The Evolution and Functional Roles of miR408 and Its Targets in Plants
Abstract
:1. Introduction
2. Evolution Analysis of the miR408 Family in Plants
2.1. Origin and Distribution of miR408 Family Members in Plants
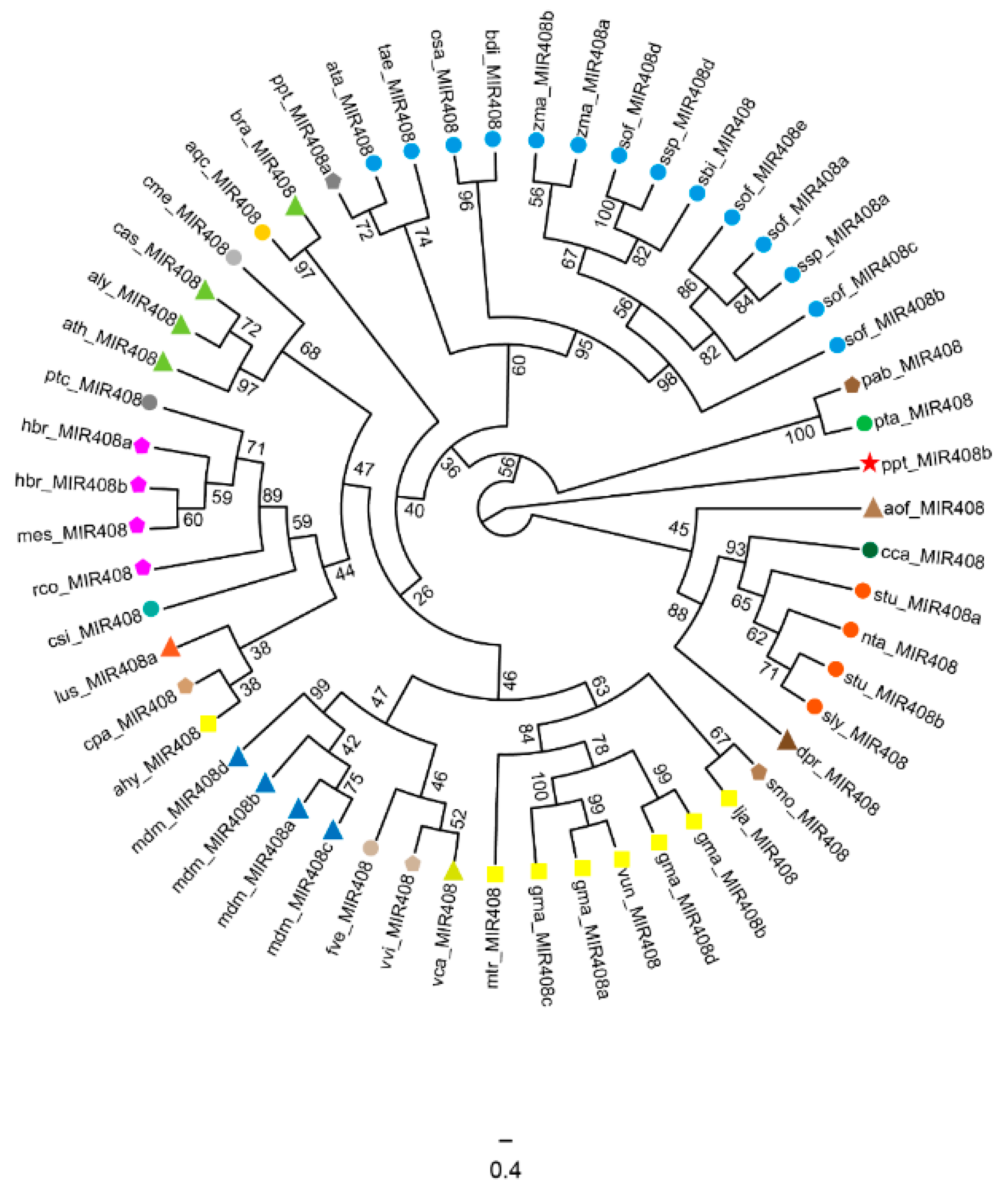
2.2. Evolution Characteristics of the miR408 Family in Plants
3. Overview of the miR408-Regulated Target Genes
3.1. Types of the miR408-Regulated Target Genes
3.2. Functions of miR408-Regulated Target Genes
3.3. Species Distribution and Splicing Sites of miR408-Regulated Target Genes
4. The Roles of miR408 and Its Targets in Plant Development
4.1. Leaf Development
4.2. Flower Development
4.3. Seed Development and Grain Yield
4.4. Seed Germination
5. Functional Roles of miR408 and Its Targets in Response to Stresses
5.1. Cold Stress
5.2. Salinity Stress
5.3. Drought Stress
5.4. Nutrient Deficiency
5.5. Other Abiotic Stresses
5.6. Biotic Stress
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu Rogers, Y.; Jia, T.; Chen, X. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 2017, 216, 1002–1017. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Wang, X.J.; Reyes, J.L.; Chua, N.H.; Gaasterland, T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004, 5, R65. [Google Scholar] [CrossRef] [Green Version]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [Green Version]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [Green Version]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [Green Version]
- Axtell, M.J.; Bartel, D.P. Antiquity of microRNAs and their targets in land plants. Plant Cell 2005, 17, 1658–1673. [Google Scholar] [CrossRef] [Green Version]
- Fattash, I.; Voss, B.; Reski, R.; Hess, W.R.; Frank, W. Evidence for the rapid expansion of microRNA-mediated regulation in early land plant evolution. BMC Plant Biol. 2007, 7, 13. [Google Scholar] [CrossRef] [Green Version]
- Alaba, S.; Piszczalka, P.; Pietrykowska, H.; Pacak, A.M.; Sierocka, I.; Nuc, P.W.; Singh, K.; Plewka, P.; Sulkowska, A.; Jarmolowski, A.; et al. The liverwort Pellia endiviifolia shares microtranscriptomic traits that are common to green algae and land plants. New Phytol. 2015, 206, 352–367. [Google Scholar] [CrossRef] [Green Version]
- Arazi, T. MicroRNAs in the moss Physcomitrella patens. Plant Mol. Biol. 2012, 80, 55–65. [Google Scholar] [CrossRef]
- Jiang, A.; Guo, Z.; Pan, J.; Yang, Y.; Zhuang, Y.; Zuo, D.; Hao, C.; Gao, Z.; Xin, P.; Chu, J.; et al. The PIF1-miR408-PLANTACY ANIN repression cascade regulates light-dependent seed germination. Plant Cell 2021, 33, 1506–1529. [Google Scholar] [CrossRef]
- Zou, H.; Guo, X.; Yang, R.; Wang, S.; Li, L.; Niu, J.; Wang, D.; Cao, X. miR408-SmLAC3 Module Participates in Salvianolic Acid B Synthesis in Salvia miltiorrhiza. Int. J. Mol. Sci. 2021, 22, 7541. [Google Scholar] [CrossRef]
- Schuetz, M.; Benske, A.; Smith, R.A.; Watanabe, Y.; Tobimatsu, Y.; Ralph, J.; Demura, T.; Ellis, B.; Samuels, A.L. Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 2014, 166, 798–807. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Deng, K.; Cheng, Y.; Zhong, Z.; Tian, L.; Tang, X.; Tang, A.; Zheng, X.; Zhang, T.; Qi, Y.; et al. CRISPR-Cas9 Based Genome Editing Reveals New Insights into MicroRNA Function and Regulation in Rice. Front. Plant. Sci. 2017, 8, 1598. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Huang, D.; Guo, Z.; Kuang, Z.; Zhang, H.; Xie, X.; Ma, Z.; Gao, S.; Lerdau, M.T.; Chu, C.; et al. Overexpression of microRNA408 enhances photosynthesis, growth, and seed yield in diverse plants. J. Integr. Plant Biol. 2018, 60, 323–340. [Google Scholar] [CrossRef]
- Song, Z.; Zhang, L.; Wang, Y.; Li, H.; Li, S.; Zhao, H.; Zhang, H. Constitutive Expression of miR408 Improves Bio Hajyzadeh ss and Seed Yield in Arabidopsis. Front. Plant Sci. 2018, 8, 2114. [Google Scholar] [CrossRef]
- Ma, C.; Burd, S.; Lers, A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef]
- Zhang, H.; Li, L. SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J. 2013, 74, 98–109. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Hong, P.; Wu, J.Y.; Chen, X.B.; Ye, X.G.; Pan, Y.Y.; Wang, J.; Zhang, X.S. The tae-miR408-Mediated Control of TaTOC1 Genes Transcription Is Required for the Regulation of Heading Time in Wheat. Plant Physiol. 2016, 170, 1578–1594. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.P.; Yu, Y.; Feng, Y.Z.; Zhou, Y.F.; Zhang, F.; Yang, Y.W.; Lei, M.Q.; Zhang, Y.C.; Chen, Y.Q. miR408 Regulates Grain Yield and Photosynthesis via a Phytocyanin Protein. Plant Physiol. 2017, 175, 1175–1185. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, Y.C.; Zhang, J.P.; Yu, Y.; Zhou, Y.F.; Feng, Y.Z.; Yang, Y.W.; Lei, M.Q.; He, H.; Lian, J.P.; et al. UCL8, a plantacyanin gene targeted by miR408, regulates fertility by controlling pollen tube germination and growth. Rice 2018, 11, 60. [Google Scholar] [CrossRef]
- Macovei, A.; Tuteja, N. Different expression of miRNAs targeting helicases in rice in response to low and high dose rate gamma-ray treatments. Plant Signal Behav. 2013, 8, e25128. [Google Scholar] [CrossRef] [Green Version]
- Liang, G.; Ai, Q.; Yu, D. Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis. Sci. Rep. 2015, 5, 11813. [Google Scholar] [CrossRef] [Green Version]
- Liu, A.; Zhou, Z.; Yi, Y.; Chen, G. Transcriptome analysis reveals the roles of stem nodes in cadmium transport to rice grain. BMC Genom. 2020, 21, 127. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.; Tiwari, M.; Lakhwani, D.; Tripathi, R.D.; Trivedi, P.K. Differential expression of microRNAs by arsenate and arsenite stress in natural accessions of rice. Metallomics 2015, 7, 174–187. [Google Scholar] [CrossRef]
- Djami-Tchatchou, A.T.; Dubery, I.A. Lipopolysaccharide perception leads to dynamic alterations in the microtranscriptome of Arabidopsis thaliana cells and leaf tissues. BMC Plant Biol. 2015, 15, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, H.; Zhang, Q.; Wang, Q.; Wang, X.; Liu, J.; Li, M.; Huang, L.; Kang, Z. Target of tae-miR408, a chemocyanin-like protein gene (TaCLP1), plays positive roles in wheat response to high-salinity, heavy cupric stress and stripe rust. Plant Mol. Biol. 2013, 83, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Nickrent, D.L.; Parkinson, C.L.; Palmer, J.D.; Duff, R.J. Multigene phylogeny of land plants with special reference to bryophytes and the earliest land plants. Mol. Biol. Evol. 2000, 17, 1885–1895. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Cruz, J.; Kressler, D.; Tollervey, D.; Linder, P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3’ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998, 17, 1128–1140. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Ghany, S.E.; Pilon, M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 2008, 283, 15932–15945. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhao, X.; Li, J.; Cai, H.; Deng, X.W.; Li, L. MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell 2014, 26, 4933–4953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunkar, R.; Girke, T.; Jain, P.K.; Zhu, J.K. Cloning and characterization of microRNAs from rice. Plant Cell 2005, 17, 1397–1411. [Google Scholar] [CrossRef] [Green Version]
- Mutum, R.D.; Balyan, S.C.; Kansal, S.; Agarwal, P.; Kumar, S.; Kumar, M.; Raghuvanshi, S. Evolution of variety-specific regulatory schema for expression of osa-miR408 in indica rice varieties under drought stress. FEBS J. 2013, 280, 1717–1730. [Google Scholar] [CrossRef] [PubMed]
- Macovei, A.; Tuteja, N. microRNAs targeting DEAD-box helicases are involved in salinity stress response in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 183. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Li, D.; Zhou, H.; Li, J.; Lu, S. Analysis of the laccase gene family and miR397-/miR408-mediated posttranscriptional regulation in Salvia miltiorrhiza. PeerJ 2019, 7, e7605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trindade, I.; Capitão, C.; Dalmay, T.; Fevereiro, M.P.; Santos, D.M. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2010, 231, 705–716. [Google Scholar] [CrossRef]
- Thatcher, S.R.; Burd, S.; Wright, C.; Lers, A.; Green, P.J. Differential expression of miRNAs and their target genes in senescing leaves and siliques: Insights from deep sequencing of small RNAs and cleaved target RNAs. Plant Cell Environ. 2015, 38, 188–200. [Google Scholar] [CrossRef]
- Kuo, Y.W.; Lin, J.S.; Li, Y.C.; Jhu, M.Y.; King, Y.C.; Jeng, S.T. MicroR408 regulates defense response upon wounding in sweet potato. J. Exp. Bot. 2019, 70, 469–483. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Mollet, J.C.; Dong, J.; Zhang, K.; Park, S.Y.; Lord, E.M. Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc. Natl. Acad. Sci. USA 2003, 100, 16125–16130. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Kim, S.T.; Lord, E.M. Plantacyanin plays a role in reproduction in Arabidopsis. Plant Physiol. 2005, 138, 778–789. [Google Scholar] [CrossRef] [Green Version]
- Khan, J.A.; Wang, Q.; Sjölund, R.D.; Schulz, A.; Thompson, G.A. An early nodulin-like protein accumulates in the sieve element plasma membrane of Arabidopsis. Plant Physiol. 2007, 143, 1576–1589. [Google Scholar] [CrossRef] [Green Version]
- Yoshizaki, M.; Furumoto, T.; Hata, S.; Shinozaki, M.; Izui, K. Characterization of a novel gene encoding a phytocyanin-related protein in morning glory (Pharbitis nil). Biochem. Biophys. Res. Commun. 2000, 268, 466–470. [Google Scholar] [CrossRef]
- Luo, S.; Hu, W.; Wang, Y.; Liu, B.; Yan, H.; Xiang, Y. Genome-wide identification, classification, and expression of phytocyanins in Populus trichocarpus trichocarpa. Planta 2018, 247, 1133–1148. [Google Scholar] [CrossRef]
- Chae, K.; Lord, E.M. Pollen tube growth and guidance: Roles of small, secreted proteins. Ann. Bot. 2011, 108, 627–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rydén, L.G.; Hunt, L.T. Evolution of protein complexity: The blue copper-containing oxidases and related proteins. J. Mol. Evol. 1993, 36, 41–66. [Google Scholar] [CrossRef] [PubMed]
- Nersissian Ma, A.M.; Immoos, C.; Hill, M.G.; Hart, P.J.; Williams, G.; Herrmann, R.G.; Valentine, J.S. Uclacyanins, stellacyanins, and plantacyanins are distinct subfamilies of phytocyanins: Plant-specific mononuclear blue copper proteins. Protein Sci. 1998, 7, 1915–1929. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Zhao, H.; Liu, Z.; Zhao, J. The phytocyanin gene family in rice (Oryza sativa L.): Genome-wide identification, classification and transcriptional analysis. PLoS ONE 2011, 6, e25184. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Świderska-Burek, U.; Polak, J.; Sulej, J.; Jarosz-Wilkołazka, A.; Paszczyński, A. Laccase Properties, Physiological Functions, and Evolution. Int. J. Mol. Sci. 2020, 21, 966. [Google Scholar] [CrossRef] [Green Version]
- Ranocha, P.; Chabannes, M.; Chamayou, S.; Danoun, S.; Jauneau, A.; Boudet, A.M.; Goffner, D. Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol. 2002, 129, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Sterjiades, R.; Dean, J.F.; Eriksson, K.E. Laccase from Sycamore Maple (Acer pseudoplatanus) Polymerizes Monolignols. Plant Physiol. 1992, 99, 1162–1168. [Google Scholar] [CrossRef] [Green Version]
- Liang, M.; Davis, E.; Gardner, D.; Cai, X.; Wu, Y. Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 2006, 224, 1185–1196. [Google Scholar] [CrossRef]
- Hoopes, J.T.; Dean, J.F. Ferroxidase activity in a laccase-like multicopper oxidase from Liriodendron tulipifera. Plant Physiol Biochem. 2004, 42, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Hippler, M.; Reichert, J.; Sutter, M.; Zak, E.; Altschmied, L.; Schröer, U.; Herrmann, R.G.; Haehnel, W. The plastocyanin binding domain of photosystem I. EMBO J. 1996, 15, 6374–6384. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Davidson, V.L. Cupredoxins--a study of how proteins may evolve to use metals for bioenergetic processes. Metallomics 2011, 3, 140–151. [Google Scholar] [CrossRef]
- Alabadí, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Más, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Persson, S.; Caffall, K.H.; Freshour, G.; Hilley, M.T.; Bauer, S.; Poindexter, P.; Hahn, M.G.; Mohnen, D.; Somerville, C. The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 2007, 19, 237–255. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Ji, X.; Nie, X.; Qu, M.; Zheng, L.; Tan, Z.; Zhao, H.; Huo, L.; Liu, S.; Zhang, B.; et al. Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 2015, 207, 692–709. [Google Scholar] [CrossRef]
- Thiebaut, F.; Rojas, C.A.; Grativol, C.; Calixto, E.P.D.R.; Motta, M.R.; Ballesteros, H.G.F.; Peixoto, B.; de Lima, B.N.S.; Vieira, L.M.; Walter, M.E.; et al. Roles of Non-Coding RNA in Sugarcane-Microbe Interaction. Noncoding RNA 2017, 3, 25. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Chen, X.; Chen, Y.; Zhang, Q.; Su, L.; Chen, X.; Chen, Y.; Zhang, Z.; Lin, Y.; Lai, Z. Genome-wide identification of miRNAs and their targets during early somatic embryogenesis in Dimocarpus longan Lour. Sci. Rep. 2020, 10, 4626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burkhead, J.L.; Reynolds, K.A.; Abdel-Ghany, S.E.; Cohu, C.M.; Pilon, M. Copper homeostasis. New Phytol. 2009, 182, 799–816. [Google Scholar] [CrossRef]
- Agarwal, P.; Reddy, M.P.; Chikara, J. WRKY: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 2011, 38, 3883–3896. [Google Scholar] [CrossRef]
- Esmaeiliz, F.; Shiran, B.; Fallahi, H.; Mirakhorli, N.; Budak, H.; Martínez-Gómez, P. In silico search and biological validation of microRNAs related to drought response in peach and almond. Funct. Integr. Genomics 2017, 17, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Ghorecha, V.; Zheng, Y.; Liu, L.; Sunkar, R.; Krishnayya, N.S.R. MicroRNA dynamics in a wild and cultivated species of Convolvulaceae exposed to drought stress. Physiol. Mol. Biol. Plants 2017, 23, 291–300. [Google Scholar] [CrossRef] [Green Version]
- Trindade, V.; Patel, K.; Ingle, S.; Sunkar, R.; Krishnayya, N.S. Analysis of biochemical variations and microRNA expression in wild (Ipomoea campanulata) and cultivated (Jacquemontia pentantha) species exposed to in vivo water stress. Physiol. Mol. Biol. Plants 2014, 20, 57–67. [Google Scholar] [CrossRef] [Green Version]
- Balyan, S.; Kumar, M.; Mutum, R.D.; Raghuvanshi, U.; Agarwal, P.; Mathur, S.; Raghuvanshi, S. Identification of miRNA-mediated drought responsive multi-tiered regulatory network in drought tolerant rice, Nagina 22. Sci. Rep. 2017, 7, 15446. [Google Scholar] [CrossRef]
- Akdogan, G.; Tufekci, E.D.; Uranbey, S.; Unver, T. miRNA-based drought regulation in wheat. Funct. Integr. Genom. 2016, 16, 221–233. [Google Scholar] [CrossRef]
- Candar-Cakir, B.; Arican, E.; Zhang, B. Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol. J. 2016, 14, 1727–1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, M.; Yang, J.; Cai, X.; Shen, Y.; Cui, N.; Zhu, Y.; Jia, B.; Sun, X. The opposite roles of OsmiR408 in cold and drought stress responses in Oryza sativa. Mol. Breed. 2018, 38, 120. [Google Scholar] [CrossRef]
- Hajyzadeh, M.; Turktas, M.; Khawar, K.M.; Unver, T. miR408 overexpression causes increased drought tolerance in chickpea. Gene 2015, 555, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Ž.; Stanisavljević, N.; Mikić, A.; Radović, S.; Maksimović, V. Water deficit down-regulates miR398 and miR408 in pea (Pisum sativum L.). Plant Physiol. Biochem. 2014, 83, 26–31. [Google Scholar] [CrossRef]
- Kantar, M.; Unver, T.; Budak, H. Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct. Integr. Genom. 2010, 10, 493–507. [Google Scholar] [CrossRef]
- Guo, X.; Niu, J.; Cao, X. Heterologous Expression of Salvia miltiorrhiza MicroRNA408 Enhances Tolerance to Salt Stress in Nicotiana benthamiana. Int. J. Mol. Sci. 2018, 19, 3985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Zhong, S.; Li, X.; Li, W.; Rothstein, S.J.; Zhang, S.; Bi, Y.; Xie, C. Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS ONE 2011, 6, e28009. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Wang, Z.; Yang, C.; Yang, Z.; Li, H.; Wu, Y. Physiological responses and small RNAs changes in maize under nitrogen deficiency and resupply. Genes Genom. 2019, 41, 1183–1194. [Google Scholar] [CrossRef]
- Liang, G.; He, H.; Yu, D. Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS ONE 2012, 7, e48951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.B.; Qi, Y.P.; Yang, L.T.; Guo, P.; Li, Y.; Chen, L.S. Boron-deficiency-responsive microRNAs and their targets in Citrus sinensis leaves. BMC Plant Biol. 2015, 15, 271. [Google Scholar] [CrossRef] [Green Version]
- Melnikova, N.V.; Dmitriev, A.A.; Belenikin, M.S.; Speranskaya, A.S.; Krinitsina, A.A.; Rachinskaia, O.A.; Lakunina, V.A.; Krasnov, G.S.; Snezhkina, A.V.; Sadritdinova, A.F. Excess fertilizer responsive miRNAs revealed in Linum usitatissimum L. Biochimie 2015, 109, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Pant, B.D.; Musialak-Lange, M.; Nuc, P.; May, P.; Buhtz, A.; Kehr, J.; Walther, D.; Scheible, W.R. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 2009, 150, 1541–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Liu, Q.; Chen, L.; Kuang, J.; Walk, T.; Wang, J.; Liao, H. Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genom. 2013, 14, 66. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Liu, X.; Guo, C.; Gu, J.; Xiao, K. Identification and characterization of microRNAs from wheat (Triticum aestivum L.) under phosphorus deprivation. J. Plant Biochem. Biotechnol. 2013, 22, 113–123. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Shi, D.; Liu, X.; Qin, J.; Ge, Q.; Xu, L.; Pan, X.; Li, W.; Zhu, Y.; et al. Spatial-temporal analysis of zinc homeostasis reveals the response mechanisms to acute zinc deficiency in Sorghum bicolor. New Phytol. 2013, 200, 1102–1115. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, K.; Liu, G.; Li, S.; Zhao, S.; Liu, X.; Yang, X.; Xiao, K. Global identification and characterization of miRNA family members responsive to potassium deprivation in wheat (Triticum aestivum L.). Sci. Rep. 2020, 10, 15812. [Google Scholar] [CrossRef]
- Qiu, Z.; Hai, B.; Guo, J.; Li, Y.; Zhang, L. Characterization of wheat miRNAs and their target genes responsive to cadmium stress. Plant Biochem. 2016, 101, 60–67. [Google Scholar] [CrossRef] [Green Version]
- Valdés-López, O.; Yang, S.S.; Aparicio-Fabre, R.; Graham, P.H.; Reyes, J.L.; Vance, C.P.; Hernández, G. MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol. 2010, 187, 805–818. [Google Scholar] [CrossRef]
- Yin, J.; Wang, G.; Xiao, J.; Ma, F.; Zhang, H.; Sun, Y.; Diao, Y.; Huang, J.; Guo, Q.; Liu, D. Identification of genes involved in stem rust resistance from wheat mutant D51 with the cDNA-AFLP technique. Mol. Biol. Rep. 2010, 37, 1111–1117. [Google Scholar] [CrossRef]
- Liu, W.; Liu, J.; Triplett, L.; Leach, J.E.; Wang, G.L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014, 52, 213–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hang, N.; Shi, T.; Liu, Y.; Ye, W.; Taier, G.; Sun, Y.; Wang, K.; Zhang, W. Overexpression of Os-microRNA408 enhances drought tolerance in perennial ryegrass. Physiol. Plant 2021, 172, 733–747. [Google Scholar] [CrossRef]
- Gupta, O.P.; Permar, V.; Koundal, V.; Singh, U.D.; Praveen, S. MicroRNA regulated defense responses in Triticum aestivum L. during Puccinia graminis f.sp. tritici infection. Mol. Biol. Rep. 2012, 39, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Megha, S.; Basu, U.; Kav, N.N.V. Regulation of low temperature stress in plants by microRNAs. Plant Cell Environ. 2018, 41, 1–15. [Google Scholar] [CrossRef]
- Ji, H.; Wang, Y.; Cloix, C.; Li, K.; Jenkins, G.I.; Wang, S.; Shang, Z.; Shi, Y.; Yang, S.; Li, X. The Arabidopsis RCC1 Family Protein TCF1 Regulates Freezing Tolerance and Cold Acclimation through Modulating Lignin Biosynthesis. PLoS Genet. 2015, 11, e1005471. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Jin, D.; Wang, Y.; Zhao, Y.; Chen, M. MicroRNAs and their cross-talks in plant development. J. Genet. Genom. 2013, 40, 161–170. [Google Scholar] [CrossRef]
- Nath, M.; Tuteja, N. NPKS uptake, sensing, and signaling and miRNAs in plant nutrient stress. Protoplasma 2016, 253, 767–786. [Google Scholar] [CrossRef]
- Li, S.; Castillo-González, C.; Yu, B.; Zhang, X. The functions of plant small RNAs in development and in stress responses. Plant J. 2017, 90, 654–670. [Google Scholar] [CrossRef] [Green Version]
- Noman, A.; Aqeel, M. miRNA-based heavy metal homeostasis and plant growth. Environ. Sci. Pollut. Res. Int. 2017, 24, 10068–10082. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.; Fu, C.; Yun, J.; Shao, H.; Wang, X.; Wang, Z.Y.; Dixon, R.A. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 2013, 25, 3976–3987. [Google Scholar] [CrossRef] [Green Version]
- Carrió-Seguí, À.; Ruiz-Rivero, O.; Villamayor-Belinchón, L.; Puig, S.; Perea-García, A.; Peñarrubia, L. The altered expression of microRNA408 influences the Arabidopsis response to iron deficiency. Front. Plant Sci. 2019, 10, 324. [Google Scholar] [CrossRef] [PubMed]
- Talesh Sasani, S.M.; Soltani, B.; Mehrabi, R.; Fereidoun Padasht-Dehkaei, H.S. Expression Alteration of Candidate Rice MiRNAs in Response to Sheath Blight Disease. Iran J. Biotechnol. 2020, 18, e2451. [Google Scholar] [CrossRef] [PubMed]
- Peñarrubia, L.; Romero, P.; Carrió-Seguí, A.; Andrés-Bordería, A.; Moreno, J.; Sanz, A. Temporal aspects of copper homeostasis and its crosstalk with hormones. Front. Plant Sci. 2015, 6, 255. [Google Scholar] [CrossRef]




| Stresses | Species | miRNA | Target Genes | References |
|---|---|---|---|---|
| ABIOTIC | ||||
| Mild drought | Prunus persica | miR408 | Plastocyanin, | [68] |
| Copper ion | ||||
| binding protein | ||||
| Prunus dulcis | miR408 | No | ||
| Severe drought | Prunus persica | miR408 | Plantacyanin | |
| Prunus dulcis | miR408 | No | ||
| Interspecific Prunus persica–Armeniaca Prunus dulcis | miR408 | No | ||
| Drought | Convolvulacee | miR408 | No | [69] |
| (tolerant wild Ipomoea | ||||
| campanulata) | ||||
| Convolvulacee | miR408 | No | ||
| (sensitive Cultivated | ||||
| Jacquemontia Pentantha) | ||||
| Convolvulacee | miR408 | No | [70] | |
| (tolerant wild Ipomoea | ||||
| campanulata) | ||||
| Convolvulacee | miR408 | No | ||
| (sensitive Cultivated | ||||
| Jacquemontia Pentantha) | ||||
| Oryza sativa | miR408-3p | Plantacyanin, | [39,71] | |
| (tolerant Cultivars N22, dana) | plastocyanin-like | |||
| domain, | ||||
| containing | ||||
| proteins | ||||
| Oryza sativa | miR408-3p | No | ||
| (sensitive cultivars PB1, IR64) | ||||
| Triticum aestivum | miR408 | No | [72] | |
| (root) | ||||
| miR408 | No | |||
| (leaf) | ||||
| Lycopersicon esculentum | miR408a-3p | No | [73] | |
| (sensitive) | ||||
| L.esculentum var. | miR408 | No | ||
| Cerasiforme (tolerant) | ||||
| Pisum sativum | miR408 | No | [72] | |
| Oryza sativa | miR408 | plastocyanin genes (Os01g53880 and Os09g29390) | [74] | |
| Arabidopsis thaliana | miR408 | Cupredoxin | [22] | |
| Plantacyanin | ||||
| LAC3 | ||||
| Cicer arietinum | miR408 | No | [75] | |
| Water | Ipomoea campanulate | miR408 | Plantacyanin | [70] |
| deficit | (tolerate) | |||
| Jacquemontia Pentantha | miR408 | |||
| (sensitive) | ||||
| Pisum sativum | miR408 | P1B-ATPase | [76] | |
| Medicago truncatula | miR408 | Plantacyanin | [42] | |
| Dehydration | Hordeum vulgare | miR408 | Plantacyanin | [77] |
| Salinity | Oryza sativa | miR408 | DSHCT | [40] |
| Plastocyanin-like | ||||
| Salvia miltiorrhiza | miR408 | No | [78] | |
| Arabidopsis thaliana | miR408 | Cupredoxin | [22] | |
| Plantacyanin | ||||
| Uclacyanin | ||||
| LAC3 | ||||
| Cold | miR408 | Cupredoxin | ||
| Plantacyanin | ||||
| Uclacyanin | ||||
| LAC3 | ||||
| Osmotic | miR408 | Cupredoxin | ||
| Plantacyanin | ||||
| Uclacyanin | ||||
| LAC3 | ||||
| Oxidative | miR408 | Cupredoxin | ||
| Plantacyanin | ||||
| Uclacyanin | ||||
| LAC3 | ||||
| Low dose | Oryza sativa | miR408 | DSHCT | [27] |
| rate γ-ray | ||||
| High dose | miR408 | |||
| rate γ-ray | ||||
| Nutrient deprivation | ||||
| Zea mays | miR408 | Cupredoxin | [79,80] | |
| Nitrogen | SOD1A | |||
| deficiency | Arabidopsis thaliana | miR408 | Laccase | [81] |
| Plantacyanin | ||||
| Carbon, nitrogen, | miR408 | LAC3 | [28] | |
| and sulfur | LAC13 | |||
| deficiency | ||||
| Copper deficiency | miR408 | LAC3 | [22,36] | |
| LAC12 | ||||
| LAC13 | ||||
| Plantacyanin | ||||
| Iron deficiency | miR408 | LAC3 | [18] | |
| LAC12 | ||||
| LAC13 | ||||
| Plantacyanin | ||||
| Boron deficiency | miR408 | Plantacyanin | [82] | |
| LAC3 | ||||
| LAC13 | ||||
| Cu/Zn SODs | ||||
| (CSDs) | ||||
| Excess fertilizer | Linum usitatissimum | miR408 | No | [83] |
| Phosphorus deficiency | Arabidopsis thaliana | miR408 | No | [84] |
| Glycine max | miR408 | No | [85] | |
| Triticum aestivum | miR408 | No | [86] | |
| Zinc deficiency | Sorghum bicolor | miR408 | Plantacyanin | [87] |
| Potassium | Triticum aestivum | miR408 | No | [88] |
| deficiency | ||||
| Heavy metals | ||||
| Cadmium | Triticum aestivum | miR408 (12h, leaves) | Chemocyanin-like protein | [89] |
| miR408 (24h, leaves) | ||||
| miR408 (6, 12, 24 and 48h, roots) | ||||
| Oryza sativa | miR408 | bZIP, ERF, | [29] | |
| MYB, SnRK1 | ||||
| and HSPs | ||||
| Arsenate and | Oryza sativa | miR408 | No | [30] |
| arsenite | ||||
| Manganese | Phaseolus vulgaris | miR408 | No | [90] |
| BIOTIC | Triticum aestivum | miR408 | Plantacyanin | [91] |
| Puccinia graminis | ||||
| f.sp. tritici | ||||
| Rhizoctonia solani | Oryza sativa | miR408 | No | [92] |
| Lipopolysaccharide | Arabidopsis thaliana | miR408 | Plantacyanin | [31] |
| Stresses | Species | Approach | Phenotype | References |
|---|---|---|---|---|
| Cold | Arabidopsis thaliana | miR408 overexpression | More tolerant to cold tolerant, | [22] |
| Lower electrolyte leakage, | ||||
| Higher Fv/Fm value, | ||||
| Lower MDA, | ||||
| Higher chlorophyll | ||||
| T-DNA miR408 mutant | Enhanced cold sensitivity, | |||
| Enhanced electrolyte leakage, | ||||
| Lower Fv/Fmvalue, | ||||
| MDA were elevated, | ||||
| Oryza sativa | miR408 overexpression | Lower chlorophyll | [74] | |
| longer shoots and roots, | ||||
| Lower ion leakage, | ||||
| Enhanced SOD activity, | ||||
| Enhanced proline content | ||||
| Salinity | Arabidopsis thaliana | miR408 overexpression | Root development was better, | [22] |
| Lower ROS | ||||
| T-DNA miR408 mutant | Inhibited root development, | |||
| Enhanced ROS | ||||
| Triticum aestivum | TaCLP1 overexpression | In yeast enhances cell tolerance | [32] | |
| Salvia miltiorrhiza | miR408 overexpression | Improved root growth, | [78] | |
| Significantly higher fresh weights, | ||||
| Higher germination rates, | ||||
| Lower growth inhibition, | ||||
| Reduced ROS Accumulation, | ||||
| Lower accumulations of H2O2, | ||||
| Higher POD, SOD, CAT activities, | ||||
| Lower levels of O2−and H2O2 | ||||
| Oxidative | Arabidopsis thaliana | miR408 overexpression | More tolerant to oxidative stress, | [22] |
| Higher biomass, | ||||
| Higher total root length, | ||||
| Increased CSD1, CSD2, | ||||
| CCS1, GST-2U25 and SAP12 | ||||
| T-DNA miR408 mutant | Lower total root length, | |||
| CSD1, CSD2, CCS1, | ||||
| GST-U25 and SAP12 reduced | ||||
| Drought | Arabidopsis thaliana | miR408 overexpression | Retarded growth, | [22] |
| Lower FW, | ||||
| Death rate higher | ||||
| T-DNA miR408 mutant | Grew better, | |||
| Higher height, | ||||
| Death rate lower | ||||
| Cicer arietinum | miR408 overexpression | High stress tolerance, | [75] | |
| Lower height, | ||||
| Increasing number of leaves, | ||||
| BHLH23 down-regulated, | ||||
| ERF/AP reduced expression, | ||||
| DREB2A/1A genes were increased, | ||||
| Rd17 and Rd29a were increased, | ||||
| Rd22 up-regulated | ||||
| Lolium perenne | miR408 overexpression | Narrower leaves of similar length, | [93] | |
| Less vein number, | ||||
| More closely folded leaves, | ||||
| Greener, | ||||
| Higher chlorophyll, | ||||
| More bristle-like trichomes on the | ||||
| leaf surface, | ||||
| Relatively smaller and more sunken | ||||
| stomata, | ||||
| Lower stomatal conductance, | ||||
| Less tissue damage, | ||||
| Higher leaf RWC, | ||||
| Lower water loss rate, | ||||
| Higher leaf electrolyte leakage (EL), | ||||
| Higher activities of SOD, CAT and | ||||
| POD, | ||||
| Lower accumulation of H2O2 and | ||||
| MDA | ||||
| Osmotic | Arabidopsis thaliana | miR408 overexpression | Retarded growth, | [22] |
| Lower FW | ||||
| T-DNA miR408 mutant | Grow better, | |||
| Higher height | ||||
| Copper deficiency | Triticum aestivum | TaCLP1 overexpression | Higher tolerance to copper deficiency | [32] |
| Iron deficiency | Arabidopsis thaliana | T-DNA miR408 mutant | Lower chlorophyll-a content, | [18] |
| Lower lignin content, | ||||
| LAC3, LAC12, LAC13 and | ||||
| plantacyanin (ARPN) up-regulated, | ||||
| Lower lignification-related genes (F6’H1’, CCR1, B-GLU23, LAC17), | ||||
| Lower bHLH39, | ||||
| Higher phenoloxidase activity, | ||||
| Higher H2O2 levels, | ||||
| Lower MCO3 and CAT2 | ||||
| miR408 overexpression | Lower chlorophyll-a content, | |||
| Lower lignin content, | ||||
| LAC3, LAC12, LAC13 and | ||||
| plantacyanin (ARPN) down-regulated, | ||||
| lignification-related genes (F6’H1’, | ||||
| B-GLU23, LAC17) significantly | ||||
| increased, | ||||
| Lower FIT, | ||||
| Lower H2O2 levels, | ||||
| lower MCO3 and CAT2 | ||||
| Wild type | Higher Copper levels, | |||
| miR408 expression decreased, | ||||
| LAC3, LAC12, LAC13 and | ||||
| plantacyanin (ARPN) up-regulated, | ||||
| Lower FRO2, FRO3, IRT1, COPT2, | ||||
| Lower phenoloxidase activity, | ||||
| Lower ferroxidase activity, | ||||
| Higher H2O2 levels, | ||||
| Higher lignin staining of vascular | ||||
| cylinder, | ||||
| Lignification of the vascular bundles | ||||
| was more evident in the aerial part | ||||
| Puccinia striiformis f. sp. tritici | Triticum | RNAi TaCLP1 mutant | Decreased stripe rust resistance | [94] |
| aestivum |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Feng, B.; Gao, C.; Zhang, H.; Wen, F.; Tao, L.; Fu, G.; Xiong, J. The Evolution and Functional Roles of miR408 and Its Targets in Plants. Int. J. Mol. Sci. 2022, 23, 530. https://doi.org/10.3390/ijms23010530
Gao Y, Feng B, Gao C, Zhang H, Wen F, Tao L, Fu G, Xiong J. The Evolution and Functional Roles of miR408 and Its Targets in Plants. International Journal of Molecular Sciences. 2022; 23(1):530. https://doi.org/10.3390/ijms23010530
Chicago/Turabian StyleGao, Yu, Baohua Feng, Caixia Gao, Huiquan Zhang, Fengting Wen, Longxing Tao, Guanfu Fu, and Jie Xiong. 2022. "The Evolution and Functional Roles of miR408 and Its Targets in Plants" International Journal of Molecular Sciences 23, no. 1: 530. https://doi.org/10.3390/ijms23010530
APA StyleGao, Y., Feng, B., Gao, C., Zhang, H., Wen, F., Tao, L., Fu, G., & Xiong, J. (2022). The Evolution and Functional Roles of miR408 and Its Targets in Plants. International Journal of Molecular Sciences, 23(1), 530. https://doi.org/10.3390/ijms23010530







