miR-486-5p and miR-22-3p Enable Megakaryocytic Differentiation of Hematopoietic Stem and Progenitor Cells without Thrombopoietin
Abstract
1. Introduction
2. Results
2.1. The miR Content of huMkMPs Is Well Preserved among Donors, with Seven miRs Making Up 57% of the Total miR Content
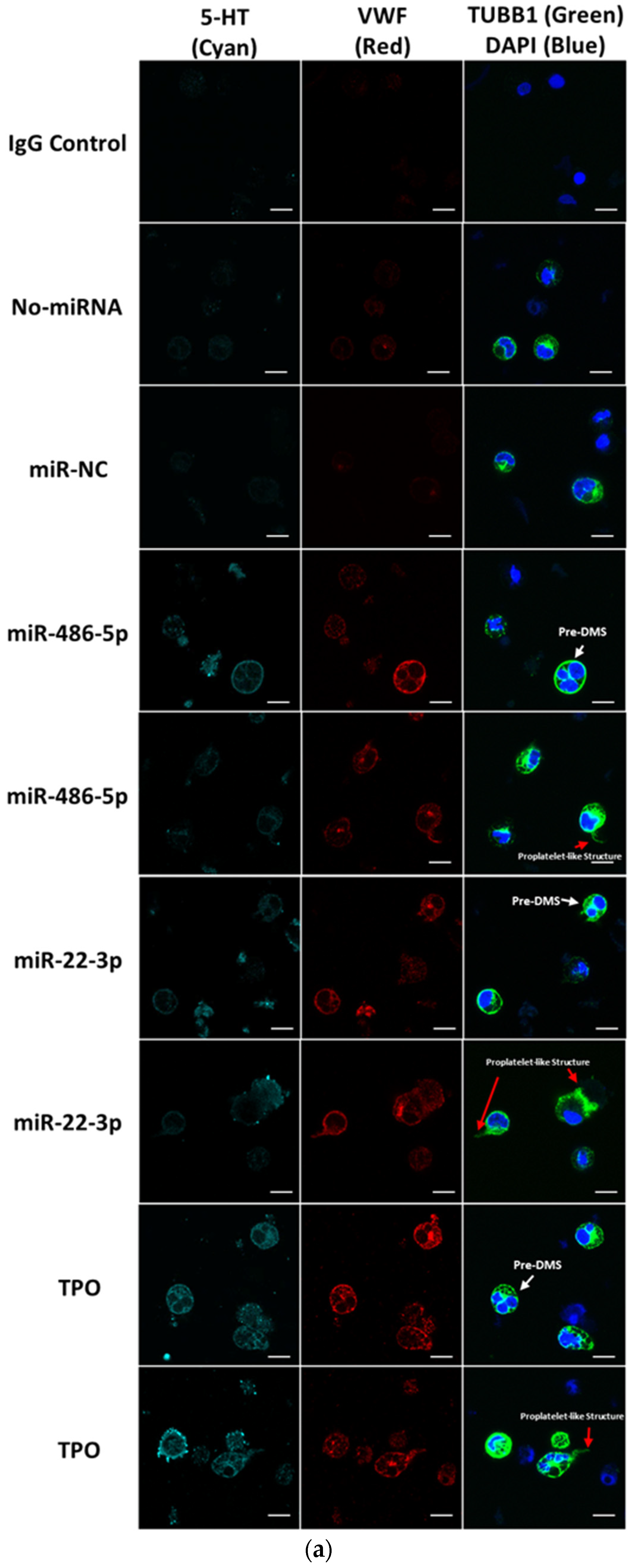
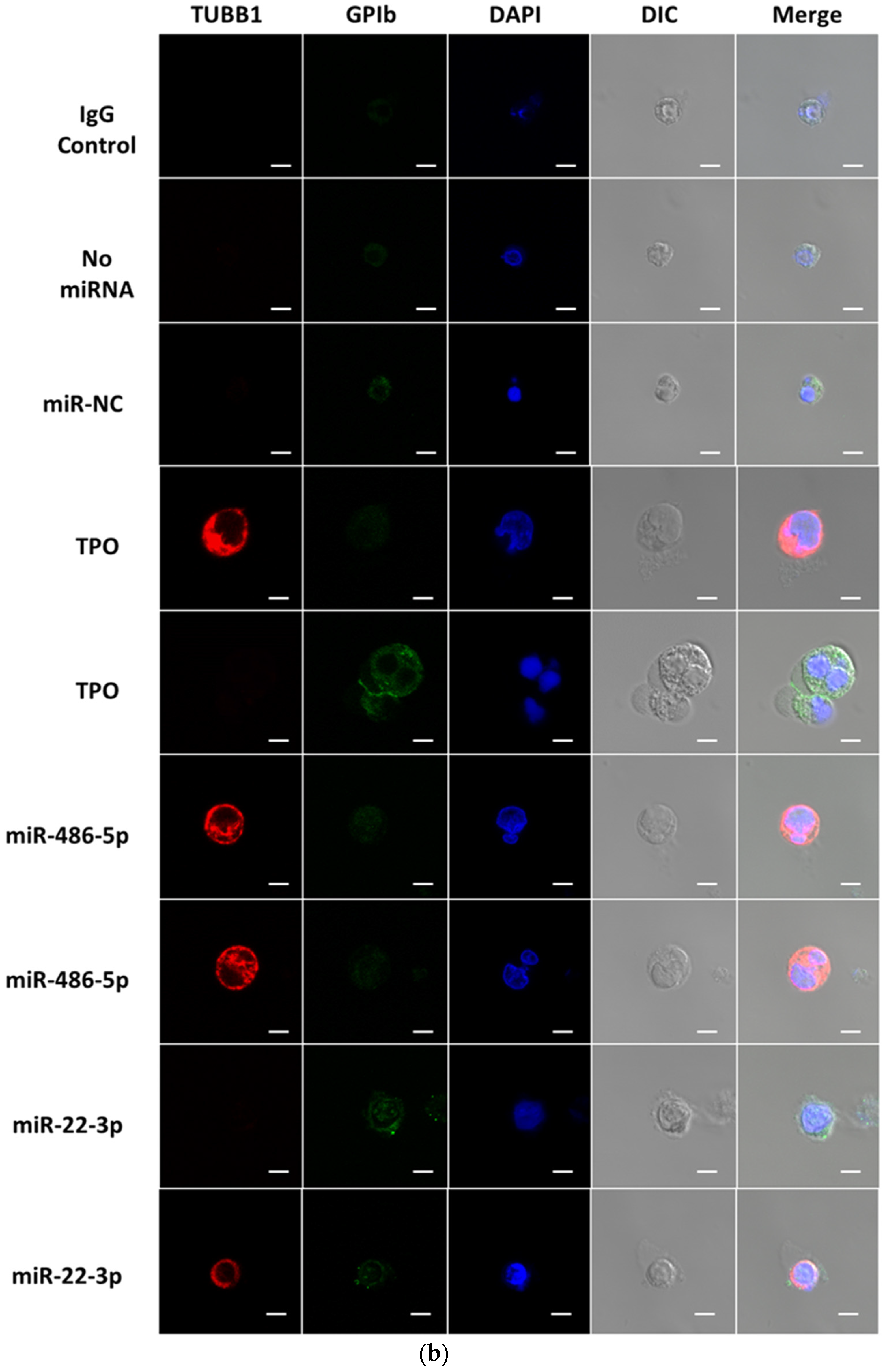
2.2. miR-486-5p in Combination with miR-22-3p Recapitulates, Partially at Least, the Megakaryopoietic Effect of TPO and MkMPs on CD34+ HSPCs
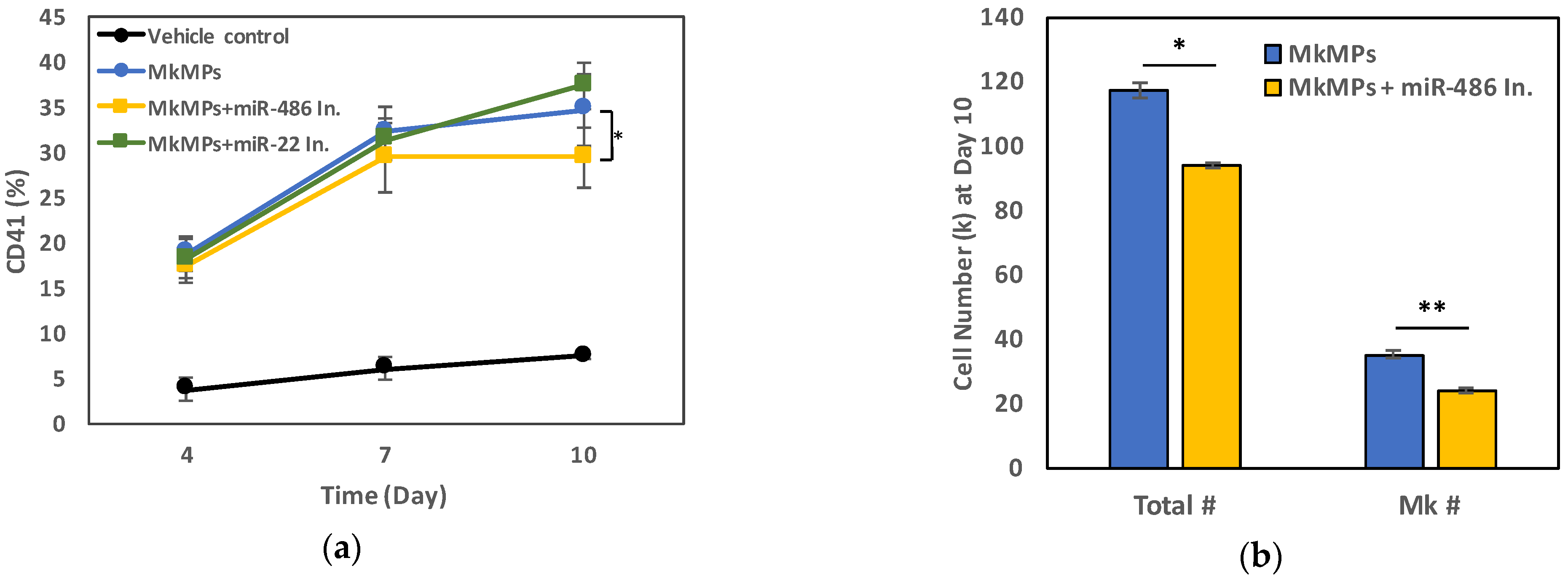
2.3. miR-486-5p in MkMPs Is an Essential Mediator of MkMP-Induced Megakaryocytic Differentiation
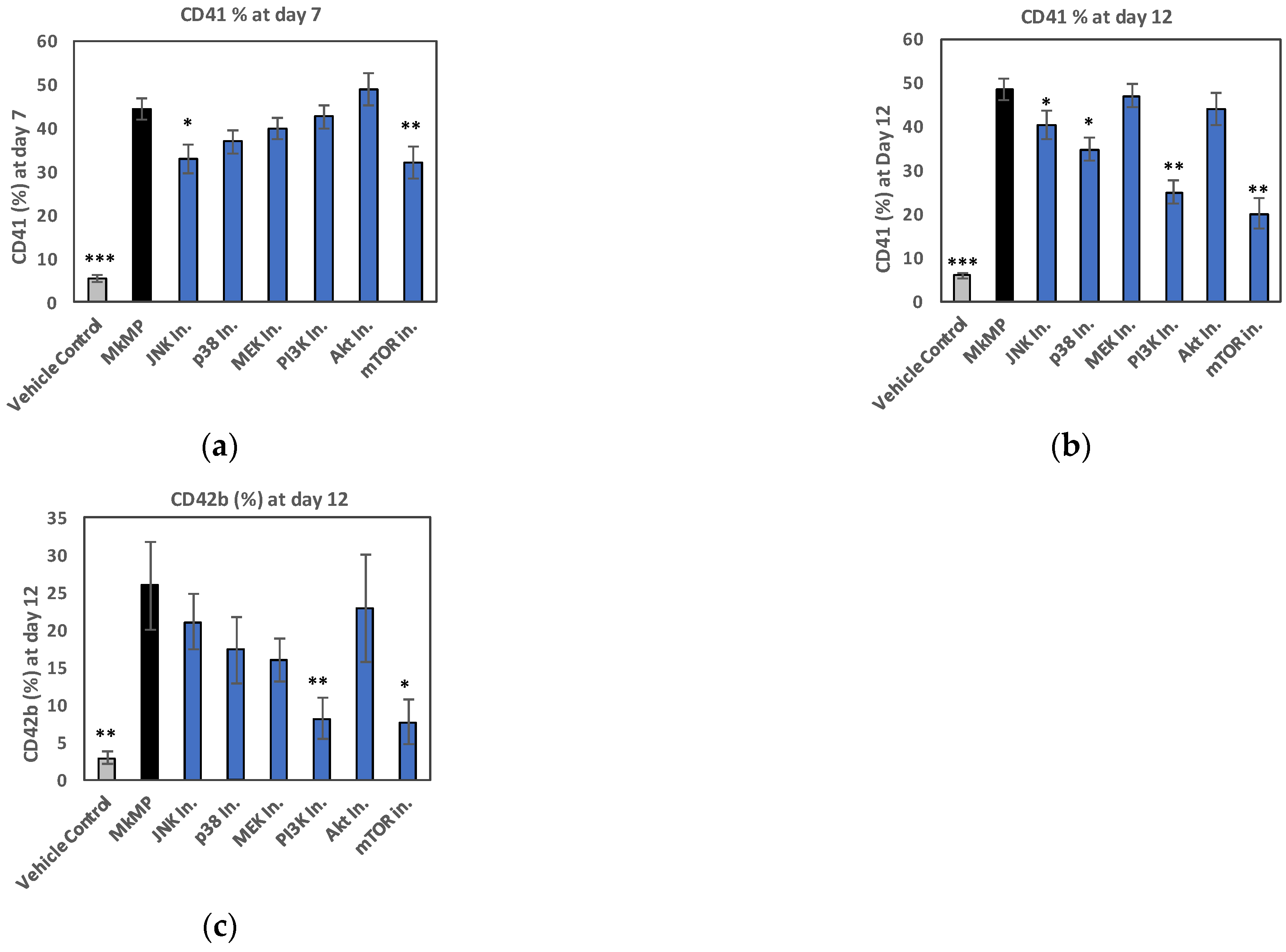
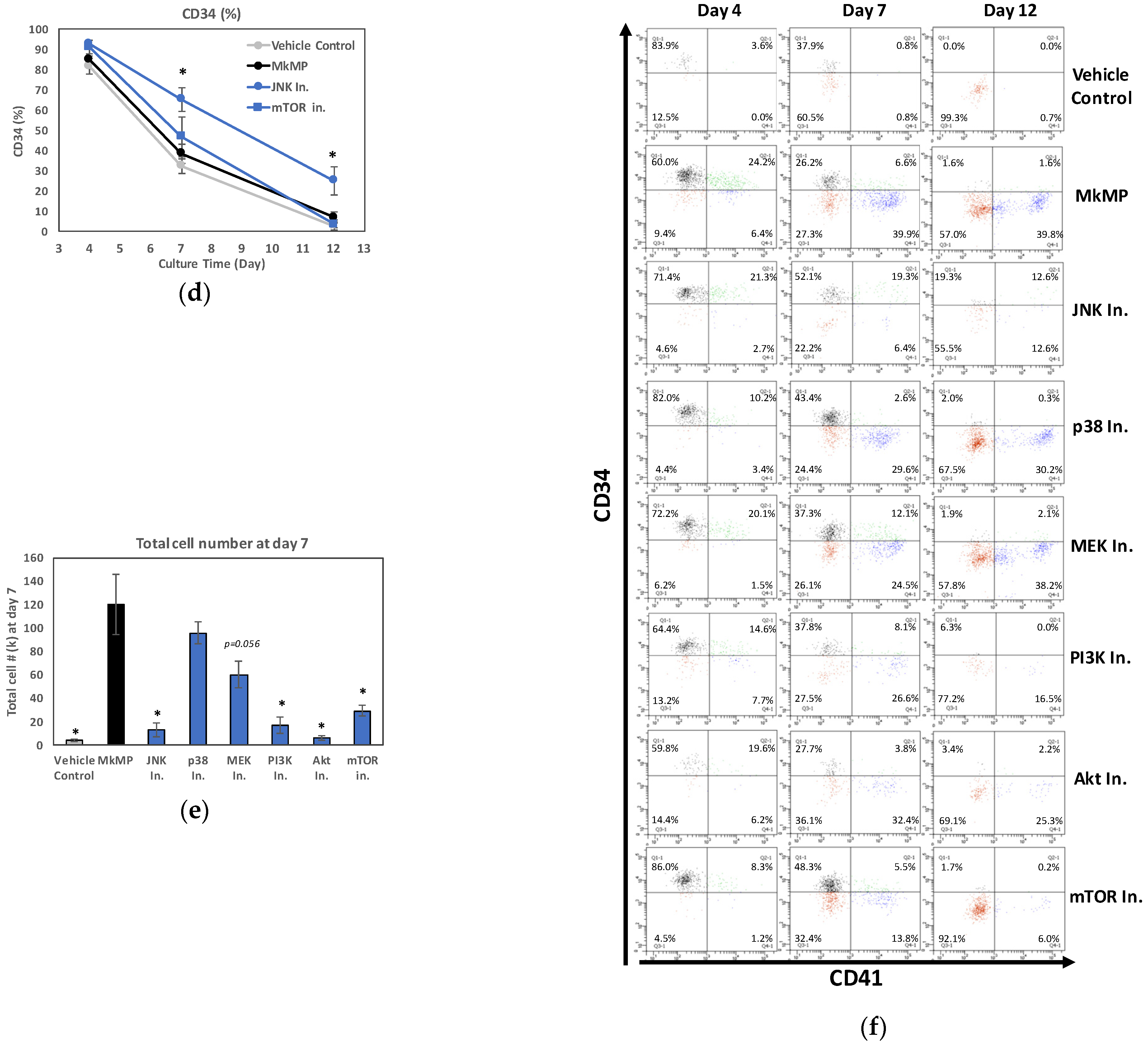
2.4. Use of Signaling-Pathway Inhibitors Suggests That JNK and PI3K/Akt/mTOR Signaling Regulate MkMP-Induced Mk Differentiation of HSPCs
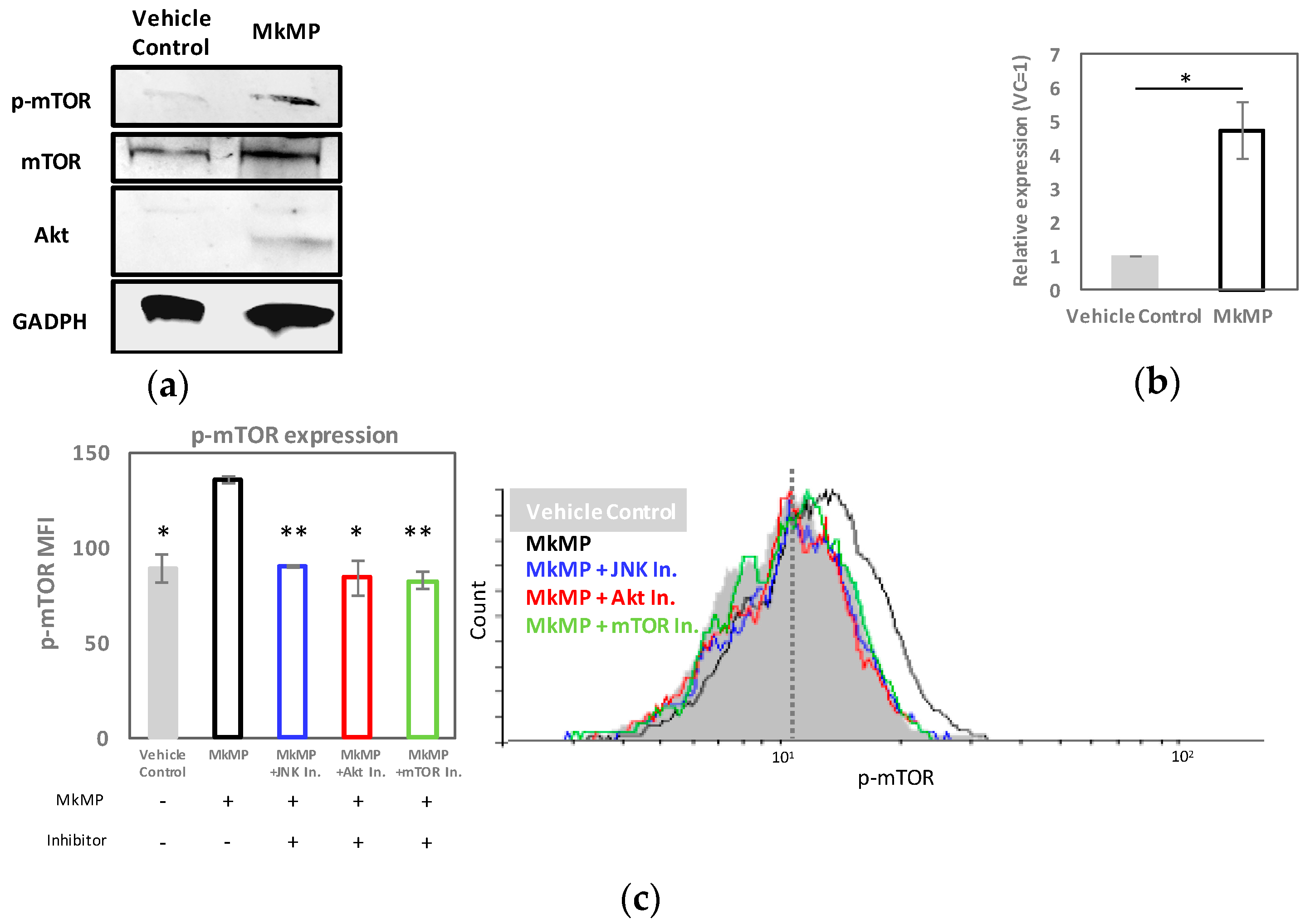
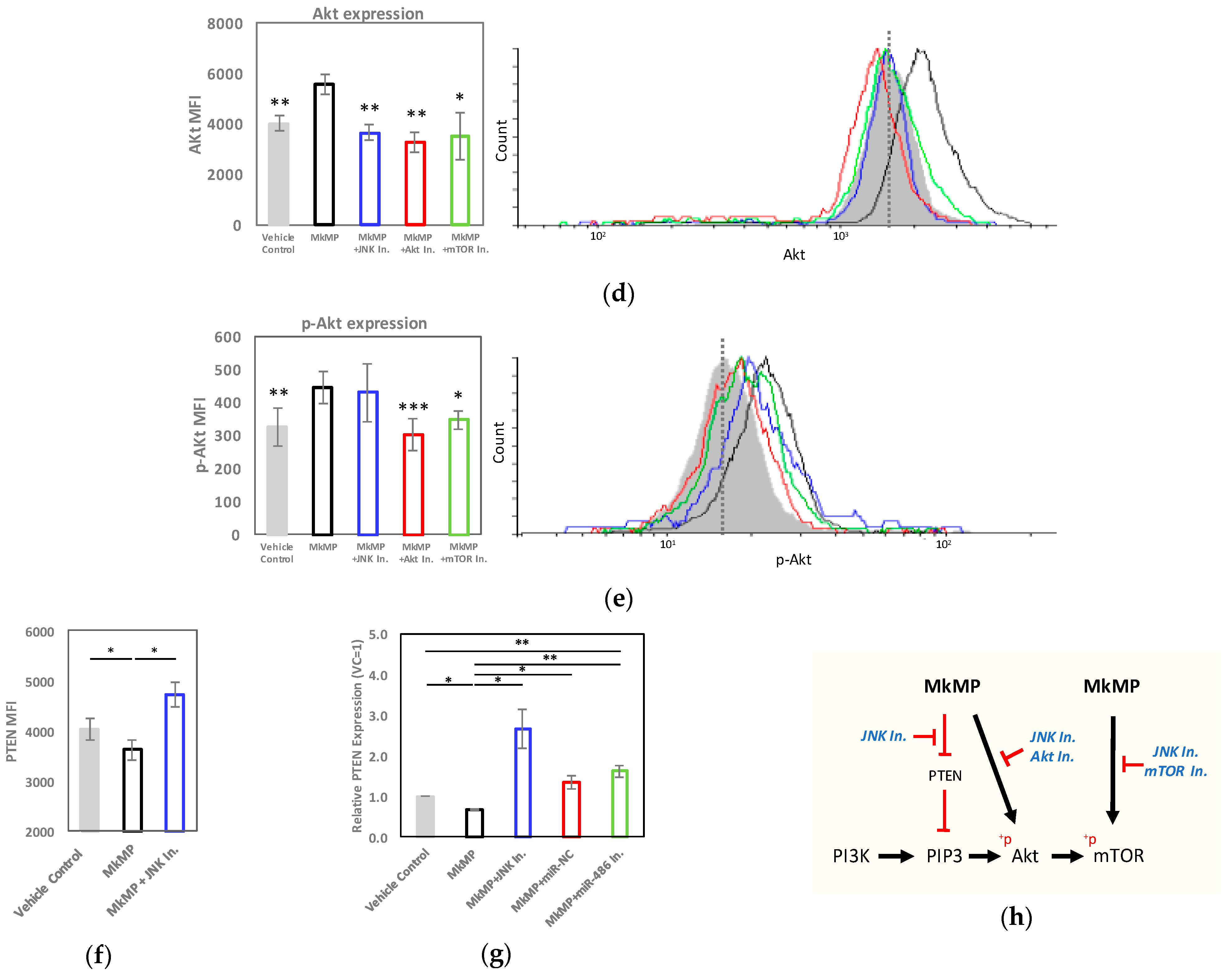
2.5. MkMPs Target JNK-Mediated PI3K/Akt/mTOR Signaling in HSPCs
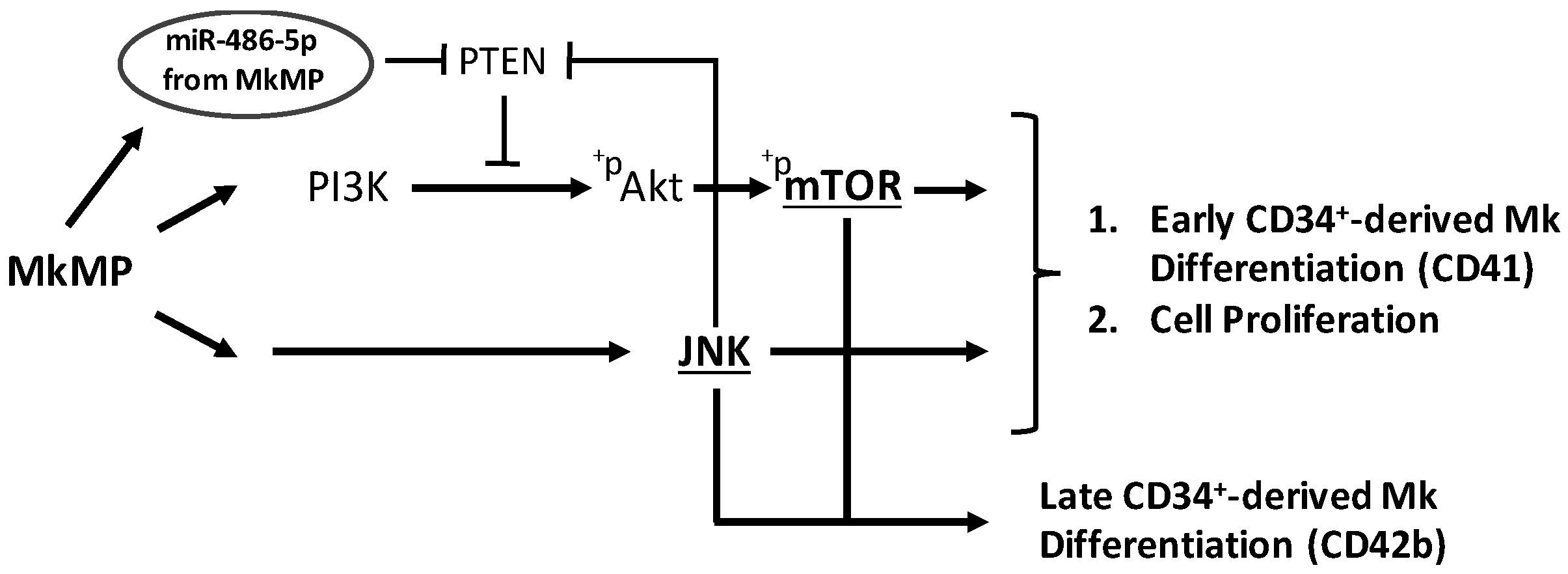
3. Discussion
3.1. Synergistic Action of Two miRs in Emulating TPO-like Signaling Leading to Megakaryocytic Differentiation of HSPCs in the Absence of TPO
3.2. JNK and Akt/mTOR Signaling in MkMP-Induced Mk Differentiation of CD34+ HSPCs
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Generation of Megakaryocytic MPs (MkMPs) from Cultured Megakaryocytes (Mks) Starting with CD34+ HSPCs
4.3. Isolation of Platelet-like Particles (PLPs) and Megakaryocytic Microparticles (MkMPs)
4.4. Human Platelets
4.5. RNA Extraction and Library Preparation for RNAseq Analysis
4.6. RNAseq Data Analysis
4.7. Transfection of CD34+ HSPCs with miR Mimics
4.8. miR-Inhibitor Experiments
4.9. Signaling-Inhibitor Experiments
4.10. Immunoblotting
4.11. Quantitative Reverse Transcription PCR (qRT-PCR)
4.12. Intracellular Protein Analysis by Flow Cytometry
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Cocucci, E.; Racchetti, G.; Meldolesi, J. Shedding microvesicles: Artefacts no more. Trends Cell Biol. 2009, 19, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Deregibus, M.C.; Cantaluppi, V.; Calogero, R.; Lo Iacono, M.; Tetta, C.; Biancone, L.; Bruno, S.; Bussolati, B.; Camussi, G. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood 2007, 110, 2440–2448. [Google Scholar] [CrossRef] [PubMed]
- Laffont, B.; Corduan, A.; Ple, H.; Duchez, A.C.; Cloutier, N.; Boilard, E.; Provost, P. Activated platelets can deliver mRNA regulatory Ago2*microRNA complexes to endothelial cells via microparticles. Blood 2013, 122, 253–261. [Google Scholar] [CrossRef]
- Bruno, S.; Grange, C.; Deregibus, M.C.; Calogero, R.A.; Saviozzi, S.; Collino, F.; Morando, L.; Busca, A.; Falda, M.; Bussolati, B.; et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J. Am. Soc. Nephrol. 2009, 20, 1053–1067. [Google Scholar] [CrossRef]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Das, S.; Halushka, M.K. Extracellular vesicle microRNA transfer in cardiovascular disease. Cardiovasc. Pathol. 2015, 24, 199–206. [Google Scholar] [CrossRef]
- Chen, J.; Hu, C.; Pan, P. Extracellular Vesicle MicroRNA Transfer in Lung Diseases. Front. Physiol. 2017, 8, 1028. [Google Scholar] [CrossRef]
- Morhayim, J.; van de Peppel, J.; Braakman, E.; Rombouts, E.W.; Ter Borg, M.N.; Dudakovic, A.; Chiba, H.; van der Eerden, B.C.; Raaijmakers, M.H.; van Wijnen, A.J.; et al. Osteoblasts secrete miRNA-containing extracellular vesicles that enhance expansion of human umbilical cord blood cells. Sci. Rep. 2016, 6, 32034. [Google Scholar] [CrossRef]
- Vinas, J.L.; Burger, D.; Zimpelmann, J.; Haneef, R.; Knoll, W.; Campbell, P.; Gutsol, A.; Carter, A.; Allan, D.S.; Burns, K.D. Transfer of microRNA-486-5p from human endothelial colony forming cell-derived exosomes reduces ischemic kidney injury. Kidney Int. 2016, 90, 1238–1250. [Google Scholar] [CrossRef]
- Shu, J.; Silva, B.; Gao, T.; Xu, Z.; Cui, J. Dynamic and Modularized MicroRNA Regulation and Its Implication in Human Cancers. Sci. Rep. 2017, 7, 13356. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Catchpoole, D.R.; Kennedy, P.J.; Li, J. Identification of lung cancer miRNA-miRNA co-regulation networks through a progressive data refining approach. J. Theor. Biol. 2015, 380, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Cursons, J.; Pillman, K.A.; Scheer, K.G.; Gregory, P.A.; Foroutan, M.; Hediyeh-Zadeh, S.; Toubia, J.; Crampin, E.J.; Goodall, G.J.; Bracken, C.P.; et al. Combinatorial Targeting by MicroRNAs Co-ordinates Post-transcriptional Control of EMT. Cell Syst. 2018, 7, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Yoo, J.J.; Atala, A.; Lee, S.J. Combination of small RNAs for skeletal muscle regeneration. FASEB J. 2016, 30, 1198–1206. [Google Scholar] [CrossRef]
- Xu, J.; Feng, Y.; Jeyaram, A.; Jay, S.M.; Zou, L.; Chao, W. Circulating Plasma Extracellular Vesicles from Septic Mice Induce Inflammation via MicroRNA- and TLR7-Dependent Mechanisms. J. Immunol. 2018, 201, 3392–3400. [Google Scholar] [CrossRef] [PubMed]
- Panuganti, S.; Schlinker, A.C.; Lindholm, P.F.; Papoutsakis, E.T.; Miller, W.M. Three-stage ex vivo expansion of high-ploidy megakaryocytic cells: Toward large-scale platelet production. Tissue Eng. Part A 2013, 19, 998–1014. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Woulfe, D.S.; Papoutsakis, E.T. Shear enhances thrombopoiesis and formation of microparticles that induce megakaryocytic differentiation of stem cells. Blood 2014, 124, 2094–2103. [Google Scholar] [CrossRef]
- Jiang, J.; Kao, C.Y.; Papoutsakis, E.T. How do megakaryocytic microparticles target and deliver cargo to alter the fate of hematopoietic stem cells? J. Control. Release 2017, 247, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Escobar, C.; Kao, C.Y.; Das, S.; Papoutsakis, E.T. Human megakaryocytic microparticles induce de novo platelet biogenesis in a wild-type murine model. Blood Adv. 2020, 4, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Kaushansky, K. Molecular mechanisms of thrombopoietin signaling. J. Thromb. Haemost. 2009, 7, 235–238. [Google Scholar] [CrossRef]
- de Graaf, C.A.; Metcalf, D. Thrombopoietin and hematopoietic stem cells. Cell Cycle 2011, 10, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Drachman, J.G.; Millett, K.M.; Kaushansky, K. Thrombopoietin signal transduction requires functional JAK2, not TYK2. J. Biol. Chem. 1999, 274, 13480–13484. [Google Scholar] [CrossRef] [PubMed]
- Geddis, A.E.; Fox, N.E.; Kaushansky, K. Phosphatidylinositol 3-kinase is necessary but not sufficient for thrombopoietin-induced proliferation in engineered Mpl-bearing cell lines as well as in primary megakaryocytic progenitors. J. Biol. Chem. 2001, 276, 34473–34479. [Google Scholar] [CrossRef] [PubMed]
- Rojnuckarin, P.; Drachman, J.G.; Kaushansky, K. Thrombopoietin-induced activation of the mitogen-activated protein kinase (MAPK) pathway in normal megakaryocytes: Role in endomitosis. Blood 1999, 94, 1273–1282. [Google Scholar] [CrossRef] [PubMed]
- Kirito, K.; Fox, N.; Kaushansky, K. Thrombopoietin stimulates Hoxb4 expression: An explanation for the favorable effects of TPO on hematopoietic stem cells. Blood 2003, 102, 3172–3178. [Google Scholar] [CrossRef]
- Miyazaki, R.; Ogata, H.; Kobayashi, Y. Requirement of thrombopoietin-induced activation of ERK for megakaryocyte differentiation and of p38 for erythroid differentiation. Ann. Hematol. 2001, 80, 284–291. [Google Scholar] [CrossRef]
- Nagata, Y.; Nishida, E.; Todokoro, K. Activation of JNK signaling pathway by erythropoietin, thrombopoietin, and interleukin-3. Blood 1997, 89, 2664–2669. [Google Scholar] [CrossRef]
- Kadmon, C.S.; Landers, C.T.; Li, H.S.; Watowich, S.S.; Rodriguez, A.; King, K.Y. MicroRNA-22 controls interferon alpha production and erythroid maturation in response to infectious stress in mice. Exp. Hematol. 2017, 56, 7–15. [Google Scholar] [CrossRef]
- Wang, L.S.; Li, L.; Li, L.; Chu, S.; Shiang, K.D.; Li, M.; Sun, H.Y.; Xu, J.; Xiao, F.J.; Sun, G.; et al. MicroRNA-486 regulates normal erythropoiesis and enhances growth and modulates drug response in CML progenitors. Blood 2015, 125, 1302–1313. [Google Scholar] [CrossRef]
- Kao, C.Y.; Papoutsakis, E.T. Engineering human megakaryocytic microparticles for targeted delivery of nucleic acids to hematopoietic stem and progenitor cells. Sci. Adv. 2018, 4, eaau6762. [Google Scholar] [CrossRef]
- Kamat, V.; Paluru, P.; Myint, M.; French, D.L.; Gadue, P.; Diamond, S.L. MicroRNA screen of human embryonic stem cell differentiation reveals miR-105 as an enhancer of megakaryopoiesis from adult CD34+ cells. Stem Cells 2014, 32, 1337–1346. [Google Scholar] [CrossRef]
- Bianchi, E.; Bulgarelli, J.; Ruberti, S.; Rontauroli, S.; Sacchi, G.; Norfo, R.; Pennucci, V.; Zini, R.; Salati, S.; Prudente, Z.; et al. MYB controls erythroid versus megakaryocyte lineage fate decision through the miR-486-3p-mediated downregulation of MAF. Cell Death Differ. 2015, 22, 1906–1921. [Google Scholar] [CrossRef] [PubMed]
- Liao, F.L.; Tan, L.; Liu, H.; Wang, J.J.; Ma, X.T.; Zhao, B.; Chen, Y.; Bihl, J.; Yang, Y.; Chen, R.L. Hematopoietic stem cell-derived exosomes promote hematopoietic differentiation of mouse embryonic stem cells in vitro via inhibiting the miR126/Notch1 pathway. Acta Pharmacol. Sin. 2018, 39, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Eckly, A.; Heijnen, H.; Pertuy, F.; Geerts, W.; Proamer, F.; Rinckel, J.Y.; Leon, C.; Lanza, F.; Gachet, C. Biogenesis of the demarcation membrane system (DMS) in megakaryocytes. Blood 2014, 123, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Fugman, D.A.; Witte, D.P.; Jones, C.L.; Aronow, B.J.; Lieberman, M.A. In vitro establishment and characterization of a human megakaryoblastic cell line. Blood 1990, 75, 1252–1261. [Google Scholar] [CrossRef] [PubMed]
- Polak, R.; Buitenhuis, M. The PI3K/PKB signaling module as key regulator of hematopoiesis: Implications for therapeutic strategies in leukemia. Blood 2012, 119, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Grindley, J.C.; Yin, T.; Jayasinghe, S.; He, X.C.; Ross, J.T.; Haug, J.S.; Rupp, D.; Porter-Westpfahl, K.S.; Wiedemann, L.M.; et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 2006, 441, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Choorapoikayil, S.; Kers, R.; Herbomel, P.; Kissa, K.; den Hertog, J. Pivotal role of Pten in the balance between proliferation and differentiation of hematopoietic stem cells in zebrafish. Blood 2014, 123, 184–190. [Google Scholar] [CrossRef]
- Weng, Z.; Li, D.; Zhang, L.; Chen, J.; Ruan, C.; Chen, G.; Gartner, T.K.; Liu, J. PTEN regulates collagen-induced platelet activation. Blood 2010, 116, 2579–2581. [Google Scholar] [CrossRef]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, e1413. [Google Scholar] [CrossRef]
- Chattapadhyaya, S.; Haldar, S.; Banerjee, S. Microvesicles promote megakaryopoiesis by regulating DNA methyltransferase and methylation of Notch1 promoter. J. Cell. Physiol. 2020, 235, 2619–2630. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sankaran, V.G.; Lodish, H.F. MicroRNAs in erythroid and megakaryocytic differentiation and megakaryocyte-erythroid progenitor lineage commitment. Leukemia 2012, 26, 2310–2316. [Google Scholar] [CrossRef] [PubMed]
- Undi, R.B.; Kandi, R.; Gutti, R.K. MicroRNAs as Haematopoiesis Regulators. Adv. Hematol. 2013, 2013, 695754. [Google Scholar] [CrossRef] [PubMed]
- Barroga, C.F.; Pham, H.; Kaushansky, K. Thrombopoietin regulates c-Myb expression by modulating micro RNA 150 expression. Exp. Hematol. 2008, 36, 1585–1592. [Google Scholar] [CrossRef]
- Lu, J.; Guo, S.; Ebert, B.L.; Zhang, H.; Peng, X.; Bosco, J.; Pretz, J.; Schlanger, R.; Wang, J.Y.; Mak, R.H.; et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev. Cell 2008, 14, 843–853. [Google Scholar] [CrossRef]
- Jiang, X.; Hu, C.; Arnovitz, S.; Bugno, J.; Yu, M.; Zuo, Z.; Chen, P.; Huang, H.; Ulrich, B.; Gurbuxani, S.; et al. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat. Commun. 2016, 7, 11452. [Google Scholar] [CrossRef]
- Wurm, A.A.; Tenen, D.G.; Behre, G. The Janus-faced Nature of miR-22 in Hematopoiesis: Is It an Oncogenic Tumor Suppressor or Rather a Tumor-Suppressive Oncogene? PLoS Genet. 2017, 13, e1006505. [Google Scholar] [CrossRef]
- Song, S.J.; Ito, K.; Ala, U.; Kats, L.; Webster, K.; Sun, S.M.; Jongen-Lavrencic, M.; Manova-Todorova, K.; Teruya-Feldstein, J.; Avigan, D.E.; et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 2013, 13, 87–101. [Google Scholar] [CrossRef]
- Weiss, C.N.; Ito, K. microRNA-22 promotes megakaryocyte differentiation through repression of its target, GFI1. Blood Adv. 2019, 3, 33–46. [Google Scholar] [CrossRef]
- Lv, K.T.; Liu, Z.; Feng, J.; Zhao, W.; Hao, T.; Ding, W.Y.; Chu, J.P.; Gao, L.J. MiR-22-3p Regulates Cell Proliferation and Inhibits Cell Apoptosis through Targeting the eIF4EBP3 Gene in Human Cervical Squamous Carcinoma Cells. Int. J. Med. Sci. 2018, 15, 142–152. [Google Scholar] [CrossRef]
- Willeit, P.; Zampetaki, A.; Dudek, K.; Kaudewitz, D.; King, A.; Kirkby, N.S.; Crosby-Nwaobi, R.; Prokopi, M.; Drozdov, I.; Langley, S.R.; et al. Circulating microRNAs as novel biomarkers for platelet activation. Circ. Res. 2013, 112, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Nagalla, S.; Shaw, C.; Kong, X.; Kondkar, A.A.; Edelstein, L.C.; Ma, L.; Chen, J.; McKnight, G.S.; Lopez, J.A.; Yang, L.; et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood 2011, 117, 5189–5197. [Google Scholar] [CrossRef] [PubMed]
- Sunderland, N.; Skroblin, P.; Barwari, T.; Huntley, R.P.; Lu, R.; Joshi, A.; Lovering, R.C.; Mayr, M. MicroRNA Biomarkers and Platelet Reactivity: The Clot Thickens. Circ. Res. 2017, 120, 418–435. [Google Scholar] [CrossRef] [PubMed]
- Kaudewitz, D.; Skroblin, P.; Bender, L.H.; Barwari, T.; Willeit, P.; Pechlaner, R.; Sunderland, N.P.; Willeit, K.; Morton, A.C.; Armstrong, P.C.; et al. Association of MicroRNAs and YRNAs With Platelet Function. Circ. Res. 2016, 118, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Juzenas, S.; Venkatesh, G.; Hubenthal, M.; Hoeppner, M.P.; Du, Z.G.; Paulsen, M.; Rosenstiel, P.; Senger, P.; Hofmann-Apitius, M.; Keller, A.; et al. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res. 2017, 45, 9290–9301. [Google Scholar] [CrossRef]
- Severin, S.; Ghevaert, C.; Mazharian, A. The mitogen-activated protein kinase signaling pathways: Role in megakaryocyte differentiation. J. Thromb. Haemost. 2010, 8, 17–26. [Google Scholar] [CrossRef]
- Xiao, X.; Lai, W.; Xie, H.; Liu, Y.; Guo, W.; Liu, Y.; Li, Y.; Li, Y.; Zhang, J.; Chen, W.; et al. Targeting JNK pathway promotes human hematopoietic stem cell expansion. Cell Discov. 2019, 5, 2. [Google Scholar] [CrossRef]
- Luff, S.A.; Papoutsakis, E.T. Megakaryocytic Maturation in Response to Shear Flow Is Mediated by the Activator Protein 1 (AP-1) Transcription Factor via Mitogen-activated Protein Kinase (MAPK) Mechanotransduction. J. Biol. Chem. 2016, 291, 7831–7843. [Google Scholar] [CrossRef]
- Raslova, H.; Baccini, V.; Loussaief, L.; Comba, B.; Larghero, J.; Debili, N.; Vainchenker, W. Mammalian target of rapamycin (mTOR) regulates both proliferation of megakaryocyte progenitors and late stages of megakaryocyte differentiation. Blood 2006, 107, 2303–2310. [Google Scholar] [CrossRef]
- Hettinger, K.; Vikhanskaya, F.; Poh, M.K.; Lee, M.K.; de Belle, I.; Zhang, J.T.; Reddy, S.A.; Sabapathy, K. c-Jun promotes cellular survival by suppression of PTEN. Cell Death Differ. 2007, 14, 218–229. [Google Scholar] [CrossRef]
- Yang, Y.; Ji, C.; Guo, S.; Su, X.; Zhao, X.; Zhang, S.; Liu, G.; Qiu, X.; Zhang, Q.; Guo, H.; et al. The miR-486-5p plays a causative role in prostate cancer through negative regulation of multiple tumor suppressor pathways. Oncotarget 2017, 8, 72835–72846. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.J.; Yuan, W.D.; Yuan, J.Q.; Yuan, K.; Wang, Y. miR-486-5p functions as an oncogene by targeting PTEN in non-small cell lung cancer. Pathol. Res. Pract. 2018, 214, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Meyers, B.C.; Green, P.J. Construction of small RNA cDNA libraries for deep sequencing. Methods 2007, 43, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Apostolidis, P.A.; Woulfe, D.S.; Chavez, M.; Miller, W.M.; Papoutsakis, E.T. Role of tumor suppressor p53 in megakaryopoiesis and platelet function. Exp. Hematol. 2012, 40, 131–142. [Google Scholar] [CrossRef]
- Giammona, L.M.; Panuganti, S.; Kemper, J.M.; Apostolidis, P.A.; Lindsey, S.; Papoutsakis, E.T.; Miller, W.M. Mechanistic studies on the effects of nicotinamide on megakaryocytic polyploidization and the roles of NAD(+) levels and SIRT inhibition. Exp. Hematol. 2009, 37, 1340–1352. [Google Scholar] [CrossRef][Green Version]
- Giammona, L.M.; Fuhrken, P.G.; Papoutsakis, E.T.; Miller, W.M. Nicotinamide (vitamin B3) increases the polyploidisation and proplatelet formation of cultured primary human megakaryocytes. Br. J. Haematol. 2006, 135, 554–566. [Google Scholar] [CrossRef]
- Lindsey, S.; Papoutsakis, E.T. The aryl hydrocarbon receptor (AHR) transcription factor regulates megakaryocytic polyploidization. Br. J. Haematol. 2011, 152, 469–484. [Google Scholar] [CrossRef]
- Baker, J.E.; Su, J.; Koprowski, S.; Dhanasekaran, A.; Aufderheide, T.P.; Gross, G.J. Thrombopoietin receptor agonists protect human cardiac myocytes from injury by activation of cell survival pathways. J. Pharmacol. Exp. Ther. 2015, 352, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, R.; Parolini, I.; Testa, U.; Samoggia, P.; Petrucci, E.; Sargiacomo, M.; Chelucci, C.; Gabbianelli, M.; Peschle, C. Inhibition of TPO-induced MEK or mTOR activity induces opposite effects on the ploidy of human differentiating megakaryocytes. J. Cell Sci. 2006, 119, 744–752. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luff, S.A.; Kao, C.Y.; Papoutsakis, E.T. Role of p53 and transcription-independent p53-induced apoptosis in shear-stimulated megakaryocytic maturation, particle generation, and platelet biogenesis. PLoS ONE 2018, 13, e0203991. [Google Scholar] [CrossRef] [PubMed]


| Rank | NCBI Gene ID | miRNA in Mks | CPM | % | NCBI Gene ID | miRNA in MkMPs | CPM | % |
| 1 | 406966 | miR-191-5p | 231,441 | 23.1 | 723876 | miR-486-5p | 164,087 | 16.4 |
| 2 | 723876 | miR-486-5p | 195,803 | 19.6 | 406966 | miR-191-5p | 117,782 | 11.8 |
| 3 | 407056 | miR-99b-5p | 164,315 | 16.4 | 407015/407016 | miR-26a-5p | 91,774 | 9.2 |
| 4 | 574447 | miR-146b-5p | 65,671 | 6.6 | 406888/406889 | let-7f-5p | 68,251 | 6.8 |
| 5 | 407015/407016 | miR-26a-5p | 39,625 | 4.0 | 407048/407049 | miR-92a-3p | 44,480 | 4.4 |
| 6 | 406888/406889 | let-7f-5p | 23,623 | 2.4 | 406913 | miR-126-5p | 44,218 | 4.4 |
| 7 | 406913 | miR-126-5p | 22,642 | 2.3 | 407004 | miR-22-3p | 43,152 | 4.3 |
| 8 | 406938 | miR-146a-5p | 21,993 | 2.2 | 406991 | miR-21-5p | 38,617 | 3.9 |
| 9 | 406991 | miR-21-5p | 20,197 | 2.0 | 574447 | miR-146b-5p | 34,455 | 3.4 |
| 10 | 406940 | miR-148a-3p | 17,054 | 1.7 | 406995/406996 | miR-181a-5p | 33,752 | 3.4 |
| Other miRNAs | 19.8 | Other miRNAs | 31.9 | |||||
| Rank | NCBI Gene ID | miRNA in PLTs | CPM | % | NCBI Gene ID | miRNA in PLPs | CPM | % |
| 1 | 406966 | miR-191-5p | 263,871 | 26.4 | 406966 | miR-191-5p | 162,495 | 16.2 |
| 2 | 723876 | miR-486-5p | 80,546 | 8.1 | 723876 | miR-486-5p | 146,874 | 14.7 |
| 3 | 406888/406889 | let-7f-5p | 76,283 | 7.6 | 407015/407016 | miR-26a-5p | 84,857 | 8.5 |
| 4 | 407056 | miR-99b-5p | 71,050 | 7.1 | 406888/406889 | let-7f-5p | 64,288 | 6.4 |
| 5 | 406902 | miR-10a-5p | 54,139 | 5.4 | 574447 | miR-146b-5p | 49,986 | 5.0 |
| 6 | 407015/407016 | miR-26a-5p | 51,712 | 5.2 | 406913 | miR-126-5p | 43,506 | 4.4 |
| 7 | 406938 | miR-146a-5p | 43,647 | 4.4 | 407056 | miR-99b-5p | 42,826 | 4.3 |
| 8 | 407048/407049 | miR-92a-3p | 43,032 | 4.3 | 406991 | miR-21-5p | 40,419 | 4.0 |
| 9 | 574447 | miR-146b-5p | 25,104 | 2.5 | 407004 | miR-22-3p | 32,727 | 3.3 |
| 10 | 406995/406996 | miR-181a-5p | 23,932 | 2.4 | 406995/406996 | miR-181a-5p | 25,520 | 2.6 |
| Other miRNAs | 26.7 | Other miRNAs | 30.7 |
| Signaling Inhibitor | Target | Pathway | Concentration |
|---|---|---|---|
| SP600125 | JNK | MAPK | 10 μM |
| SB203580 | p38 | 10 μM | |
| PD98059 | MEK | 10 μM | |
| LY-294002 | PI3K | PI3K/Akt/mTOR | 10 μM |
| Wortmannin | Akt | 10 μM | |
| Rapamycin | mTOR | 10 μM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kao, C.-Y.; Jiang, J.; Thompson, W.; Papoutsakis, E.T. miR-486-5p and miR-22-3p Enable Megakaryocytic Differentiation of Hematopoietic Stem and Progenitor Cells without Thrombopoietin. Int. J. Mol. Sci. 2022, 23, 5355. https://doi.org/10.3390/ijms23105355
Kao C-Y, Jiang J, Thompson W, Papoutsakis ET. miR-486-5p and miR-22-3p Enable Megakaryocytic Differentiation of Hematopoietic Stem and Progenitor Cells without Thrombopoietin. International Journal of Molecular Sciences. 2022; 23(10):5355. https://doi.org/10.3390/ijms23105355
Chicago/Turabian StyleKao, Chen-Yuan, Jinlin Jiang, Will Thompson, and Eleftherios T. Papoutsakis. 2022. "miR-486-5p and miR-22-3p Enable Megakaryocytic Differentiation of Hematopoietic Stem and Progenitor Cells without Thrombopoietin" International Journal of Molecular Sciences 23, no. 10: 5355. https://doi.org/10.3390/ijms23105355
APA StyleKao, C.-Y., Jiang, J., Thompson, W., & Papoutsakis, E. T. (2022). miR-486-5p and miR-22-3p Enable Megakaryocytic Differentiation of Hematopoietic Stem and Progenitor Cells without Thrombopoietin. International Journal of Molecular Sciences, 23(10), 5355. https://doi.org/10.3390/ijms23105355







