Identification of Novel Micropeptides Derived from Hepatocellular Carcinoma-Specific Long Noncoding RNA
Abstract
1. Introduction
2. Results
2.1. Identification of Hepatocellular Carcinoma-Specific lncRNAs
2.2. Identification of Micropeptide Candidates Derived from HCC-Specific lncRNAs
2.3. NONHSAT013026.2/Linc013026-68AA Is Translated into a 68 Amino Acid Long Micropeptide
2.4. Linc013026-68AA Enhances Cell Proliferation
3. Discussion
4. Materials and Methods
4.1. Cell Culture, siRNA, and Transfection
4.2. Peptide-Specific Antibodies
4.3. Wst-1 Assay
4.4. Crystal Violet Assay
4.5. Immunohistochemistry/Immunofluorescence
4.6. In Vitro Transcription/Translation
4.7. mRNA Export Assay
4.8. Polysome Profiling
4.9. Immunoblotting Procedures
4.10. Semi-Quantitative RT-PCR and qRT-PCR Analysis
4.11. Statistical Analysis
4.12. RNA Sequencing Data Analysis
4.13. GST Pull-Down Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, S.; Marais, R.; Zhu, A.X. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 2010, 29, 4989–5005. [Google Scholar] [CrossRef]
- Allaire, M.; Nault, J.C. Type 2 diabetes-associated hepatocellular carcinoma: A molecular profile. Clin. Liver Dis. 2016, 8, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Schulze, K.; Imbeaud, S.; Letouze, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Saran, S.; Tran, D.D.; Ewald, F.; Koch, A.; Hoffmann, A.; Koch, M.; Nashan, B.; Tamura, T. Depletion of three combined THOC5 mRNA export protein target genes synergistically induces human hepatocellular carcinoma cell death. Oncogene 2016, 35, 3872–3879. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.D.; Saran, S.; Koch, A.; Tamura, T. mRNA export protein THOC5 as a tool for identification of target genes for cancer therapy. Cancer Lett. 2016, 373, 222–226. [Google Scholar] [CrossRef]
- Niehus, S.E.; Allister, A.B.; Hoffmann, A.; Wiehlmann, L.; Tamura, T.; Tran, D.D.H. Myc/Max dependent intronic long antisense noncoding RNA, EVA1A-AS, suppresses the expression of Myc/Max dependent anti-proliferating gene EVA1A in a U2 dependent manner. Sci. Rep. 2019, 9, 17319. [Google Scholar] [CrossRef]
- Tran, D.D.H.; Kessler, C.; Niehus, S.E.; Mahnkopf, M.; Koch, A.; Tamura, T. Myc target gene, long intergenic noncoding RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and cell survival by titrating tumor suppressor microRNAs. Oncogene 2018, 37, 75–85. [Google Scholar] [CrossRef]
- Orr, M.W.; Mao, Y.; Storz, G.; Qian, S.B. Alternative ORFs and small ORFs: Shedding light on the dark proteome. Nucleic Acids Res. 2020, 48, 1029–1042. [Google Scholar] [CrossRef]
- Ragan, C.; Goodall, G.J.; Shirokikh, N.E.; Preiss, T. Insights into the biogenesis and potential functions of exonic circular RNA. Sci. Rep. 2019, 9, 2048. [Google Scholar] [CrossRef]
- Burbano De Lara, S.; Tran, D.D.H.; Allister, A.B.; Polenkowski, M.; Nashan, B.; Koch, M.; Tamura, T. C20orf204, a hepatocellular carcinoma-specific protein interacts with nucleolin and promotes cell proliferation. Oncogenesis 2021, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Li, Z.; Ma, M.; Wang, Z.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. LncRNA-Six1 Encodes a Micropeptide to Activate Six1 in Cis and Is Involved in Cell Proliferation and Muscle Growth. Front. Physiol. 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.A.; Hitz, B.C.; Sloan, C.A.; Chan, E.T.; Davidson, J.M.; Gabdank, I.; Hilton, J.A.; Jain, K.; Baymuradov, U.K.; Narayanan, A.K.; et al. The Encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res. 2018, 46, D794–D801. [Google Scholar] [CrossRef]
- Liu, C.; Bai, B.; Skogerbo, G.; Cai, L.; Deng, W.; Zhang, Y.; Bu, D.; Zhao, Y.; Chen, R. NONCODE: An integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005, 33, D112–D115. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Hishiki, T.; Kawamoto, S.; Morishita, S.; Okubo, K. BodyMap: A human and mouse gene expression database. Nucleic Acids Res. 2000, 28, 136–138. [Google Scholar] [CrossRef][Green Version]
- Consortium, E.P.; Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigo, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef]
- Consortium, E.P.; Moore, J.E.; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A.; et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef]
- Sonawane, A.R.; Platig, J.; Fagny, M.; Chen, C.Y.; Paulson, J.N.; Lopes-Ramos, C.M.; DeMeo, D.L.; Quackenbush, J.; Glass, K.; Kuijjer, M.L. Understanding Tissue-Specific Gene Regulation. Cell Rep. 2017, 21, 1077–1088. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Gentile, F.; Amodeo, P.; Febbraio, F.; Picaro, F.; Motta, A.; Formisano, S.; Nucci, R. SDS-resistant active and thermostable dimers are obtained from the dissociation of homotetrameric beta-glycosidase from hyperthermophilic Sulfolobus solfataricus in SDS. Stabilizing role of the A-C intermonomeric interface. J. Biol. Chem. 2002, 277, 44050–44060. [Google Scholar] [CrossRef] [PubMed]
- Lundby, A.; Lage, K.; Weinert, B.T.; Bekker-Jensen, D.B.; Secher, A.; Skovgaard, T.; Kelstrup, C.D.; Dmytriyev, A.; Choudhary, C.; Lundby, C.; et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012, 2, 419–431. [Google Scholar] [CrossRef]
- Matsumoto, A.; Pasut, A.; Matsumoto, M.; Yamashita, R.; Fung, J.; Monteleone, E.; Saghatelian, A.; Nakayama, K.I.; Clohessy, J.G.; Pandolfi, P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017, 541, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.S.; Jadiya, P.; Zhang, X.; McLendon, J.M.; Abouassaly, G.M.; Witmer, N.H.; Anderson, E.J.; Elrod, J.W.; Boudreau, R.L. Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep. 2018, 23, 3710–3720.e8. [Google Scholar] [CrossRef]
- Davenport, C.F.; Scheithauer, T.; Dunst, A.; Bahr, F.S.; Dorda, M.; Wiehlmann, L.; Tran, D.D.H. Genome-Wide Methylation Mapping Using Nanopore Sequencing Technology Identifies Novel Tumor Suppressor Genes in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 3937. [Google Scholar] [CrossRef] [PubMed]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. Gencode 2021. Nucleic Acids Res. 2021, 49, D916–D923. [Google Scholar] [CrossRef]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigo, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Tran, D.D.; Saran, S.; Dittrich-Breiholz, O.; Williamson, A.J.; Klebba-Farber, S.; Koch, A.; Kracht, M.; Whetton, A.D.; Tamura, T. Transcriptional regulation of immediate-early gene response by THOC5, a member of mRNA export complex, contributes to the M-CSF-induced macrophage differentiation. Cell Death Dis. 2013, 4, e879. [Google Scholar] [CrossRef]
- Guria, A.; Tran, D.D.; Ramachandran, S.; Koch, A.; El Bounkari, O.; Dutta, P.; Hauser, H.; Tamura, T. Identification of mRNAs that are spliced but not exported to the cytoplasm in the absence of THOC5 in mouse embryo fibroblasts. RNA 2011, 17, 1048–1056. [Google Scholar] [CrossRef]
- Tran, D.D.; Saran, S.; Williamson, A.J.; Pierce, A.; Dittrich-Breiholz, O.; Wiehlmann, L.; Koch, A.; Whetton, A.D.; Tamura, T. THOC5 controls 3'end-processing of immediate early genes via interaction with polyadenylation specific factor 100 (CPSF100). Nucleic Acids Res. 2014, 42, 12249–12260. [Google Scholar] [CrossRef] [PubMed]
- Dhamija, S.; Doerrie, A.; Winzen, R.; Dittrich-Breiholz, O.; Taghipour, A.; Kuehne, N.; Kracht, M.; Holtmann, H. IL-1-induced post-transcriptional mechanisms target overlapping translational silencing and destabilizing elements in IkappaBzeta mRNA. J. Biol. Chem. 2010, 285, 29165–29178. [Google Scholar] [CrossRef] [PubMed]
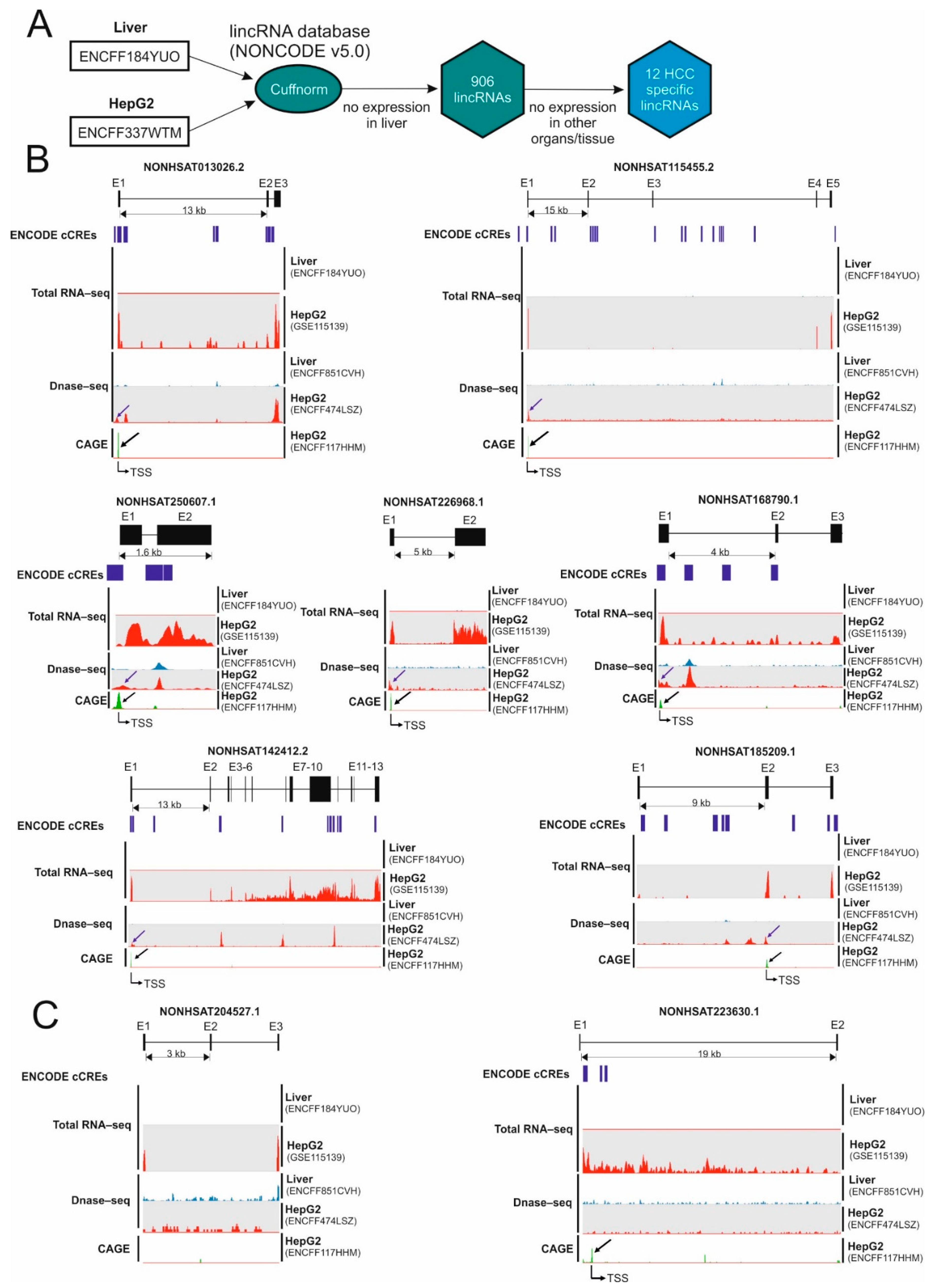
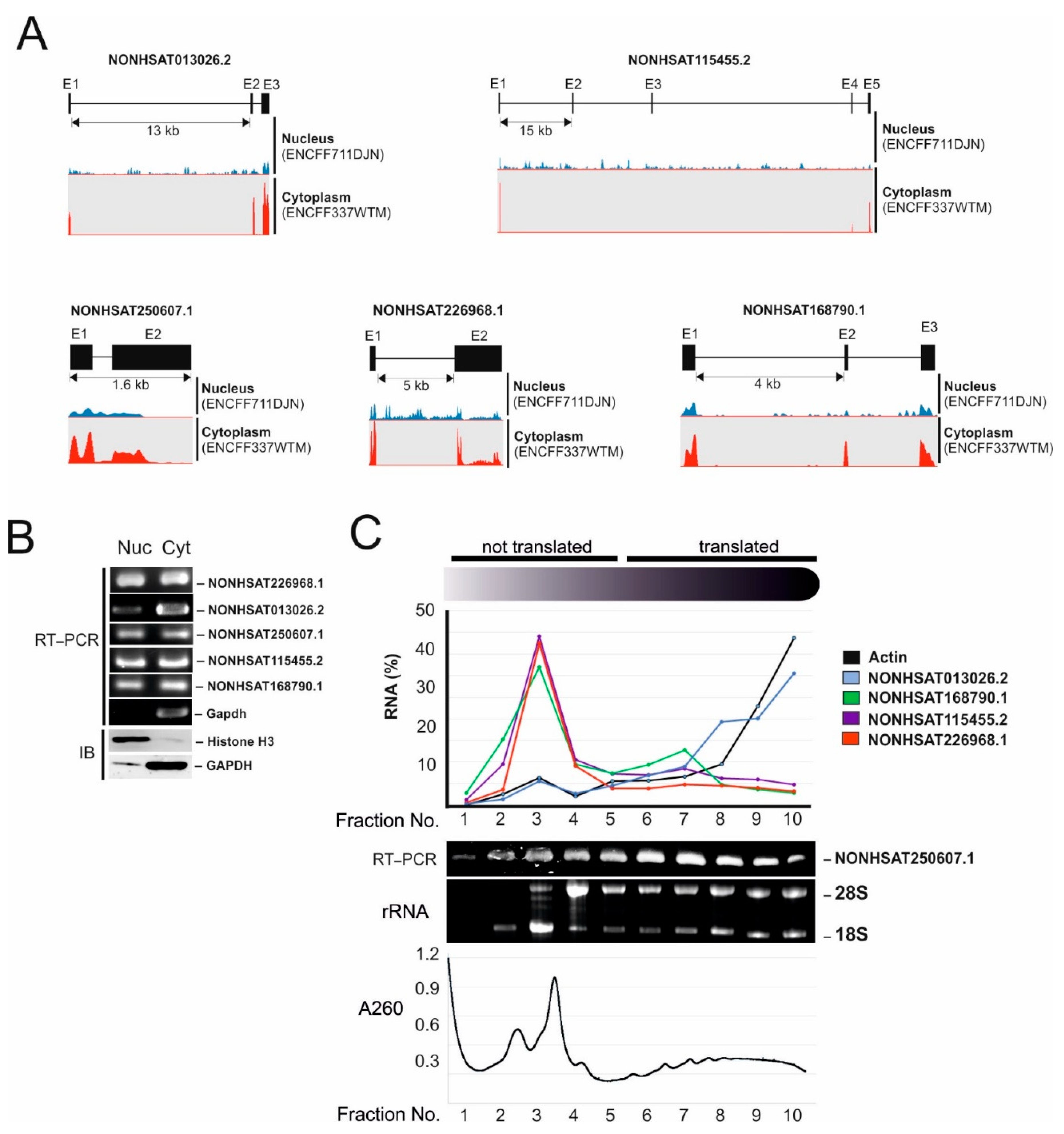
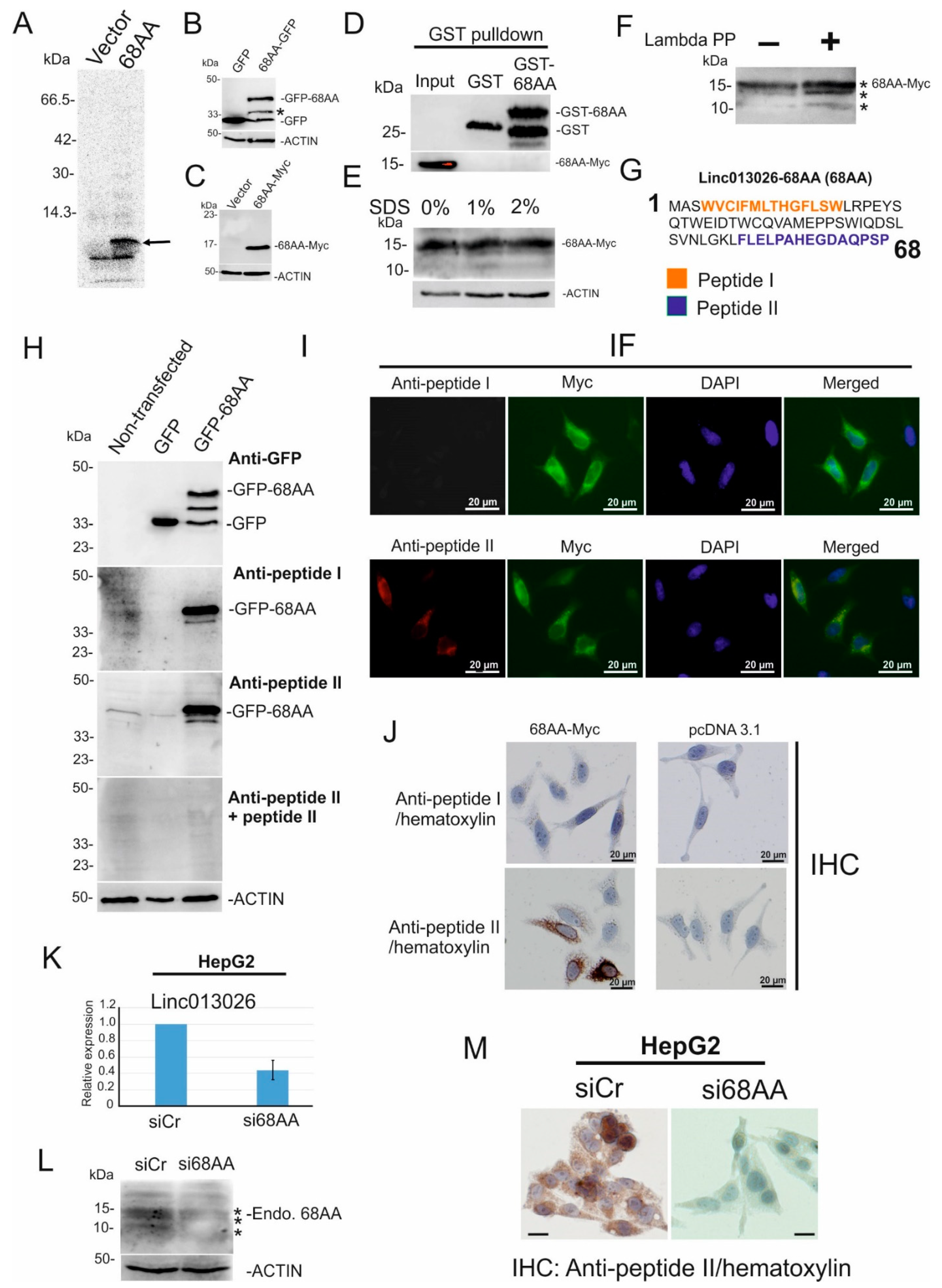
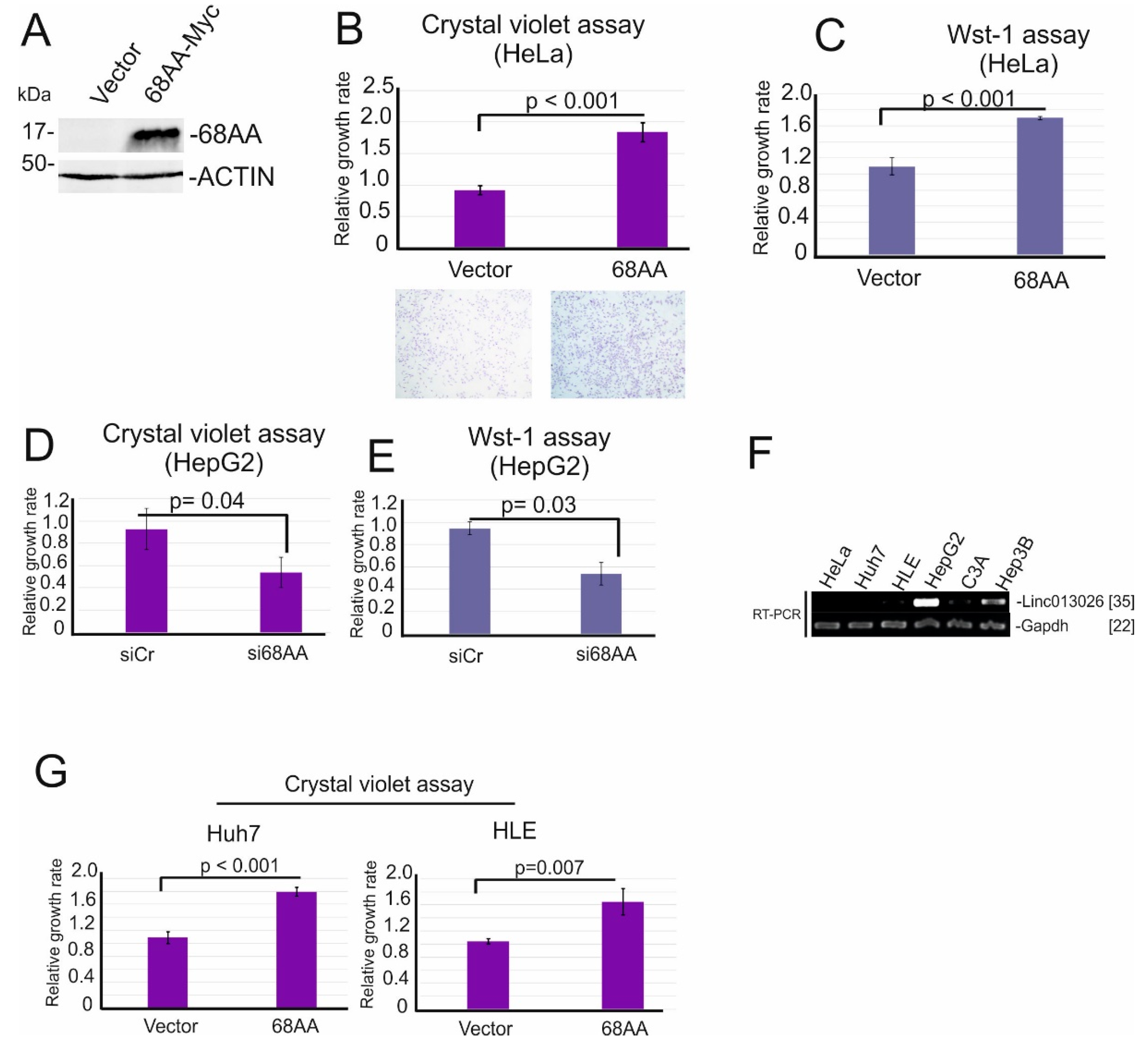
| Transcript ID | Expression in the Liver (FPKM) (ENCFF184YUO) | Expression in HepG2 (FPKM) (ENCFF337WTM) | Number of ORFs Longer Than 50AA |
|---|---|---|---|
| NONHSAT226968.1 | 0 | 104.9 | 2 |
| NONHSAT013026.2/Linc013026 | 0 | 61.3 | 2 |
| NONHSAT250607.1 | 0 | 39.3 | 2 |
| NONHSAT142412.2 | 0 | 30.5 | 12 |
| NONHSAT115455.2 | 0 | 15.0 | 2 |
| NONHSAT168790.1 | 0 | 13.7 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polenkowski, M.; Burbano de Lara, S.; Allister, A.B.; Nguyen, T.N.Q.; Tamura, T.; Tran, D.D.H. Identification of Novel Micropeptides Derived from Hepatocellular Carcinoma-Specific Long Noncoding RNA. Int. J. Mol. Sci. 2022, 23, 58. https://doi.org/10.3390/ijms23010058
Polenkowski M, Burbano de Lara S, Allister AB, Nguyen TNQ, Tamura T, Tran DDH. Identification of Novel Micropeptides Derived from Hepatocellular Carcinoma-Specific Long Noncoding RNA. International Journal of Molecular Sciences. 2022; 23(1):58. https://doi.org/10.3390/ijms23010058
Chicago/Turabian StylePolenkowski, Mareike, Sebastian Burbano de Lara, Aldrige Bernardus Allister, Thi Nhu Quynh Nguyen, Teruko Tamura, and Doan Duy Hai Tran. 2022. "Identification of Novel Micropeptides Derived from Hepatocellular Carcinoma-Specific Long Noncoding RNA" International Journal of Molecular Sciences 23, no. 1: 58. https://doi.org/10.3390/ijms23010058
APA StylePolenkowski, M., Burbano de Lara, S., Allister, A. B., Nguyen, T. N. Q., Tamura, T., & Tran, D. D. H. (2022). Identification of Novel Micropeptides Derived from Hepatocellular Carcinoma-Specific Long Noncoding RNA. International Journal of Molecular Sciences, 23(1), 58. https://doi.org/10.3390/ijms23010058






