Specific Inhibition of VanZ-Mediated Resistance to Lipoglycopeptide Antibiotics
Abstract
:1. Introduction
2. Results
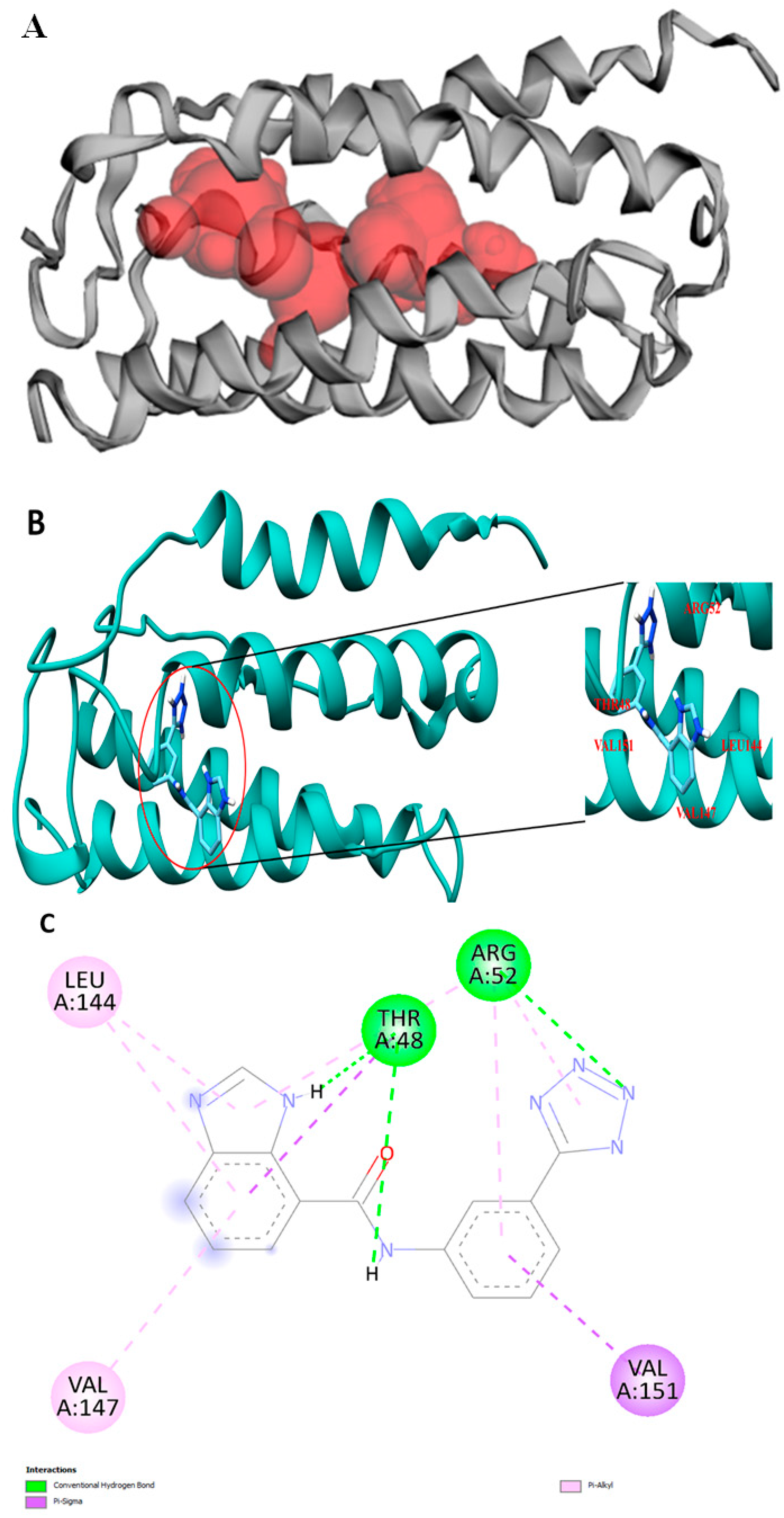
2.1. VanZ Modeling and Prediction of the VanZ Interaction with a Derivative of Benzimidazole
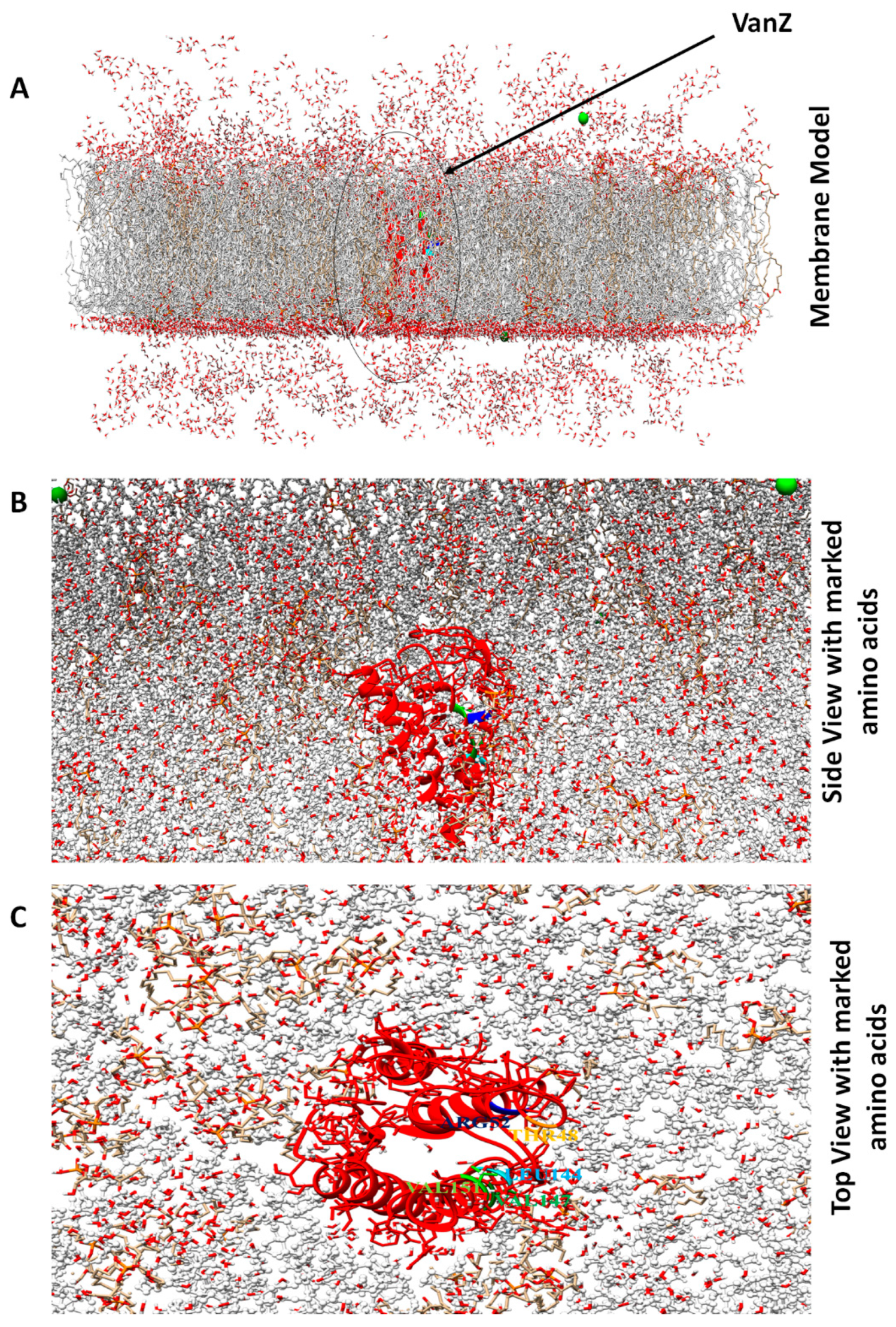
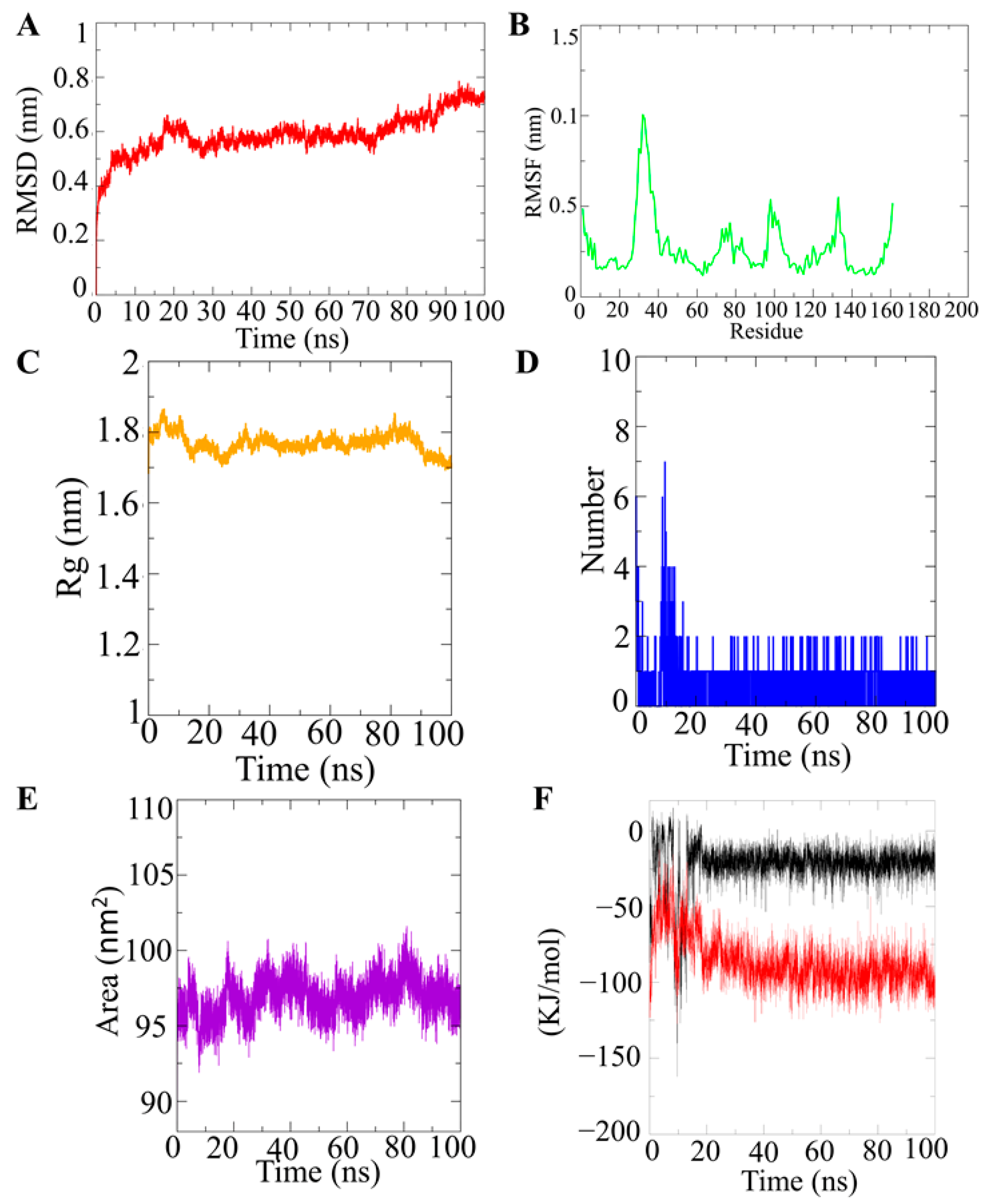
2.2. Molecular Dynamic Simulation of VanZ–G3K and Analysis
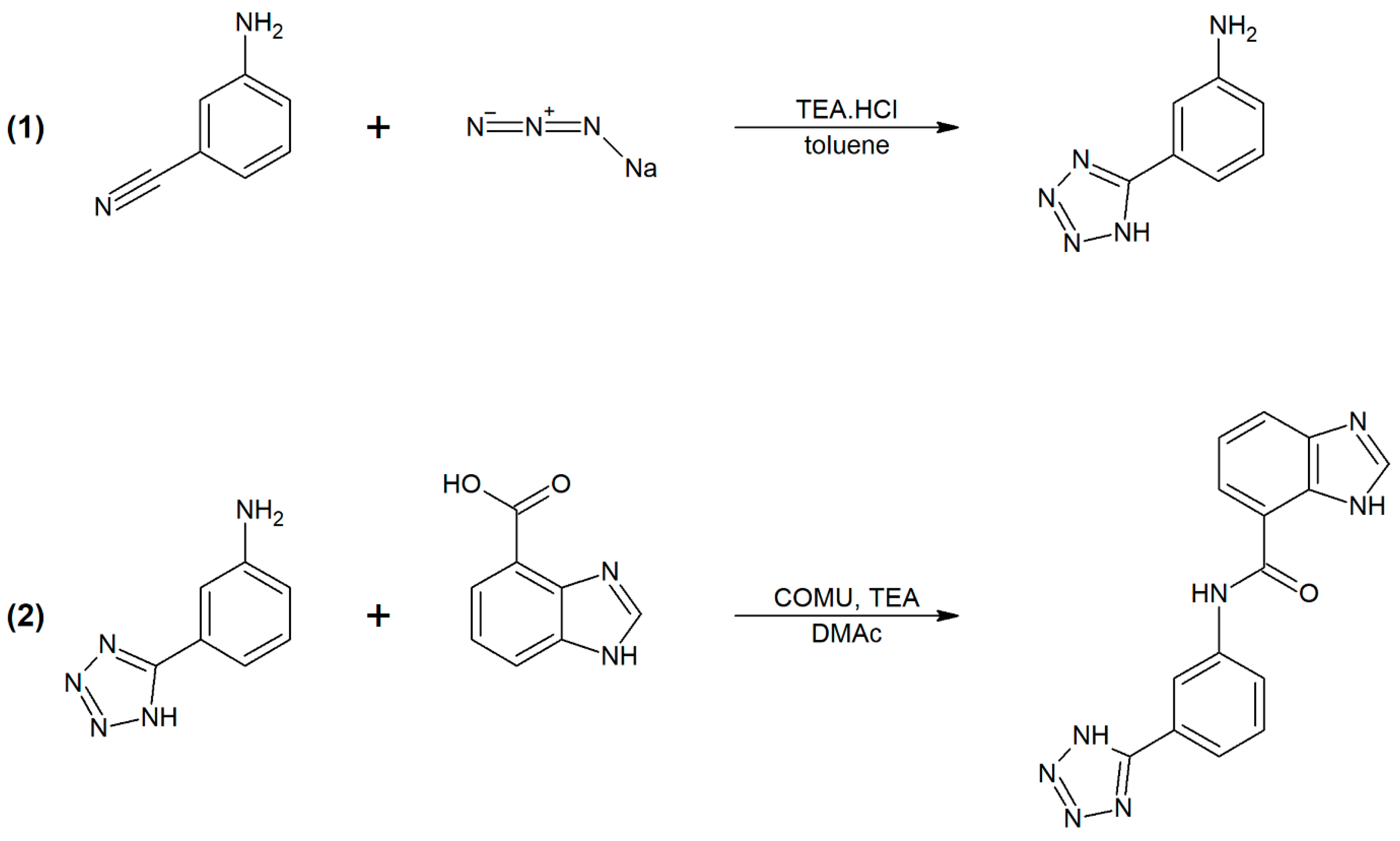
2.3. Synthesis and Characterization of G3K
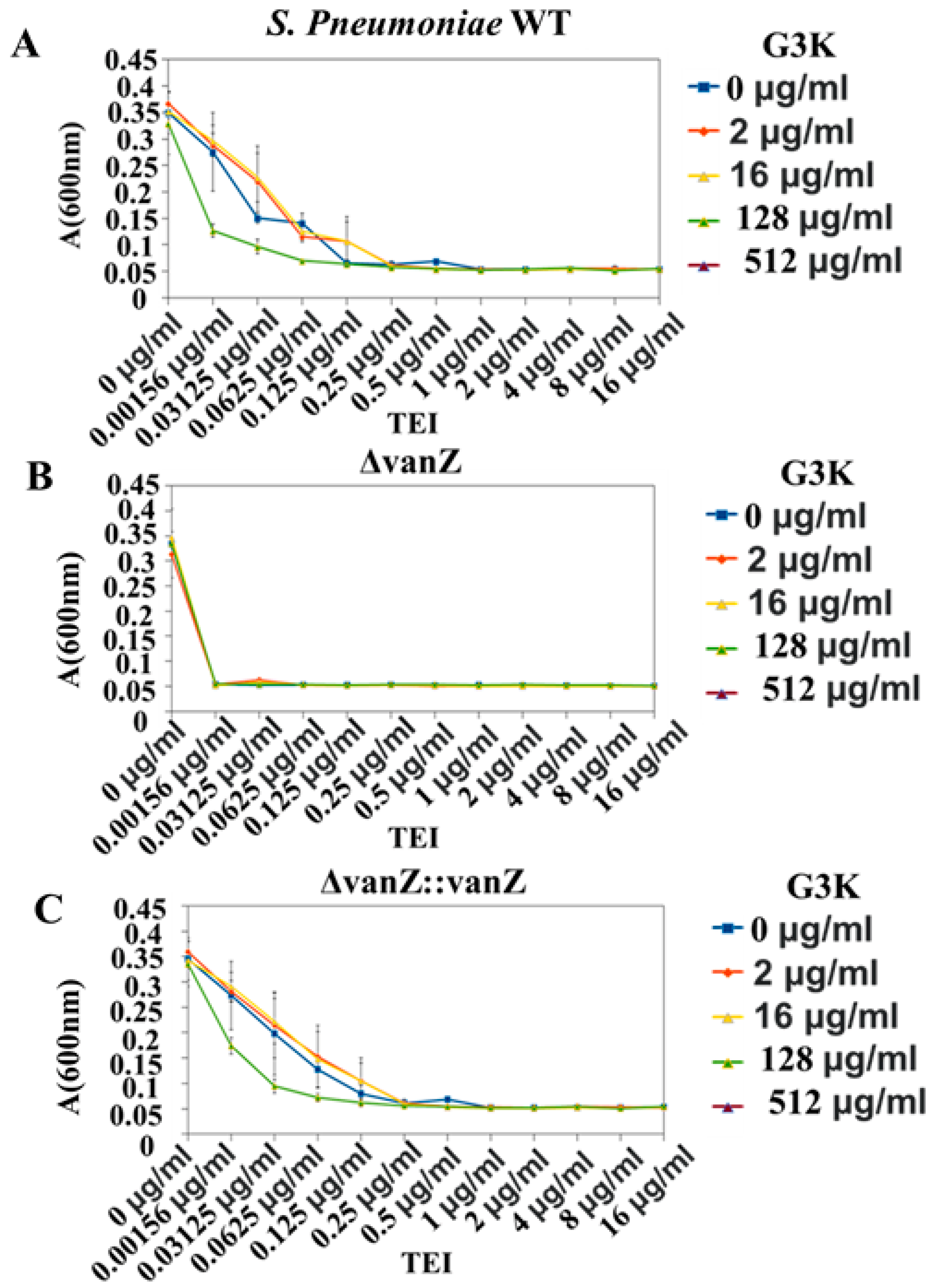
2.4. In Vivo Validation of VanZ–G3K Interaction
3. Discussion
4. Materials and Methods
4.1. VanZ Protein Structure Prediction and Validation
4.2. Chemical Synthesis of G3K and Characterization
4.2.1. Chemicals
4.2.2. Synthesis
4.2.3. Nuclear Magnetic Resonance (NMR) Spectroscopy
4.2.4. High-Performance Liquid Chromatography (HPLC)
4.3. Protein Pocket Analysis and Molecular Docking
4.4. Molecular Dynamic Simulation (MDS) Analysis
4.5. In Vivo Investigation
4.5.1. Bacterial Strains
4.5.2. Activity of G3K In Vivo against VanZ Expressing Bacterial Strains
G3K’s Effect against Gram-Positive Bacteria
The Effect of G3K in Combination with TEI on VanZ-Mediated Teicoplanin Resistance in S. aureus
G3K in Combination with TEI against S. pneumonia Strains and Clinical Variants of E. faecium and Susceptibility Testing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Mazumdar, A.; Haddad, Y.; Milosavljevic, V.; Michalkova, H.; Guran, R.; Bhowmick, S.; Moulick, A. Peptide-carbon quantum dots conjugate, derived from human retinoic acid receptor responder protein 2, against antibiotic-resistant gram positive and gram negative pathogenic bacteria. Nanomaterials 2020, 10, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazumdar, A.; Adam, V. Antimicrobial peptides-An alternative candidates to antibiotics against Staphylococcus aureus and its antibiotic-resistant strains. J. Mol. Clin. Med. 2021, 4, 1–17. [Google Scholar] [CrossRef]
- Mazumdar, A.; Haddad, Y.; Sur, V.P.; Milosavljevic, V.; Bhowmick, S.; Michalkova, H.; Guran, R.; Vesely, R.; Moulick, A. Characterization and in vitro analysis of probiotic-derived peptides against multi drug resistance bacterial infections. Front. Microbiol. 2020, 11, 1963. [Google Scholar] [CrossRef]
- Sur, V.P.; Mazumdar, A.; Kopel, P.; Mukherjee, S.; Vítek, P.; Michalkova, H.; Vaculovičová, M.; Moulick, A. A novel ruthenium based coordination compound against pathogenic bacteria. Int. J. Mol. Sci. 2020, 21, 2656. [Google Scholar] [CrossRef]
- Sur, V.P.; Mazumdar, A.; Ashrafi, A.; Mukherjee, A.; Milosavljevic, V.; Michalkova, H.; Kopel, P.; Richtera, L.; Moulick, A. A Novel Biocompatible Titanium–Gadolinium Quantum Dot as a Bacterial Detecting Agent with High Antibacterial Activity. Nanomaterials 2020, 10, 778. [Google Scholar] [CrossRef] [Green Version]
- Sur, V.P.; Kominkova, M.; Buchtova, Z.; Dolezelikova, K.; Zitka, O.; Moulick, A. CdSe QD Biosynthesis in Yeast Using Tryptone-Enriched Media and Their Conjugation with a Peptide Hecate for Bacterial Detection and Killing. Nanomaterials 2019, 9, 1463. [Google Scholar] [CrossRef] [Green Version]
- Nikaido, H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef] [Green Version]
- Xie, O.; Slavin, M.A.; Teh, B.W.; Bajel, A.; Douglas, A.P.; Worth, L.J. Epidemiology, treatment and outcomes of bloodstream infection due to vancomycin-resistant enterococci in cancer patients in a vanB endemic setting. BMC Infect. Dis. 2020, 20, 228. [Google Scholar] [CrossRef]
- Seol, C.A.; Park, J.S.; Sung, H.; Kim, M.-N. Co-colonization of vanA and vanB Enterococcus faecium of clonal complex 17 in a patient with bacteremia due to vanA E. faecium. Diagn. Microbiol. Infect. Dis. 2014, 79, 141–143. [Google Scholar] [CrossRef]
- Jelinkova, P.; Splichal, Z.; Jimenez, A.M.J.; Haddad, Y.; Mazumdar, A.; Sur, V.P.; Milosavljevic, V.; Kopel, P.; Buchtelova, H.; Guran, R. Novel vancomycin–peptide conjugate as potent antibacterial agent against vancomycin-resistant Staphylococcus aureus. Infect. Drug Resist. 2018, 11, 1807. [Google Scholar] [CrossRef] [Green Version]
- Barna, J.C.J.; Williams, D.H.; Stone, D.J.M.; Leung, T.W.C.; Doddrell, D.M. Structure elucidation of the teicoplanin antibiotics. J. Am. Chem. Soc. 1984, 106, 4895–4902. [Google Scholar] [CrossRef]
- Coşkun, F.A.; Mumcuoğlu, I.; Aksu, N.; Karahan, Z.C.; Us, E.; Tekeli, F.A.; Baran, I.; Kanyılmaz, D.; Kurşun, S. Phenotypic and genotypic traits of vancomycin-resistant enterococci in a public hospital: The first vanB-positive Enterococcus faecium isolates. Mikrobiyoloji Bul. 2012, 46, 276–282. [Google Scholar]
- Arthur, M.; Depardieu, F.; Molinas, C.; Reynolds, P.; Courvalin, P. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 1995, 154, 87–92. [Google Scholar] [CrossRef]
- Mazumdar, A. Studies on the Effect of Antimicrobial Peptides Conjugated with Nanoparticles on Pathogenic Bacteria. Ph.D. Thesis, Mendelova univerzita v Brně, Agronomická Fakulta, Brno-sever-Černá Pole, Czech Republic, 2020. [Google Scholar]
- Lai, L.; Dai, J.; Tang, H.; Zhang, S.; Wu, C.; Qiu, W.; Lu, C.; Yao, H.; Fan, H.; Wu, Z. Streptococcus suis serotype 9 strain GZ0565 contains a type VII secretion system putative substrate EsxA that contributes to bacterial virulence and a vanZ-like gene that confers resistance to teicoplanin and dalbavancin in Streptococcus agalactiae. Vet. Microbiol. 2017, 205, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Woods, E.C.; Wetzel, D.; Mukerjee, M.; McBride, S.M. Examination of the Clostridioides (Clostridium) difficile VanZ ortholog, CD1240. Anaerobe 2018, 53, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Ligozzi, M.; Lo Cascio, G.; Fontana, R. vanA gene cluster in a vancomycin-resistant clinical isolate of Bacillus circulans. Antimicrob. Agents Chemother. 1998, 42, 2055–2059. [Google Scholar] [CrossRef] [Green Version]
- Nygaard, M.; Kragelund, B.B.; Papaleo, E.; Lindorff-Larsen, K. An efficient method for estimating the hydrodynamic radius of disordered protein conformations. Biophys. J. 2017, 113, 550–557. [Google Scholar] [CrossRef] [Green Version]
- Prakash, A.; Baer, M.D.; Mundy, C.J.; Pfaendtner, J. Peptoid backbone flexibility dictates its interaction with water and surfaces: A molecular dynamics investigation. Biomacromolecules 2018, 19, 1006–1015. [Google Scholar] [CrossRef]
- Mostofian, B.; Zhuang, T.; Cheng, X.; Nickels, J.D. Branched-Chain Fatty Acid Content Modulates Structure, Fluidity, and Phase in Model Microbial Cell Membranes. J. Phys. Chem. B 2019, 123, 5814–5821. [Google Scholar] [CrossRef]
- Sirobhushanam, S.; Galva, C.; Sen, S.; Wilkinson, B.J.; Gatto, C. Broad substrate specificity of phosphotransbutyrylase from Listeria monocytogenes: A potential participant in an alternative pathway for provision of acyl CoA precursors for fatty acid biosynthesis. Biochim Biophys Acta 2016, 1861, 1102–1110. [Google Scholar] [CrossRef] [Green Version]
- Vimberg, V.; Zieglerová, L.; Buriánková, K.; Branny, P.; Balíková Novotná, G. VanZ Reduces the Binding of Lipoglycopeptide Antibiotics to Staphylococcus aureus and Streptococcus pneumoniae Cells. Front. Microbiol. 2020, 11, 566. [Google Scholar] [CrossRef] [PubMed]
- Goud, N.S.; Kumar, P.; Bharath, R.D. Recent Developments of Target-Based Benzimidazole Derivatives as Potential Anticancer Agents. In Heterocycles-Synthesis and Biological Activities; IntechOpen: London, UK, 2020. [Google Scholar]
- Sur, V.P. Metal Based Coordinate Compounds and Nanoparticles as an Alternative of Antibiotics against Resistant Bacterial Strains. Ph.D. Thesis, Mendelova Univerzita v Brně, Agronomická Fakulta, Brno-sever-Černá Pole, Czech Republic, 2020. [Google Scholar]
- Nichols, D.A.; Jaishankar, P.; Larson, W.; Smith, E.; Liu, G.; Beyrouthy, R.; Bonnet, R.; Renslo, A.R.; Chen, Y. Structure-based design of potent and ligand-efficient inhibitors of CTX-M class A β-lactamase. J. Med. Chem. 2012, 55, 2163–2172. [Google Scholar] [CrossRef] [PubMed]
- de Niederhäusern, S.; Bondi, M.; Messi, P.; Iseppi, R.; Sabia, C.; Manicardi, G.; Anacarso, I. Vancomycin-resistance Transferability from VanA Enterococci to Staphylococcus aureus. Curr. Microbiol. 2011, 62, 1363–1367. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Omasits, U.; Ahrens, C.H.; Müller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Rullmann, J.A.C.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Srivastava, R.; Prakash, A.; Lynn, A.M. Structure-based virtual screening, molecular dynamics simulation and MM-PBSA toward identifying the inhibitors for two-component regulatory system protein NarL of Mycobacterium Tuberculosis. J. Biomol. Struct. Dyn. 2020, 38, 3396–3410. [Google Scholar] [CrossRef]
- Sur, V.P.; Sen, M.K.; Komrskova, K. In Silico Identification and Validation of Organic Triazole Based Ligands as Potential Inhibitory Drug Compounds of SARS-CoV-2 Main Protease. Molecules 2021, 26, 6199. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sur, V.P.; Mazumdar, A.; Vimberg, V.; Stefani, T.; Androvic, L.; Kracikova, L.; Laga, R.; Kamenik, Z.; Komrskova, K. Specific Inhibition of VanZ-Mediated Resistance to Lipoglycopeptide Antibiotics. Int. J. Mol. Sci. 2022, 23, 97. https://doi.org/10.3390/ijms23010097
Sur VP, Mazumdar A, Vimberg V, Stefani T, Androvic L, Kracikova L, Laga R, Kamenik Z, Komrskova K. Specific Inhibition of VanZ-Mediated Resistance to Lipoglycopeptide Antibiotics. International Journal of Molecular Sciences. 2022; 23(1):97. https://doi.org/10.3390/ijms23010097
Chicago/Turabian StyleSur, Vishma Pratap, Aninda Mazumdar, Vladimir Vimberg, Tommaso Stefani, Ladislav Androvic, Lucie Kracikova, Richard Laga, Zdenek Kamenik, and Katerina Komrskova. 2022. "Specific Inhibition of VanZ-Mediated Resistance to Lipoglycopeptide Antibiotics" International Journal of Molecular Sciences 23, no. 1: 97. https://doi.org/10.3390/ijms23010097
APA StyleSur, V. P., Mazumdar, A., Vimberg, V., Stefani, T., Androvic, L., Kracikova, L., Laga, R., Kamenik, Z., & Komrskova, K. (2022). Specific Inhibition of VanZ-Mediated Resistance to Lipoglycopeptide Antibiotics. International Journal of Molecular Sciences, 23(1), 97. https://doi.org/10.3390/ijms23010097







