Molecular Pathways Modulating Sensory Hair Cell Regeneration in Adult Mammalian Cochleae: Progress and Perspectives
Abstract
:1. Introduction
2. Targeting Signaling Pathways for Hair Cell Regeneration
3. Targeting Notch Signaling for Hair Cell Regeneration
4. Targeting Wnt Signaling for Hair Cell Regeneration
5. Combinational Approaches for Hair Cell Regeneration
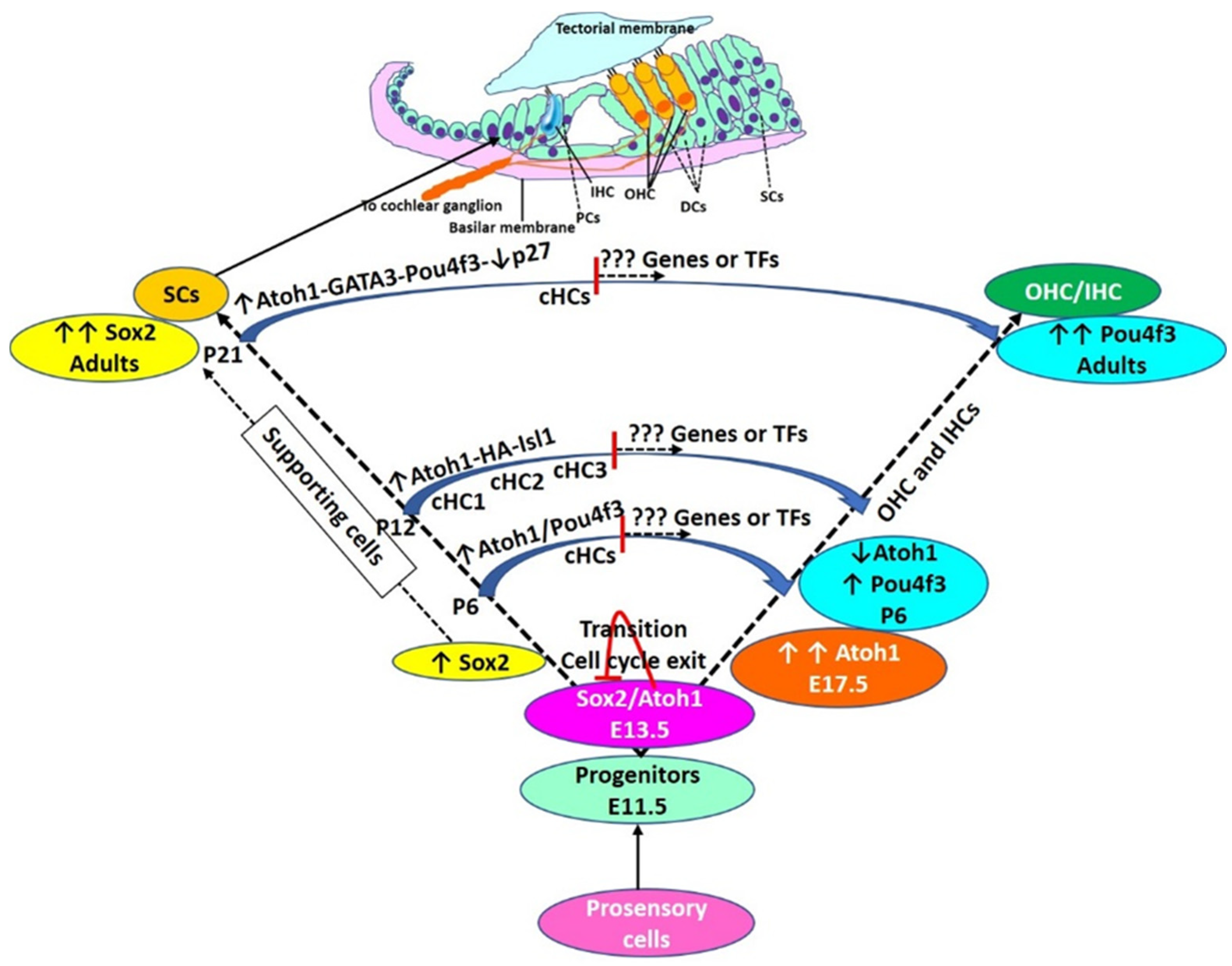
6. Modulating Transcription Factors for Hair Cell Regeneration
7. SC Subpopulations and Hair Cell Regeneration
8. Finding Additional Transcription Factors as Novel Targets for Hair Cell Regeneration
9. Investigating Epigenetic Regulation of Hair Cell Development and Regeneration
10. In Silico Approaches to Finding Novel Gene Targets
11. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davis, A.C. Hearing disorders in the population: First phase findings of the MRC National Study of Hearing. In Hearing Science and Hearing Disorders; Elsevier: Amsterdam, The Netherlands, 1983; pp. 35–60. [Google Scholar]
- Groves, A.K. The challenge of hair cell regeneration. Exp. Biol. Med. 2010, 235, 434–446. [Google Scholar] [CrossRef] [Green Version]
- Cox, B.C.; Chai, R.; Lenoir, A.; Liu, Z.; Zhang, L.; Nguyen, D.H.; Chalasani, K.; Steigelman, K.A.; Fang, J.; Rubel, E.W.; et al. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development 2014, 141, 816–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, J.S.; Cotanche, D.A. Hair cell regeneration in the avian auditory epithelium. Int. J. Dev. Biol. 2007, 51, 633–647. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Cai, Q.; West, M.B.; Youm, I.; Huang, X.; Li, W.; Cheng, W.; Nakmali, D.; Ewert, D.L.; Kopke, R.D. Regeneration of Cochlear Hair Cells and Hearing Recovery through Hes1 Modulation with siRNA Nanoparticles in Adult Guinea Pigs. Mol. Ther. J. Am. Soc. Gene Ther. 2018, 26, 1313–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLean, W.J.; Yin, X.; Lu, L.; Lenz, D.R.; McLean, D.; Langer, R.; Karp, J.M.; Edge, A.S. Clonal expansion of Lgr5-positive cells from mammalian cochlea and high-purity generation of sensory hair cells. Cell Rep. 2017, 18, 1917–1929. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Liu, D.; Dong, Y.; Zhang, Z.; Zhang, Y.; Zhou, H.; Guo, L.; Qi, J.; Qiang, R.; Tang, M.; et al. Frizzled-9+ Supporting Cells Are Progenitors for the Generation of Hair Cells in the Postnatal Mouse Cochlea. Front. Mol. Neurosci. 2019, 12, 184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, B.J.; Coak, E.; Dearman, J.; Bailey, G.; Yamashita, T.; Kuo, B.; Zuo, J. In Vivo Interplay between p27(Kip1), GATA3, ATOH1, and POU4F3 Converts Non-sensory Cells to Hair Cells in Adult Mice. Cell Rep. 2017, 19, 307–320. [Google Scholar] [CrossRef]
- Mizutari, K.; Fujioka, M.; Hosoya, M.; Bramhall, N.; Okano, H.J.; Okano, H.; Edge, A.S. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron 2013, 77, 58–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, T.; Zheng, F.; Finkelstein, D.; Kellard, Z.; Carter, R.; Rosencrance, C.D.; Sugino, K.; Easton, J.; Gawad, C.; Zuo, J. High-resolution transcriptional dissection of in vivo Atoh1-mediated hair cell conversion in mature cochleae identifies Isl1 as a co-reprogramming factor. PLoS Genet. 2018, 14, e1007552. [Google Scholar] [CrossRef]
- Shu, Y.; Li, W.; Huang, M.; Quan, Y.-Z.; Scheffer, D.; Tian, C.; Tao, Y.; Liu, X.; Hochedlinger, K.; Indzhykulian, A.A. Renewed proliferation in adult mouse cochlea and regeneration of hair cells. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Dearman, J.A.; Cox, B.C.; Walters, B.J.; Zhang, L.; Ayrault, O.; Zindy, F.; Gan, L.; Roussel, M.F.; Zuo, J. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters’ cells to immature hair cells by Atoh1 ectopic expression. J. Neurosci. 2012, 32, 6600–6610. [Google Scholar] [CrossRef] [Green Version]
- Querleu, D.; Renard, X.; Versyp, F.; Paris-Delrue, L.; Crèpin, G. Fetal hearing. Eur. J. Obstet. Gynecol. Reprod. Biol. 1988, 28, 191–212. [Google Scholar] [CrossRef]
- Edge, A.S.; Chen, Z.-Y. Hair cell regeneration. Curr. Opin. Neurobiol. 2008, 18, 377–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, A.; Sanchez-Guardado, L.; Juniat, S.; Gale, J.E.; Daudet, N.; Henrique, D. Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development 2015, 142, 1948–1959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, B.R.; Baldwin, E.M.; Layman, W.S.; Taketo, M.M.; Zuo, J. In vivo cochlear hair cell generation and survival by coactivation of β-catenin and Atoh1. J. Neurosci. 2015, 35, 10786–10798. [Google Scholar] [CrossRef]
- Ni, W.; Zeng, S.; Li, W.; Chen, Y.; Zhang, S.; Tang, M.; Sun, S.; Chai, R.; Li, H. Wnt activation followed by Notch inhibition promotes mitotic hair cell regeneration in the postnatal mouse cochlea. Oncotarget 2016, 7, 66754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkinson, P.J.; Najarro, E.H.; Sayyid, Z.N.; Cheng, A.G. Sensory hair cell development and regeneration: Similarities and differences. Development 2015, 142, 1561–1571. [Google Scholar] [CrossRef] [Green Version]
- Driver, E.C.; Pryor, S.P.; Hill, P.; Turner, J.; Rüther, U.; Biesecker, L.G.; Griffith, A.J.; Kelley, M.W. Hedgehog signaling regulates sensory cell formation and auditory function in mice and humans. J. Neurosci. 2008, 28, 7350–7358. [Google Scholar] [CrossRef]
- Tateya, T.; Imayoshi, I.; Tateya, I.; Hamaguchi, K.; Torii, H.; Ito, J.; Kageyama, R. Hedgehog signaling regulates prosensory cell properties during the basal-to-apical wave of hair cell differentiation in the mammalian cochlea. Development 2013, 140, 3848–3857. [Google Scholar] [CrossRef] [Green Version]
- Benito-Gonzalez, A.; Doetzlhofer, A. Hey1 and Hey2 control the spatial and temporal pattern of mammalian auditory hair cell differentiation downstream of Hedgehog signaling. J. Neurosci. 2014, 34, 12865–12876. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.-X.; Hou, C.-C.; Yang, W.-X. Control of hair cell development by molecular pathways involving Atoh1, Hes1 and Hes5. Gene 2015, 558, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Zuo, J. Cochlear hair cell regeneration after noise-induced hearing loss: Does regeneration follow development? Hear. Res. 2017, 349, 182–196. [Google Scholar] [CrossRef]
- Lush, M.E.; Diaz, D.C.; Koenecke, N.; Baek, S.; Boldt, H.; St Peter, M.K.; Gaitan-Escudero, T.; Romero-Carvajal, A.; Busch-Nentwich, E.M.; Perera, A.G. scRNA-Seq reveals distinct stem cell populations that drive hair cell regeneration after loss of Fgf and Notch signaling. Elife 2019, 8, e44431. [Google Scholar] [CrossRef] [PubMed]
- Millimaki, B.B.; Sweet, E.M.; Dhason, M.S.; Riley, B.B. Zebrafish atoh1 genes: Classic proneural activity in the inner ear and regulation by Fgf and Notch. Development 2007, 134, 295–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cafaro, J.; Lee, G.S.; Stone, J.S. Atoh1 expression defines activated progenitors and differentiating hair cells during avian hair cell regeneration. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2007, 236, 156–170. [Google Scholar] [CrossRef]
- Bermingham, N.A.; Hassan, B.A.; Price, S.D.; Vollrath, M.A.; Ben-Arie, N.; Eatock, R.A.; Bellen, H.J.; Lysakowski, A.; Zoghbi, H.Y. Math1: An essential gene for the generation of inner ear hair cells. Science 1999, 284, 1837–1841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radosevic, M.; Fargas, L.; Alsina, B. The role of her4 in inner ear development and its relationship with proneural genes and Notch signalling. PLoS ONE 2014, 9, e109860. [Google Scholar] [CrossRef] [Green Version]
- Chrysostomou, E.; Gale, J.E.; Daudet, N. Delta-like 1 and lateral inhibition during hair cell formation in the chicken inner ear: Evidence against cis-inhibition. Development 2012, 139, 3764–3774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Xu, J.; Jin, R.; Bai, H.; Zhang, M.; Yang, S.; Zhang, X.; Zhang, X.; Han, Z.; Zeng, S. Transcriptomic analysis of chicken cochleae after gentamicin damage and the involvement of four signaling pathways (Notch, FGF, Wnt and BMP) in hair cell regeneration. Hear. Res. 2018, 361, 66–79. [Google Scholar] [CrossRef]
- Ku, Y.-C.; Renaud, N.A.; Veile, R.A.; Helms, C.; Voelker, C.C.; Warchol, M.E.; Lovett, M. The transcriptome of utricle hair cell regeneration in the avian inner ear. J. Neurosci. 2014, 34, 3523–3535. [Google Scholar] [CrossRef] [Green Version]
- Zine, A.; Aubert, A.; Qiu, J.; Therianos, S.; Guillemot, F.; Kageyama, R.; de Ribaupierre, F. Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J. Neurosci. 2001, 21, 4712–4720. [Google Scholar] [CrossRef] [Green Version]
- Neves, J.; Parada, C.; Chamizo, M.; Giráldez, F. Jagged 1 regulates the restriction of Sox2 expression in the developing chicken inner ear: A mechanism for sensory organ specification. Development 2011, 138, 735–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aibar, S.; González-Blas, C.B.; Moerman, T.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.-C.; Geurts, P.; Aerts, J.; van den Oord, J. SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, X.-X.; Yan, J.; Li, Y.; Shi, J.-P. Wnt/β-catenin signaling was activated in supporting cells during exposure of the zebrafish lateral line to cisplatin. Ann. Anat.-Anat. Anz. 2019, 226, 48–56. [Google Scholar] [CrossRef]
- Jacques, B.E.; Montgomery IV, W.H.; Uribe, P.M.; Yatteau, A.; Asuncion, J.D.; Resendiz, G.; Matsui, J.I.; Dabdoub, A. The role of Wnt/β-catenin signaling in proliferation and regeneration of the developing basilar papilla and lateral line. Dev. Neurobiol. 2014, 74, 438–456. [Google Scholar] [CrossRef]
- Stevens, C.B.; Davies, A.L.; Battista, S.; Lewis, J.H.; Fekete, D.M. Forced activation of Wnt signaling alters morphogenesis and sensory organ identity in the chicken inner ear. Dev. Biol. 2003, 261, 149–164. [Google Scholar] [CrossRef] [Green Version]
- Hammond, K.L.; van Eeden, F.J.; Whitfield, T.T. Repression of Hedgehog signalling is required for the acquisition of dorsolateral cell fates in the zebrafish otic vesicle. Development 2010, 137, 1361–1371. [Google Scholar] [CrossRef] [Green Version]
- Son, E.J.; Ma, J.-H.; Ankamreddy, H.; Shin, J.-O.; Choi, J.Y.; Wu, D.K.; Bok, J. Conserved role of Sonic Hedgehog in tonotopic organization of the avian basilar papilla and mammalian cochlea. Proc. Natl. Acad. Sci. USA 2015, 112, 3746–3751. [Google Scholar] [CrossRef] [Green Version]
- Maier, E.C.; Whitfield, T.T. RA and FGF signalling are required in the zebrafish otic vesicle to pattern and maintain ventral otic identities. PLoS Genet. 2014, 10, e1004858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacques, B.E.; Dabdoub, A.; Kelley, M.W. Fgf signaling regulates development and transdifferentiation of hair cells and supporting cells in the basilar papilla. Hear. Res. 2012, 289, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Bermingham-McDonogh, O.; Stone, J.S.; Reh, T.A.; Rubel, E.W. FGFR3 expression during development and regeneration of the chick inner ear sensory epithelia. Dev. Biol. 2001, 238, 247–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirvola, U.; Ylikoski, J.; Trokovic, R.; Hébert, J.M.; McConnell, S.K.; Partanen, J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron 2002, 35, 671–680. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, T.; Ray, C.A.; Younkins, C.; Bermingham-McDonogh, O. Expression patterns of FGF receptors in the developing mammalian cochlea. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2010, 239, 1019–1026. [Google Scholar] [CrossRef] [Green Version]
- Mueller, K.L.; Jacques, B.E.; Kelley, M.W. Fibroblast growth factor signaling regulates pillar cell development in the organ of corti. J. Neurosci. 2002, 22, 9368–9377. [Google Scholar] [CrossRef]
- Jacques, B.E.; Montcouquiol, M.E.; Layman, E.M.; Lewandoski, M.; Kelley, M.W. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development 2007, 134, 3021–3029. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, T.; Ray, C.A.; Bermingham-McDonogh, O. Fgf20 is required for sensory epithelial specification in the developing cochlea. J. Neurosci. 2008, 28, 5991–5999. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Cunningham, D.; Bermingham-McDonogh, O. Loss of Fgfr3 leads to excess hair cell development in the mouse organ of Corti. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2007, 236, 525–533. [Google Scholar] [CrossRef]
- Kelley, M.W. Regulation of cell fate in the sensory epithelia of the inner ear. Nature reviews. Neuroscience 2006, 7, 837–849. [Google Scholar] [CrossRef] [PubMed]
- Maass, J.C.; Gu, R.; Basch, M.L.; Waldhaus, J.; Lopez, E.M.; Xia, A.; Oghalai, J.S.; Heller, S.; Groves, A.K. Changes in the regulation of the Notch signaling pathway are temporally correlated with regenerative failure in the mouse cochlea. Front. Cell. Neurosci. 2015, 9, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, B.J.; Yamashita, T.; Zuo, J. Sox2-CreER mice are useful for fate mapping of mature, but not neonatal, cochlear supporting cells in hair cell regeneration studies. Sci. Rep. 2015, 5, 11621. [Google Scholar] [CrossRef] [Green Version]
- McGovern, M.M.; Zhou, L.; Randle, M.R.; Cox, B.C. Spontaneous hair cell regeneration is prevented by increased notch signaling in supporting cells. Front. Cell. Neurosci. 2018, 12, 120. [Google Scholar] [CrossRef] [Green Version]
- Jansson, L.; Kim, G.S.; Cheng, A.G. Making sense of Wnt signaling—linking hair cell regeneration to development. Front. Cell. Neurosci. 2015, 9, 66. [Google Scholar] [CrossRef] [Green Version]
- Chai, R.; Kuo, B.; Wang, T.; Liaw, E.J.; Xia, A.; Jan, T.A.; Liu, Z.; Taketo, M.M.; Oghalai, J.S.; Nusse, R. Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc. Natl. Acad. Sci. USA 2012, 109, 8167–8172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, F.; Hu, L.; Edge, A.S. Generation of hair cells in neonatal mice by β-catenin overexpression in Lgr5-positive cochlear progenitors. Proc. Natl. Acad. Sci. USA 2013, 110, 13851–13856. [Google Scholar] [CrossRef] [Green Version]
- Jan, T.A.; Chai, R.; Sayyid, Z.N.; van Amerongen, R.; Xia, A.; Wang, T.; Sinkkonen, S.T.; Zeng, Y.A.; Levin, J.R.; Heller, S. Tympanic border cells are Wnt-responsive and can act as progenitors for postnatal mouse cochlear cells. Development 2013, 140, 1196–1206. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Li, W.; Lin, C.; Chen, Y.; Cheng, C.; Sun, S.; Tang, M.; Chai, R.; Li, H. Co-regulation of the Notch and Wnt signaling pathways promotes supporting cell proliferation and hair cell regeneration in mouse utricles. Sci. Rep. 2016, 6, 1–16. [Google Scholar] [CrossRef]
- Li, W.; Wu, J.; Yang, J.; Sun, S.; Chai, R.; Chen, Z.-Y.; Li, H. Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc. Natl. Acad. Sci. USA 2015, 112, 166–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, W.; Lin, C.; Guo, L.; Wu, J.; Chen, Y.; Chai, R.; Li, W.; Li, H. Extensive supporting cell proliferation and mitotic hair cell generation by in vivo genetic reprogramming in the neonatal mouse cochlea. J. Neurosci. 2016, 36, 8734–8745. [Google Scholar] [CrossRef] [Green Version]
- Romero-Carvajal, A.; Acedo, J.N.; Jiang, L.; Kozlovskaja-Gumbrienė, A.; Alexander, R.; Li, H.; Piotrowski, T. Regeneration of sensory hair cells requires localized interactions between the Notch and Wnt pathways. Dev. Cell 2015, 34, 267–282. [Google Scholar] [CrossRef] [Green Version]
- Menendez, L.; Trecek, T.; Gopalakrishnan, S.; Tao, L.; Markowitz, A.L.; Haoze, V.Y.; Wang, X.; Llamas, J.; Huang, C.; Lee, J. Generation of inner ear hair cells by direct lineage conversion of primary somatic cells. eLife 2020, 9, e55249. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Qian, W.; Jiang, G.; Ma, Y. Transcription factors in the development of inner ear hair cells. Front. Biosci. 2016, 21, 1118–1125. [Google Scholar] [CrossRef] [Green Version]
- Gálvez, H.; Abelló, G.; Giraldez, F. Signaling and transcription factors during inner ear development: The generation of hair cells and otic neurons. Front. Cell Dev. Biol. 2017, 5, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkatesh, I.; Blackmore, M.G. Selecting optimal combinations of transcription factors to promote axon regeneration: Why mechanisms matter. Neurosci. Lett. 2017, 652, 64–73. [Google Scholar] [CrossRef] [Green Version]
- Dhara, S.P.; Rau, A.; Flister, M.J.; Recka, N.M.; Laiosa, M.D.; Auer, P.L.; Udvadia, A.J. Cellular reprogramming for successful CNS axon regeneration is driven by a temporally changing cast of transcription factors. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begeman, I.J.; Kang, J. Transcriptional Programs and Regeneration Enhancers Underlying Heart Regeneration. J. Cardiovasc. Dev. Dis. 2019, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Kirjavainen, A.; Sulg, M.; Heyd, F.; Alitalo, K.; Ylä-Herttuala, S.; Möröy, T.; Petrova, T.V.; Pirvola, U. Prox1 interacts with Atoh1 and Gfi1, and regulates cellular differentiation in the inner ear sensory epithelia. Dev. Biol. 2008, 322, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Sayyid, Z.N.; Wang, T.; Chen, L.; Jones, S.M.; Cheng, A.G. Atoh1 directs regeneration and functional recovery of the mature mouse vestibular system. Cell Rep. 2019, 28, 312–324.e4. [Google Scholar] [CrossRef] [Green Version]
- Chonko, K.T.; Jahan, I.; Stone, J.; Wright, M.C.; Fujiyama, T.; Hoshino, M.; Fritzsch, B.; Maricich, S.M. Atoh1 directs hair cell differentiation and survival in the late embryonic mouse inner ear. Dev. Biol. 2013, 381, 401–410. [Google Scholar] [CrossRef]
- Walters, B.J.; Zuo, J. A Sox10 rtTA/+ Mouse Line Allows for Inducible Gene Expression in the Auditory and Balance Organs of the Inner Ear. J. Assoc. Res. Otolaryngol. 2015, 16, 331–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, B.C.; Dearman, J.A.; Brancheck, J.; Zindy, F.; Roussel, M.F.; Zuo, J. Generation of Atoh1-rtTA transgenic mice: A tool for inducible gene expression in hair cells of the inner ear. Sci. Rep. 2014, 4, 6885. [Google Scholar] [CrossRef] [Green Version]
- Wiwatpanit, T.; Lorenzen, S.M.; Cantu, J.A.; Foo, C.Z.; Hogan, A.K.; Marquez, F.; Clancy, J.C.; Schipma, M.J.; Cheatham, M.A.; Duggan, A. Trans-differentiation of outer hair cells into inner hair cells in the absence of INSM1. Nature 2018, 563, 691–695. [Google Scholar] [CrossRef]
- Chessum, L.; Matern, M.S.; Kelly, M.C.; Johnson, S.L.; Ogawa, Y.; Milon, B.; McMurray, M.; Driver, E.C.; Parker, A.; Song, Y. Helios is a key transcriptional regulator of outer hair cell maturation. Nature 2018, 563, 696–700. [Google Scholar] [CrossRef]
- Xu, Z.; Rai, V.; Zuo, J. TUB and ZNF532 Promote the Atoh1-Mediated Hair Cell Regeneration in Mouse Cochleae. Front. Cell. Neurosci. 2021, 15, 759223. [Google Scholar] [CrossRef] [PubMed]
- Hoa, M.; Olszewski, R.; Li, X.; Taukulis, I.; Gu, S.; DeTorres, A.; Lopez, I.A.; Linthicum Jr, F.H.; Ishiyama, A.; Martin, D. Characterizing Adult cochlear supporting cell transcriptional diversity using single-cell RNA-Seq: Validation in the adult mouse and translational implications for the adult human cochlea. Front. Mol. Neurosci. 2020, 13, 13. [Google Scholar] [CrossRef]
- Cheng, C.; Wang, Y.; Guo, L.; Lu, X.; Zhu, W.; Muhammad, W.; Zhang, L.; Lu, L.; Gao, J.; Tang, M. Age-related transcriptome changes in Sox2+ supporting cells in the mouse cochlea. Stem Cell Res. Ther. 2019, 10, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGovern, M.M.; Randle, M.R.; Cuppini, C.L.; Graves, K.A.; Cox, B.C. Multiple supporting cell subtypes are capable of spontaneous hair cell regeneration in the neonatal mouse cochlea. Development 2019, 146, dev171009. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Fang, J.; Dearman, J.; Zhang, L.; Zuo, J. In vivo generation of immature inner hair cells in neonatal mouse cochleae by ectopic Atoh1 expression. PLoS ONE 2014, 9, e89377. [Google Scholar] [CrossRef] [Green Version]
- Kempfle, J.S.; Turban, J.L.; Edge, A.S. Sox2 in the differentiation of cochlear progenitor cells. Sci. Rep. 2016, 6, 23293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, T.; Jen, H.-I.; Kang, H.; Klisch, T.J.; Zoghbi, H.Y.; Groves, A.K. Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J. Neurosci. 2015, 35, 5870–5883. [Google Scholar] [CrossRef] [Green Version]
- Jen, H.-I.; Hill, M.C.; Tao, L.; Sheng, K.; Cao, W.; Zhang, H.; Haoze, V.Y.; Llamas, J.; Zong, C.; Martin, J.F. Transcriptomic and epigenetic regulation of hair cell regeneration in the mouse utricle and its potentiation by Atoh1. Elife 2019, 8, e44328. [Google Scholar] [CrossRef]
- Li, H. Epigenetic and Signaling Pathway Regulation in Hair Cell Regeneration; iMedPub LTD: London, UK, 2015. [Google Scholar]
- Layman, W.S.; Zuo, J. Epigenetic regulation in the inner ear and its potential roles in development, protection, and regeneration. Front. Cell. Neurosci. 2015, 8, 446. [Google Scholar] [CrossRef] [Green Version]
- Doetzlhofer, A.; Avraham, K.B. Insights into inner ear-specific gene regulation: Epigenetics and non-coding RNAs in inner ear development and regeneration. Semin. Cell Dev. Biol. 2017, 65, 69–79. [Google Scholar] [CrossRef] [Green Version]
- Stojanova, Z.P.; Kwan, T.; Segil, N. Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea. Development 2016, 143, 1632. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Hu, Z. Epigenetic DNA demethylation causes inner ear stem cell differentiation into hair cell-like cells. Front. Cell. Neurosci. 2016, 10, 185. [Google Scholar] [CrossRef] [Green Version]
- Layman, W.; Williams, D.; Dearman, J.; Sauceda, M.; Zuo, J. Histone deacetylase inhibition protects hearing against acute ototoxicity by activating the Nf-κ B pathway. Cell Death Discov. 2015, 1, 1–7. [Google Scholar] [CrossRef]
- Matys, V.; Kel-Margoulis, O.V.; Fricke, E.; Liebich, I.; Land, S.; Barre-Dirrie, A.; Reuter, I.; Chekmenev, D.; Krull, M.; Hornischer, K. TRANSFAC® and its module TRANSCompel®: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006, 34, D108–D110. [Google Scholar] [CrossRef] [Green Version]
- Portales-Casamar, E.; Thongjuea, S.; Kwon, A.T.; Arenillas, D.; Zhao, X.; Valen, E.; Yusuf, D.; Lenhard, B.; Wasserman, W.W.; Sandelin, A. JASPAR 2010: The greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010, 38, D105–D110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulakovskiy, I.V.; Medvedeva, Y.A.; Schaefer, U.; Kasianov, A.S.; Vorontsov, I.E.; Bajic, V.B.; Makeev, V.J. HOCOMOCO: A comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 2013, 41, D195–D202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weirauch, M.T.; Yang, A.; Albu, M.; Cote, A.G.; Montenegro-Montero, A.; Drewe, P.; Najafabadi, H.S.; Lambert, S.A.; Mann, I.; Cook, K. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014, 158, 1431–1443. [Google Scholar] [CrossRef] [Green Version]
- Janky, R.; Verfaillie, A.; Imrichova, H.; Van de Sande, B.; Standaert, L.; Christiaens, V.; Hulselmans, G.; Herten, K.; Naval Sanchez, M.; Potier, D.; et al. iRegulon: From a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 2014, 10, e1003731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, S.; Li, S.; Luo, Z.; Ren, M.; He, S.; Wang, G.; Liu, Z. Dual expression of Atoh1 and Ikzf2 promotes transformation of adult cochlear supporting cells into outer hair cells. Elife 2021, 10, e66547. [Google Scholar] [CrossRef]
- Qian, D.; Radde-Gallwitz, K.; Kelly, M.; Tyrberg, B.; Kim, J.; Gao, W.Q.; Chen, P. Basic helix–loop–helix gene Hes6 delineates the sensory hair cell lineage in the inner ear. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2006, 235, 1689–1700. [Google Scholar] [CrossRef] [Green Version]
- Sölter, M.; Locker, M.; Boy, S.; Taelman, V.; Bellefroid, E.J.; Perron, M.; Pieler, T. Characterization and function of the bHLH-O protein XHes2: Insight into the mechanisms controlling retinal cell fate decision. Development 2006, 133, 4097–4108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, S.; Bessho, Y.; Hojo, M.; Kageyama, R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development 2000, 127, 2933–2943. [Google Scholar] [CrossRef]
- Kousa, Y.A.; Zhu, H.; Fakhouri, W.D.; Lei, Y.; Kinoshita, A.; Roushangar, R.R.; Patel, N.K.; Agopian, A.; Yang, W.; Leslie, E.J. The TFAP2A–IRF6–GRHL3 genetic pathway is conserved in neurulation. Hum. Mol. Genet. 2019, 28, 1726–1737. [Google Scholar] [CrossRef] [PubMed]
- Klisch, T.J.; Xi, Y.; Flora, A.; Wang, L.; Li, W.; Zoghbi, H.Y. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc. Natl. Acad. Sci. USA 2011, 108, 3288–3293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, A.; Valerius, M.T.; Mugford, J.W.; Carroll, T.J.; Self, M.; Oliver, G.; McMahon, A.P. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 2008, 3, 169–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
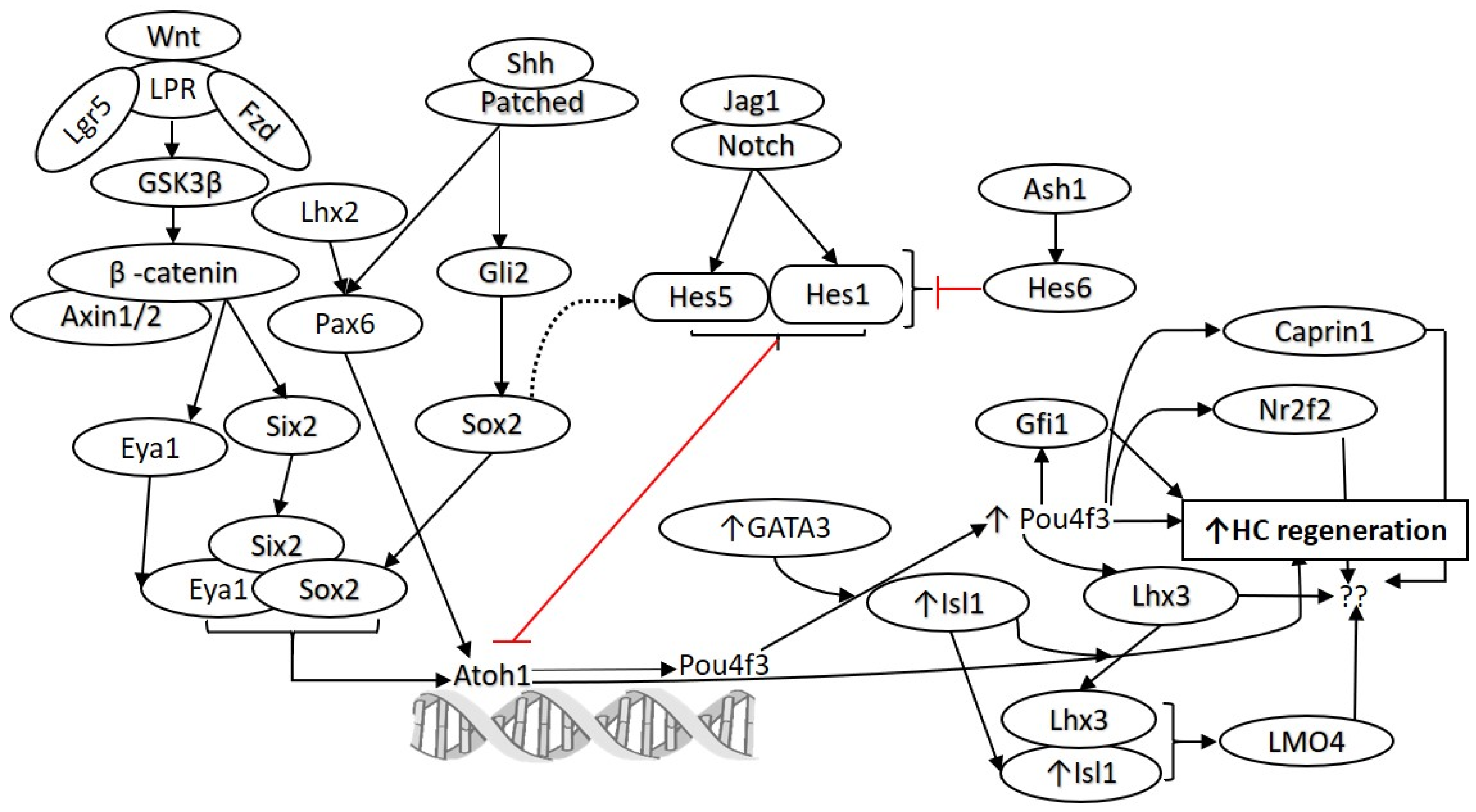
| Target | Zebrafish | Birds | Mice | Inference |
|---|---|---|---|---|
| Notch Atoh1 | atoh1a is expressed in all differentiating hair cells [24] Addition of exogenous atoh1a mRNA results in HC overproduction [25] | Atoh1 is immediately upregulated in Sox2+ SCs of the avian BP following HC loss [26]; Atoh1 protein expression is not detectable in mature HCs or SCs in the absence of damage [26] | Math1-null mice fail to develop cochlear and vestibular HCs [27] Atoh1 overexpression can convert SCs into HC-like cells in neonatal mouse cochleae [12] | Atoh1 modulation promotes regeneration in juvenile and adult mice, hence being a potential therapeutic target |
| Notch Hes5 | Does not affect atoh1a expression [28] Hes5 morphants do not generate supernumerary HCs [28] | Notch signaling activates Hes5 expression which inhibits hair cell fate [29] Hes5 is downregulated in SCs following HC damage and loss [30,31] | Beginning at postnatal ages, Hes5 is restricted to supporting cells [32], Hes5 deletion results primarily in supernumerary OHC formation, but also some supernumerary IHC formation [32] | Hes5 inhibition might be a therapeutic target in HC regeneration. |
| Notch Hey1/Hey 2 | Hey1 is downregulated in hair cells [24] | Hey1 and Hey2 are activated by Notch signaling in the basilar papilla and inhibit HC fate [33] | Hey1 and Hey2 negatively regulate Atoh1 to prevent premature HC differentiation [21] Exogenous Shh increases Hey1 and Hey2 mRNA levels in cochlear explants [21] | Hey1/2 inhibition may help regenerate HC-like cells to adopt a more HC-like phenotype. |
| Wnt β-catenin | Wnt/β-catenin inhibition in embryonic zebrafish reduces proliferation of sox2+ SCs in the developing neuromast [34]; Wnt/β-catenin is upregulated in SCs following HC loss [35] but is not sufficient for regeneration | Increases the proliferation of SCs following HC damage and regulates the number of HCs that form in the embryonic basilar papilla [36], Forced expression of β-catenin and Wnt3a results in the formation of ectopic sensory patches within the embryonic basilar papilla [37] | Activation of Wnt/β -catenin results in proliferation of Sox2+ SCs [17]; Lgr5+ SCs exhibit increased proliferation and differentiation into HCs in vivo in mice which overexpress β-catenin and Atoh1 [16] | β-catenin is a key therapeutic target for expansion of the HC progenitor pool. Wnt/β-catenin is conserved between species and plays a role in HC development and proliferation |
| Shh | Modifying hedgehog signaling interferes with axial patterning of the zebrafish otic vesicle [38] | Ectopic Shh signaling induces apical hair cell identities in the basal and middle regions of the avian basilar papilla [39] | Constitutive activation of Shh signaling hinders HC differentiation in developing murine cochleae [20] Inhibition of hedgehog signaling in cochlear explants results in an expanded sensory domain and formation of ectopic hair cells [19] | Modifying Shh signaling does not seem to be an effective strategy to promote regeneration |
| FGF | Fgf signaling is required for Atoh1 expression and hair cell development [25] During development, weak Fgf inhibition expands the sox2+ prosensory domain while strong Fgf inhibition reduces the sox2+ prosensory domain [40] While Fgf inhibition hinders HC differentiation, by expanding the sox2+ prosensory domain, it ultimately results in the overproduction of hair cells Fgfr3 knockout results in supernumerary HC formation [40] Fgf8 knockout results in reduced HC formation [40] Fgf3 overexpression results in reduced HC number [40] | Inhibition of FGF signaling in E5-E9 chicks results in overproduction of HCs through non-proliferative mechanisms. FGF inhibition increases the number of Sox2+ HCs in the embryonic basilar papilla, suggesting that the formation of extra hair cells is due to transdifferentiation [41] Fgfr3 is restricted to supporting cells in the mature basilar papilla [42] Fgfr3 expression is downregulated in the mature basilar papilla following damage to hair cells [42] | Fgfr1 hypomorphs lack 3rd-row OHCs [43] Fgfr1 plays a role in prosensory specification [44], In the embryo, Fgfr3 is expressed in the area of the cochlear duct that gives rise to pillar cells, OHCs, and Deiter’s cells, but Fgfr3 is confined to pillar cells by birth [45], Activation of Fgfr3 with Fgf17 inhibits OHC differentiation without affecting IHCs [46], Pan Fgf inhibition decreases expression of Atoh1 in murine cochlear explants [47], Fgfr3-/- mice lack a row of pillar cells, but have an ectopic additional row of Deiters cells and an additional row of OHCs which appear to have normal bundle morphology [48] | FGF signaling seems to be important signaling to modulate to promote HC regeneration, however, the results seem to be receptor-specific and different receptors have different effects of modulating FGF signaling |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rai, V.; Tu, S.; Frank, J.R.; Zuo, J. Molecular Pathways Modulating Sensory Hair Cell Regeneration in Adult Mammalian Cochleae: Progress and Perspectives. Int. J. Mol. Sci. 2022, 23, 66. https://doi.org/10.3390/ijms23010066
Rai V, Tu S, Frank JR, Zuo J. Molecular Pathways Modulating Sensory Hair Cell Regeneration in Adult Mammalian Cochleae: Progress and Perspectives. International Journal of Molecular Sciences. 2022; 23(1):66. https://doi.org/10.3390/ijms23010066
Chicago/Turabian StyleRai, Vikrant, Shu Tu, Joseph R. Frank, and Jian Zuo. 2022. "Molecular Pathways Modulating Sensory Hair Cell Regeneration in Adult Mammalian Cochleae: Progress and Perspectives" International Journal of Molecular Sciences 23, no. 1: 66. https://doi.org/10.3390/ijms23010066
APA StyleRai, V., Tu, S., Frank, J. R., & Zuo, J. (2022). Molecular Pathways Modulating Sensory Hair Cell Regeneration in Adult Mammalian Cochleae: Progress and Perspectives. International Journal of Molecular Sciences, 23(1), 66. https://doi.org/10.3390/ijms23010066







