg, R.J.T.; Stenzel, W.; Kaindl, A.M.; Schuelke, M. Recessive mutation in EXOSC3 associates with mitochondrial dysfunction and pontocerebellar hypoplasia. Mitochondrion 2017, 37, 46–54. [Google Scholar] [CrossRef]
- Boczonadi, V.; Müller, J.S.; Pyle, A.; Munkley, J.; Dor, T.; Quartararo, J.; Ferrero, I.; Karcagi, V.; Giunta, M.; Polvikoski, T.; et al. EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat. Commun. 2014, 5, 4287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eggens, V.R.C.; Barth, P.G.; Niermeijer, J.M.F.; Berg, J.N.; Darin, N.; Dixit, A.; Fluss, J.; Foulds, N.; Fowler, D.; Hortobágyi, T.; et al. EXOSC3 mutations in pontocerebellar hypoplasia type 1: Novel mutations and genotype-phenotype correlations. Orphanet J. Rare Dis. 2014, 9, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, C.; Fraga de Andrade, I.; Matson, D.R.; Dewey, C.N.; Bresnick, E.H. RNA-regulatory exosome complex confers cellular survival to promote erythropoiesis. Nucleic Acids Res. 2021, 49, 9007–9025. [Google Scholar] [CrossRef]
- Müller, J.S.; Burns, D.T.; Griffin, H.; Wells, G.R.; Zendah, R.A.; Munro, B.; Schneider, C.; Horvath, R. RNA exosome mutations in pontocerebellar hypoplasia alter ribosome biogenesis and p53 levels. Life Sci. Alliance 2020, 3, e202000678. [Google Scholar] [CrossRef]
- Chiu, A.C.; Suzuki, H.I.; Wu, X.; Mahat, D.B.; Kriz, A.J.; Sharp, P.A. Transcriptional Pause Sites Delineate Stable Nucleosome-Associated Premature Polyadenylation Suppressed by U1 snRNP. Mol. Cell 2018, 69, 648–663.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pefanis, E.; Wang, J.; Rothschild, G.; Lim, J.; Kazadi, D.; Sun, J.; Federation, A.; Chao, J.; Elliott, O.; Liu, Z.P.; et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 2015, 161, 774–789. [Google Scholar] [CrossRef] [Green Version]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304.e6. [Google Scholar] [CrossRef] [Green Version]
- Campbell, P.J.; Getz, G.; Korbel, J.O.; Stuart, J.M.; Jennings, J.L.; Stein, L.D.; Perry, M.D.; Nahal-Bose, H.K.; Ouellette, B.F.F.; Li, C.H.; et al. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.W.; Shi, Z.Z.; Shen, T.Y.; Che, X.; Wang, Z.; Shi, S.S.; Xu, X.; Cai, Y.; Zhao, P.; Wang, C.F.; et al. Identification of genomic alterations in pancreatic cancer using array-based comparative genomic hybridization. PLoS ONE 2014, 9, e114616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rausch, V.; Krieg, A.; Camps, J.; Behrens, B.; Beier, M.; Wangsa, D.; Heselmeyer-Haddad, K.; Baldus, S.E.; Knoefel, W.T.; Ried, T.; et al. Array comparative genomic hybridization of 18 pancreatic ductal adenocarcinomas and their autologous metastases. BMC Res. Notes 2017, 10, 560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.; Tong, J.H.M.; Kang, W.; Lung, R.W.M.; Chak, W.P.; Chung, L.Y.; Wu, F.; Li, H.; Yu, J.; Chan, A.W.H.; et al. EXOSC4 functions as a potential oncogene in development and progression of colorectal cancer. Mol. Carcinog. 2018, 57, 1780–1791. [Google Scholar] [CrossRef] [PubMed]
- Witkiewicz, A.K.; McMillan, E.A.; Balaji, U.; Baek, G.H.; Lin, W.C.; Mansour, J.; Mollaee, M.; Wagner, K.U.; Koduru, P.; Yopp, A.; et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015, 6, 6744. [Google Scholar] [CrossRef]
- Park, M.; Kim, M.; Hwang, D.; Park, M.; Kim, W.K.; Kim, S.K.; Shin, J.; Park, E.S.; Kang, C.M.; Paik, Y.-K.; et al. Characterization of gene expression and activated signaling pathways in solid-pseudopapillary neoplasm of pancreas. Mod. Pathol. 2014, 27, 580–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voss, A.K.; Strasser, A. The essentials of developmental apoptosis. F1000Research 2020, 9, 148. [Google Scholar] [CrossRef]
- Chinnadurai, G.; Vijayalingam, S.; Rashmi, R. BIK, the founding member of the BH3-only family proteins: Mechanisms of cell death and role in cancer and pathogenic processes. Oncogene 2008, 27, S20–S29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; An, S.; Ro, S.H.; Teixeira, F.; Jin Park, G.; Kim, C.; Cho, C.S.; Kim, J.S.; Jakob, U.; Lee, J.H.; et al. Janus-faced Sestrin2 controls ROS and mTOR signalling through two separate functional domains. Nat. Commun. 2015, 6, 10025. [Google Scholar] [CrossRef] [Green Version]
- Ding, B.; Parmigiani, A.; Yang, C.; Budanov, A.V. Sestrin2 facilitates death receptor-induced apoptosis in lung adenocarcinoma cells through regulation of XIAP degradation. Cell Cycle 2015, 14, 3231–3241. [Google Scholar] [CrossRef] [Green Version]
- Pasha, M.; Eid, A.H.; Eid, A.A.; Gorin, Y.; Munusamy, S. Sestrin2 as a Novel Biomarker and Therapeutic Target for Various Diseases. Oxidative Med. Cell. Longev. 2017, 2017, 3296294. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Li, Y.Z.; Zhao, W.L.; Zhang, Z.Y.; Qian, X.L.; He, G.Y. STX2 drives colorectal cancer proliferation via upregulation of EXOSC4. Life Sci. 2020, 263, 118597. [Google Scholar] [CrossRef]
- Yoshino, S.; Matsui, Y.; Fukui, Y.; Seki, M.; Yamaguchi, K.; Kanamori, A.; Saitoh, Y.; Shimamura, T.; Suzuki, Y.; Furukawa, Y.; et al. EXOSC9 depletion attenuates P-body formation, stress resistance, and tumorigenicity of cancer cells. Sci. Rep. 2020, 10, 9275. [Google Scholar] [CrossRef]
- Marshansky, V.; Wang, X.; Bertrand, R.; Luo, H.; Duguid, W.; Chinnadurai, G.; Kanaan, N.; Vu, M.D.; Wu, J. Proteasomes Modulate Balance Among Proapoptotic and Antiapoptotic Bcl-2 Family Members and Compromise Functioning of the Electron Transport Chain in Leukemic Cells. J. Immunol. 2001, 166, 3130–3142. [Google Scholar] [CrossRef]
- Nikrad, M.; Johnson, T.; Puthalalath, H.; Coultas, L.; Adams, J.; Kraft, A.S. The proteasome inhibitor bortezomib sensitizes cells to killing by death receptor ligand TRAIL via BH3-only proteins Bik and Bim. Mol. Cancer Ther. 2005, 4, 443–449. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, L.; Dong, F.; Guo, W.; Wu, S.; Teraishi, F.; Davis, J.J.; Chiao, P.J.; Fang, B. Bik/NBK accumulation correlates with apoptosis-induction by bortezomib (PS-341, Velcade) and other proteasome inhibitors. Oncogene 2005, 24, 4993–4999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathai, J.P.; Germain, M.; Shore, G.C. BH3-only BIK regulates BAX,BAK-dependent release of Ca2+ from endoplasmic reticulum stores and mitochondrial apoptosis during stress-induced cell death. J. Biol. Chem. 2005, 280, 23829–23836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathai, J.P.; Germain, M.; Marcellus, R.C.; Shore, G.C. Induction and endoplasmic reticulum location of bik/nbk in response to apoptotic signaling by e1a and p53. Oncogene 2002, 21, 2534–2544. [Google Scholar] [CrossRef] [Green Version]
- Fennelly, C.; Amaravadi, R.K. Lysosomal biology in cancer. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2017; Volume 1594, pp. 293–308. [Google Scholar]
- Piao, S.; Amaravadi, R.K. Targeting the lysosome in cancer. Ann. N. Y. Acad. Sci. 2016, 1371, 45–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appelqvist, H.; Wäster, P.; Kågedal, K.; Öllinger, K. The lysosome: From waste bag to potential therapeutic target. J. Mol. Cell Biol. 2013, 5, 214–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoka, V.; Turk, V.; Turk, B. Lysosomal cysteine cathepsins: Signaling pathways in apoptosis. Biol. Chem. 2007, 388, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, S.; Repnik, U.; Boji, L.; Petelin, A.; Turk, V.; Turk, B. Chapter Nine Lysosomes in Apoptosis. Methods Enzymol. 2008, 442, 183–199. [Google Scholar]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef]
- Wolfer, A.; Ramaswamy, S. MYC and metastasis. Cancer Res. 2011, 71, 2034–2037. [Google Scholar] [CrossRef] [Green Version]
- Borgi, A.; Baldetorpm, B.; Ferno, H.; Olsson, H.; Helgi, S. c-myc Amplification is an independent prognostic factor in postmenopausal breast cancer. Int. J. Cancer 1992, 51, 687–691. [Google Scholar] [CrossRef]
- Bourhis, J.; Lê, M.G.; Barrois, M.; Gerbaulet, A.; Jeannel, D.; Duvillard, P.; Le Doussal, V.; Chassagne, D.; Riou, G. Prognostic value of c-myc proto-oncogene overexpression in early invasive carcinoma of the cervix. J. Clin. Oncol. 1990, 8, 1789–1796. [Google Scholar] [CrossRef]
- Stefanska, B.; Cheishvili, D.; Suderman, M.; Arakelian, A.; Huang, J.; Hallett, M.; Han, Z.G.; Al-Mahtab, M.; Akbar, S.M.F.; Khan, W.A.; et al. Genome-wide study of hypomethylated and induced genes in patients with liver cancer unravels novel anticancer targets. Clin. Cancer Res. 2014, 20, 3118–3132. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Dunning, M.J.; Smith, M.; Ritchie, M.; Tavaré, S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 2007, 23, 2183–2184. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taniue, K.; Tanu, T.; Shimoura, Y.; Mitsutomi, S.; Han, H.; Kakisaka, R.; Ono, Y.; Tamamura, N.; Takahashi, K.; Wada, Y.; et al. RNA Exosome Component EXOSC4 Amplified in Multiple Cancer Types Is Required for the Cancer Cell Survival. Int. J. Mol. Sci. 2022, 23, 496. https://doi.org/10.3390/ijms23010496
Taniue K, Tanu T, Shimoura Y, Mitsutomi S, Han H, Kakisaka R, Ono Y, Tamamura N, Takahashi K, Wada Y, et al. RNA Exosome Component EXOSC4 Amplified in Multiple Cancer Types Is Required for the Cancer Cell Survival. International Journal of Molecular Sciences. 2022; 23(1):496. https://doi.org/10.3390/ijms23010496
Chicago/Turabian StyleTaniue, Kenzui, Tanzina Tanu, Yuki Shimoura, Shuhei Mitsutomi, Han Han, Rika Kakisaka, Yusuke Ono, Nobue Tamamura, Kenji Takahashi, Youichiro Wada, and et al. 2022. "RNA Exosome Component EXOSC4 Amplified in Multiple Cancer Types Is Required for the Cancer Cell Survival" International Journal of Molecular Sciences 23, no. 1: 496. https://doi.org/10.3390/ijms23010496
APA StyleTaniue, K., Tanu, T., Shimoura, Y., Mitsutomi, S., Han, H., Kakisaka, R., Ono, Y., Tamamura, N., Takahashi, K., Wada, Y., Mizukami, Y., & Akimitsu, N. (2022). RNA Exosome Component EXOSC4 Amplified in Multiple Cancer Types Is Required for the Cancer Cell Survival. International Journal of Molecular Sciences, 23(1), 496. https://doi.org/10.3390/ijms23010496





