Class III Peroxidases PRX01, PRX44, and PRX73 Control Root Hair Growth in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
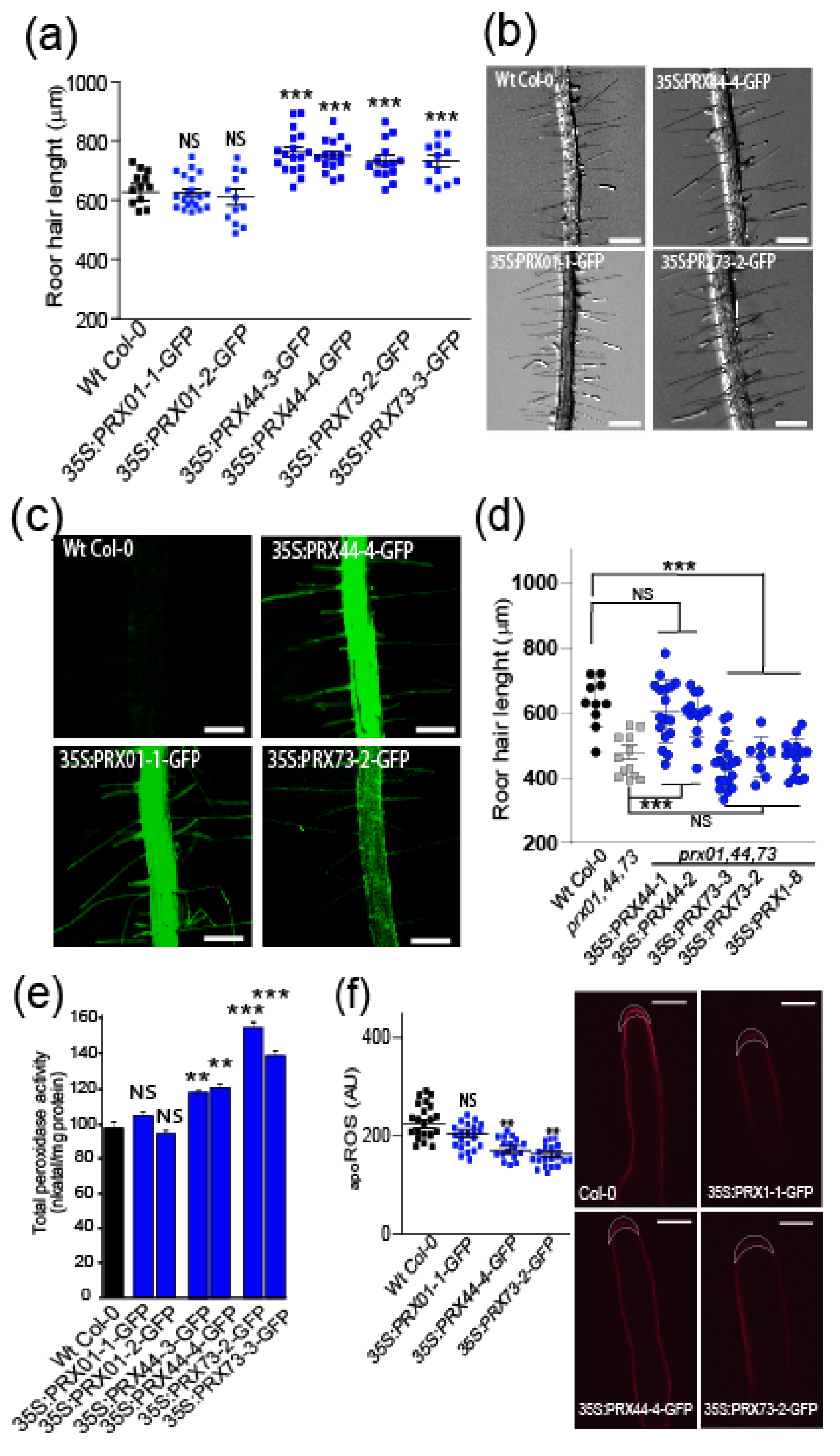
2.1. Class-III PRX01, PRX44, and PRX73 Are Required for ROS-Mediated Root Hair Growth
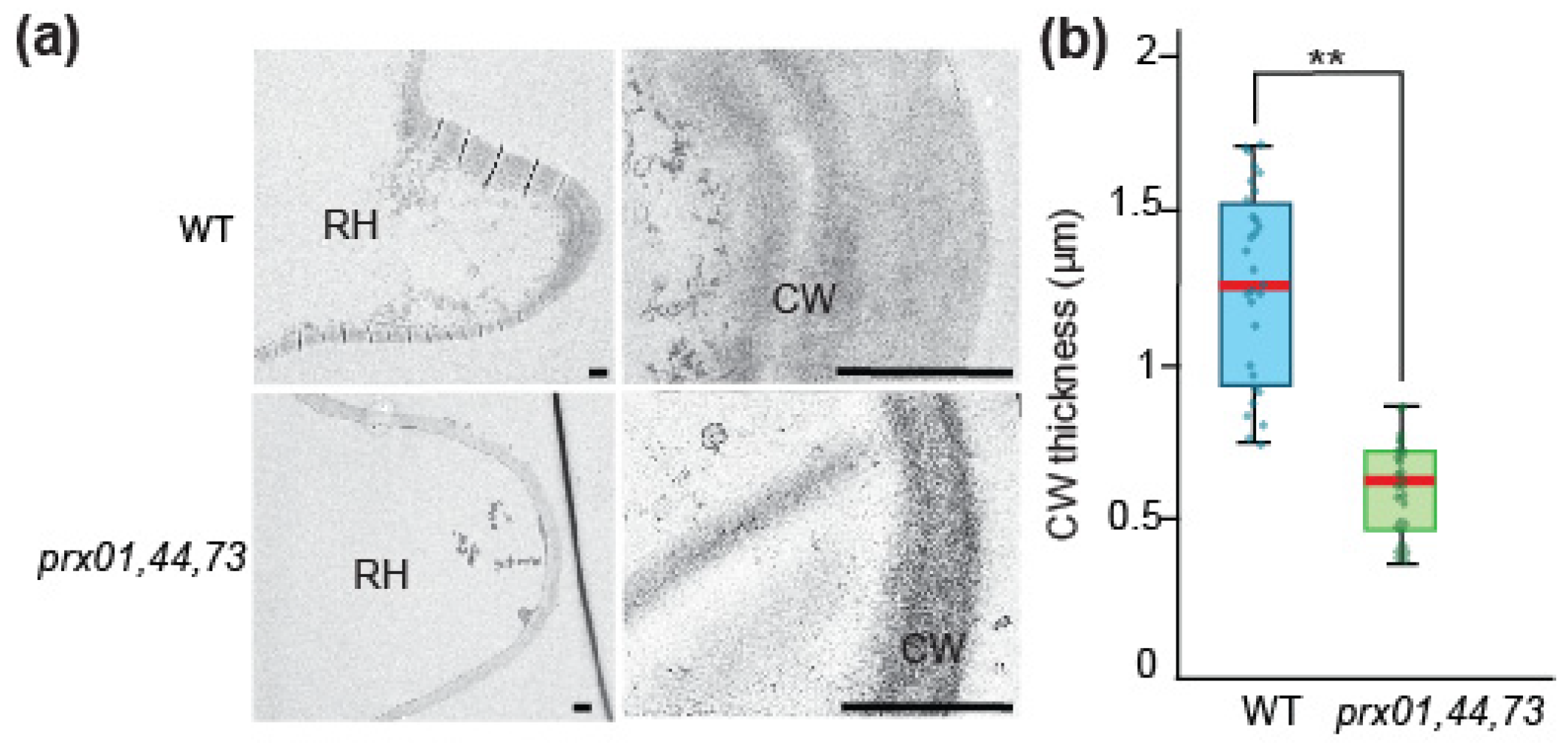
2.2. Changes in prx01,44,73 Affect Cell Wall Structure and EXT Retention in Root Hairs
2.3. PRX01, PRX44, and PRX73 Might Interact with EXTs
2.4. Phylogenetic Insights of PRX01, PRX44, and PRX73
3. Discussion
3.1. Proposal of how PRX01, PRX44, and PRX73 Functions in the Root Hair Cell Walls
3.2. Unique Functions of the EXT-PRXs in the Root Hair Cell Walls
4. Materials and Methods
4.1. Plant and Growth Conditions
4.2. PRX:GFP and 35S:PRX-GFP Lines
4.3. SS-TOM and SS-TOM-EXT-LONG Constructs
4.4. Root Hair Phenotype
4.5. Confocal Imaging
4.6. Peroxidase Activity
4.7. Cytoplasmic ROS (cytROS) Measurements
4.8. Apoplastic ROS (apoROS) Measurements
4.9. Phylogenetic Analysis
4.10. Co-Expression Analysis Network
4.11. Tyr-Crosslinking Analysis
4.12. Immunoblot Analysis
4.13. Transmission Electron Microscopy of Root Hair Cell Walls
4.14. Modeling and Molecular Docking between PRXs and EXTs
4.15. EXT Conformational Coarse-Grained Model
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lamport, D.T.A.; Kieliszewski, M.J.; Chen, Y.; Cannon, M.C. Role of the Extensin Superfamily in Primary Cell Wall Architecture. Plant Physiol. 2011, 156, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marzol, E.; Borassi, C.; Bringas, M.; Sede, A.; Rodríguez Garcia, D.R.; Capece, L. Filling the Gaps to Solve the Extensin Puzzle. Mol. Plant 2018, 11, 645–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumberger, N.; Ringli, C.; Keller, B. The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 2001, 15, 1128–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumberger, N.; Steiner, M.; Ryser, U.; Keller, B.; Ringli, C. Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J. 2003, 35, 71–81. [Google Scholar] [CrossRef]
- Ringli, C. The hydroxyproline-rich glycoprotein domain of the Arabidopsis LRX1 requires Tyr for function but not for insolubilization in the cell wall. Plant J. 2010, 63, 662–669. [Google Scholar] [CrossRef]
- Held, M.A.; Tan, L.; Kamyab, A.; Hare, M.; Shpak, E.; Kieliszewski, M.J. Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. J. Biol. Chem. 2004, 279, 55474–55482. [Google Scholar] [CrossRef] [Green Version]
- Velasquez, S.M.; Marzol, E.; Borassi, C.; Pol-Fachin, L.; Ricardi, M.M.; Mangano, S.; Juarez, S.P.D.; Salter, J.D.S.; Dorosz, J.G.; Marcus, S.E.; et al. Low Sugar is not Always Good: Impact of Specific O-Glycan Defects on Tip Growth in Arabidopsis. Plant Physiol. 2015, 168, 808–813. [Google Scholar] [CrossRef] [Green Version]
- Brady, J.D.; Sadler, I.H.; Fry, S.C. Pulcherosine, an oxidatively coupled trimer of tyrosine in plant cell walls: Its role in cross-link formation. Phytochemistry 1998, 47, 349–353. [Google Scholar] [CrossRef]
- Brady, J.D.; Sadler, I.H.; Fry, S.C. Di-isodityrosine, a novel tetrametric derivative of tyrosine in plant cell wall proteins: A new potential cross-link. Biochem. J. 1996, 315, 323–327. [Google Scholar] [CrossRef] [Green Version]
- Nuñez, A.; Fishman, M.L.; Fortis, L.L.; Cooke, P.H.; Hotchkiss, A.T. Identification of Extensin Protein Associated with Sugar Beet Pectin. J. Agric. Food Chem. 2009, 57, 10951–10958. [Google Scholar] [CrossRef]
- Valentin, R.; Cerclier, C.; Geneix, N.; Aguié-Béghin, V.; Gaillard, C.; Ralet, M.C.; Cathala, B. Elaboration of extensin-pectin thin film model of primary plant cell wall. Langmuir 2010, 26, 9891–9898. [Google Scholar] [CrossRef] [PubMed]
- Hall, Q.; Cannon, M.C. The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 2002, 14, 1161–1172. [Google Scholar] [CrossRef] [Green Version]
- Cannon, M.C.; Terneus, K.; Hall, Q.; Tan, L.; Wang, Y.; Wegenhart, B.L.; Chen, L.; Lamport, D.T.; Chen, Y.; Kieliszewski, M.J. Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 2008, 105, 2226–2231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, F.; Suyama, A.; Oka, T.; Yoko-o, T.; Matsuoka, K.; Jigami, Y.; Shimma, Y.I. Identification of Novel Peptidyl Serine α-Galactosyltransferase Gene Family in Plants. J. Biol. Chem. 2014, 289, 20405–20420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Møller, S.R.; Yi, X.; Velásquez, S.M.; Gille, S.; Hansen, P.L.M.; Poulsen, C.P.; Olsen, C.E.; Rejzek, M.; Parsons, H.; Yang, Z.; et al. Identification and evolution of a plant cell wall specific glycoprotein glycosyl transferase, ExAD. Sci. Rep. 2017, 7, 45341. [Google Scholar] [CrossRef]
- Velasquez, S.M.; Ricardi, M.M.; Dorosz, J.G.; Fernandez, P.V.; Nadra, A.D.; Pol-Fachin, L.; Egelund, J.; Gille, S.; Harholt, J.; Ciancia, M.; et al. O-glycosylated cell wall proteins are essential in root hair growth. Science 2011, 332, 1401–1403. [Google Scholar] [CrossRef]
- Velasquez, M.; Salter, J.S.; Dorosz, J.G.; Petersen, B.L.; Estevez, J.M. Recent Advances on the Posttranslational Modifications of EXTs and Their Roles in Plant Cell Walls. Front. Plant Sci. 2012, 3, 93. [Google Scholar] [CrossRef] [Green Version]
- Velasquez, S.M.; Ricardi, M.M.; Poulsen, C.P.; Oikawa, A.; Dilokpimol, A.; Halim, A.; Mangano, S.; Juarez, S.P.D.; Marzol, E.; Salter, J.D.S.; et al. Complex regulation of prolyl-4-hydroxylases impacts root hair expansion. Mol. Plant. 2015, 8, 734–746. [Google Scholar] [CrossRef] [Green Version]
- Fabrice, T.N.; Vogler, H.; Draeger, C.; Munglani, G.; Gupta, S.; Herger, A.G.; Knox, P.; Grossniklaus, U.; Ringli, C. LRX Proteins Play a Crucial Role in Pollen Grain and Pollen Tube Cell Wall Development. Plant Physiol. 2018, 176, 1981–1992. [Google Scholar] [CrossRef] [Green Version]
- Sede, A.R.; Borassi, C.; Wengier, D.L.; Mecchia, M.A.; Estevez, J.M.; Muschietti, J.P. Arabidopsis pollen extensins LRX are required for cell wall integrity during pollen tube growth. FEBS Lett. 2018, 592, 233–243. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, K.; Yin, G.; Liu, X.; Liu, M.; Cao, N.; Duan, Y.; Gao, H.; Wang, W.; Ge, W.; et al. Pollen-Expressed Leucine-Rich Repeat Extensins Are Essential for Pollen Germination and Growth. Plant Physiol. 2018, 176, 1993–2006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Chapple, C. The origin and evolution of lignin biosynthesis. New Phytologist. 2010, 187, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Dunand, C.; Crèvecoeur, M.; Penel, C. Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: Possible interaction with peroxidases. New Phytol. 2007, 174, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Cosio, C.; Vuillemin, L.; De Meyer, M.; Kevers, C.; Penel, C.; Dunand, C. An anionic class III peroxidase from zucchini may regulate hypocotyl elongation through its auxin oxidase activity. Planta 2009, 229, 823–836. [Google Scholar] [CrossRef] [Green Version]
- Kunieda, T.; Shimada, T.; Kondo, M.; Nishimura, M.; Nishitani, K.; Hara-Nishimura, I. Spatiotemporal Secretion of PEROXIDASE36 Is Required for Seed Coat Mucilage Extrusion in Arabidopsis. Plant Cell 2013, 25, 1355–1367. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Rubio, M.C.; Alassimone, J.; Geldner, N. A mechanism for localized lignin deposition in the endodermis. Cell 2013, 153, 402–412. [Google Scholar] [CrossRef] [Green Version]
- Roppolo, D.; De Rybel, B.; Tendon, V.D.; Pfister, A.; Alassimone, J.; Vermeer, J.E.; Yamazaki, M.; Stierhof, Y.D.; Beeckman, T.; Geldner, N. A novel protein family mediates Casparian strip formation in the endodermis. Nature 2011, 473, 380–383. [Google Scholar] [CrossRef]
- Francoz, E.; Ranocha, P.; Le Ru, A.; Martinez, Y.; Fourquaux, I.; Jauneau, A.; Dunand, C.; Burlat, V. Pectin Demethylesterification Generates Platforms that Anchor Peroxidases to Remodel Plant Cell Wall Domains. Dev. Cell 2019, 48, 261–276. [Google Scholar] [CrossRef] [Green Version]
- Cosio, C.; Ranocha, P.; Francoz, E.; Burlat, V.; Zheng, Y.; Perry, S.E.; Ripoll, J.J.; Yanofsky, M.; Dunand, C. The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol. 2017, 213, 250–263. [Google Scholar] [CrossRef]
- Herrero, J.; Fernández-Pérez, F.; Yebra, T.; Novo-Uzal, E.; Pomar, F.; Pedreño, M.; Cuello, J.; Guéra, A.; Esteban-Carrasco, A.; Zapata, J.M. Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 2013, 237, 1599–1612. [Google Scholar] [CrossRef] [PubMed]
- Bernards, M.A.; Fleming, W.D.; Llewellyn, D.B.; Priefer, R.; Yang, X.; Sabatino, A.; Plourde, G.L. Biochemical Characterization of the Suberization-Associated Anionic Peroxidase of Potato. Plant Physiol. 1999, 121, 135–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnabelrauch, L.S.; Kieliszewski, M.; Upham, B.L.; Alizedeh, H.; Lamport, D.T. Isolation of pl 4.6 extensin peroxidase from tomato cell suspension cultures and identification of Val-Tyr-Lys as putative intermolecular cross-link site. Plant J. 1996, 9, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Jackson, P.A.; Galinha, C.I.; Pereira, C.S.; Fortunato, A.; Soares, N.C.; Amâncio, S.B.; Ricardo, C.P.P. Rapid deposition of extensin during the elicitation of grapevine callus cultures is specifically catalyzed by a 40-kilodalton peroxidase. Plant Physiol. 2001, 127, 1065–1076. [Google Scholar] [CrossRef]
- Wojtaszek, P.; Trethowan, J.; Bolwell, G.P. Reconstitution in vitro of the components and conditions required for the oxidative cross-linking of extracellular proteins in French bean (Phaseolus vulgaris L.). FEBS Lett. 1997, 405, 95–98. [Google Scholar] [CrossRef] [Green Version]
- Price, N.J.; Pinheiro, C.; Soares, C.M.; Ashford, D.A.; Ricardo, C.P.; Jackson, P.A. A Biochemical and Molecular Characterization of LEP1, an Extensin Peroxidase from Lupin. J. Biol. Chem. 2003, 278, 41389–41399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graças, J.P.; Lima, J.E.; Peres, L.E.P.; Jamet, E.; Dunand, C.; Vitorello, V.A.; Chervin, C. Ethylene Signaling Causing Tolerance of Arabidopsis thaliana Roots to Low pH Stress is Linked to Class III Peroxidase Activity. J. Plant Growth Regul. 2021, 40, 116–125. [Google Scholar] [CrossRef]
- Dong, W.; Kieliszewski, M.; Held, M.A. Identification of the pI 4.6 extensin peroxidase from Lycopersicon esculentum using proteomics and reverse-genomics. Phytochemistry 2015, 112, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Jacobowitz, J.R.; Weng, J.-K. PRX9 and PRX40 are extensin peroxidases essential for maintaining tapetum and microspore cell wall integrity during Arabidopsis anther development. Plant Cell 2019, 31, 848–861. [Google Scholar] [CrossRef] [Green Version]
- Pacheco, J.M.; Ranocha, P.; Kasulin, L.; Fusari, C.M.; Servi, L.; Aptekmann, A.; Gabarain, V.B.; Peralta, J.M.; Borassi, C.; Marzol, E.; et al. Apoplastic class III peroxidases PRX62 and PRX69 promote Arabidopsis root hair growth at low temperature. Nat. Commun. 2022, 13, 1310. [Google Scholar] [CrossRef]
- Martin, R.E.; Marzol, E.; Estevez, J.M.; Muday, G.K. Ethylene signaling increases reactive oxygen species accumulation to drive root hair initiation in Arabidopsis. BioRxiv 2022. [Google Scholar] [CrossRef]
- Mangano, S.; Denita-Juarez, S.P.; Choi, H.S.; Marzol, E.; Hwang, Y.; Ranocha, P.; Velasquez, S.M.; Borassi, C.; Barberini, M.L.; Aptekmann, A.A.; et al. The molecular link between auxin and ROS-Mediated polar root hair growth. Proc. Natl. Acad. Sci. USA 2017, 114, 5289–5294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, C.S.; Ribeiro, J.M.L.; Vatulescu, A.D.; Findlay, K.; MacDougall, A.J.; Jackson, P.A.P. Extensin network formation in Vitis vinifera callus cells is an essential and causal event in rapid and H2O2-induced reduction in primary cell wall hydration. BMC Plant Biol. 2011, 11, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, K.; Menand, B.; Bell, E.; Dolan, L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 2010, 42, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Dynowski, M.; Schaaf, G.; Loque, D.; Moran, O.; Ludewig, U. Plant plasma membrane water channels conduct the signalling molecule H2O2. Biochem. J. 2008, 414, 53–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooijmaijers, C.; Rhee, J.Y.; Kwak, K.J.; Chung, G.C.; Horie, T.; Katsuhara, M.; Kang, H. Hydrogen peroxide permeability of plasma membrane aquaporins of Arabidopsis thaliana. J. Plant Res. 2012, 125, 147–153. [Google Scholar] [CrossRef]
- Rodrigues, O.; Reshetnyak, G.; Grondin, A.; Saijo, Y.; Leonhardt, N.; Maurel, C.; Verdoucq, L. Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proc. Natl. Acad. Sci. USA 2017, 114, 9200–9205. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Rosendale, M.; Campbell, R.E.; Perrais, D. pHuji, a pH-sensitive red fluorescent protein for imaging of exo- and endocytosis. J. Cell Biol. 2014, 207, 419–432. [Google Scholar] [CrossRef] [Green Version]
- Stoddard, A.; Rolland, V. I see the light! Fluorescent proteins suitable for cell wall/apoplast targeting in leaves. Plant Direct. 2019, 3, e00112. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Dong, W.; Tan, L.; Held, M.A.; Kieliszewski, M.J. Arabinosylation Plays a Crucial Role in Extensin Cross-linking In Vitro. Biochem. Insights 2015, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Borassi, C.; Sede, A.R.; Mecchia, M.A.; Salgado Salter, J.D.; Marzol, E.; Muschietti, J.P.; Estevez, J.M. An update on cell surface proteins containing extensin-motifs. J. Exp. Bot. 2016, 67, 477–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stafstrom, J.P.; Andrew Staehelin, L. The Role of Carbohydrate in Maintaining Extensin in an Extended Conformation. Plant Physiol. 1986, 81, 242–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owens, N.W.; Stetefeld, J.; Lattová, E.; Schweizer, F. Contiguous O-galactosylation of 4(R)-hydroxy-l-proline residues forms very stable polyproline II helices. J. Am. Chem. Soc. 2010, 132, 5036–5042. [Google Scholar] [CrossRef] [PubMed]
- Ishiwata, A.; Kaeothip, S.; Takeda, Y.; Ito, Y. Synthesis of the Highly Glycosylated Hydrophilic Motif of Extensins. Angew. Chem. Int. Ed. 2014, 53, 9812–9816. [Google Scholar] [CrossRef] [PubMed]
- Monshausen, G.B.; Bibikova, T.N.; Messerli, M.A.; Shi, C.; Gilroy, S. Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc. Natl. Acad. Sci. USA 2007, 104, 20996–21001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, S.; Gapper, C.; Kaya, H.; Bell, E.; Kuchitsu, K.; Dolan, L. Local Positive Feedback Regulation Determines Cell Shape in Root Hair Cells. Science 2008, 319, 1241–1244. [Google Scholar] [CrossRef]
- Orman-Ligeza, B.; Parizot, B.; De Rycke, R.; Fernandez, A.; Himschoot, E.; Van Breusegem, F.; Bennett, M.J.; Périlleux, C.; Beeckman, T.; Draye, X. RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development 2016, 143, 3328–3339. [Google Scholar] [CrossRef] [Green Version]
- Egelund, J.; Obel, N.; Ulvskov, P.; Geshi, N.; Pauly, M.; Bacic, A.; Petersen, B.L. Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol. Biol. 2007, 64, 439–451. [Google Scholar] [CrossRef]
- Vanacore, R.; Ham, A.J.L.; Voehler, M.; Sanders, C.R.; Conrads, T.P.; Veenstra, T.D.; Sharpless, K.B.; Dawson, P.E.; Hudson, B.G. A Sulfilimine Bond Identified in Collagen IV. Science 2009, 325, 1230–1234. [Google Scholar] [CrossRef] [Green Version]
- Bhave, G.; Cummings, C.F.; Vanacore, R.M.; Kumagai-Cresse, C.; Ero-Tolliver, I.A.; Rafi, M.; Kang, J.S.; Pedchenko, V.; Fessler, L.I.; Fessler, J.H.; et al. Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nat. Chem. Biol. 2012, 8, 784–790. [Google Scholar] [CrossRef] [Green Version]
- Brown, K.L.; Hudson, B.G.; Voziyan, P.A. Halogens are key cofactors in building of collagen IV scaffolds outside the cell. Curr. Opin. Nephrol. Hypertens. 2018, 27, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7, Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savelli, B.; Li, Q.; Webber, M.; Jemmat, A.M.; Robitaille, A.; Zamocky, M.; Mathé, C.; Dunand, C. RedoxiBase: A database for ROS homeostasis regulated proteins. Redox Biol. 2019, 26, 101247. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Darré, L.; Machado, M.R.; Brandner, A.F.; González, H.C.; Ferreira, S.; Pantano, S. SIRAH: A structurally unbiased coarse-grained force field for proteins with aqueous solvation and long-range electrostatics. J. Chem. Theory Comput. 2015, 11, 723–739. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- McGibbon, R.T.; Beauchamp, K.A.; Harrigan, M.P.; Klein, C.; Swails, J.M.; Hernández, C.X.; Schwantes, C.R.; Wang, L.P.; Lane, T.J.; Pande, V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015, 109, 1528–1532. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Oliphant, T.E. A Guide to NumPy; Trelgol Publishing: Austin, TX, USA, 2006; 376p. [Google Scholar]
- Mutwil, M.; Klie, S.; Tohge, T.; Giorgi, F.M.; Wilkins, O.; Campbell, M.M.; Fernie, A.R.; Usadel, B.; Nikoloski, Z.; Persson, S. PlaNet: Combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 2011, 23, 895–910. [Google Scholar] [CrossRef] [Green Version]


| ng Tyr/μg CW (STD) | ng IDT/μg CW (STD) | |
|---|---|---|
| Wt Col-0 | 7.799 ± 0.26 | 0.853 ± 0.08 |
| prx01,44,73 | 9.588 ± 0.31 ** | 0.963 ± 0.02 |
| 35S:PRX44 | 8.649 ± 0.07 | 0.953 ± 0.04 |
| 35S:PRX73 | 8.700 ± 0.12 | 1.042 ± 0.02 ** |
| under O-glycosylated EXTs | ||
| sergt1-1 rra3 | 3.530 ± 0.08 *** | 0.235 ± 0.01 *** |
| p4h5 sergt1-1 | 3.766 ± 0.06 *** | 0.225 ± 0.02 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marzol, E.; Borassi, C.; Carignani Sardoy, M.; Ranocha, P.; Aptekmann, A.A.; Bringas, M.; Pennington, J.; Paez-Valencia, J.; Martínez Pacheco, J.; Rodríguez-Garcia, D.R.; et al. Class III Peroxidases PRX01, PRX44, and PRX73 Control Root Hair Growth in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 5375. https://doi.org/10.3390/ijms23105375
Marzol E, Borassi C, Carignani Sardoy M, Ranocha P, Aptekmann AA, Bringas M, Pennington J, Paez-Valencia J, Martínez Pacheco J, Rodríguez-Garcia DR, et al. Class III Peroxidases PRX01, PRX44, and PRX73 Control Root Hair Growth in Arabidopsis thaliana. International Journal of Molecular Sciences. 2022; 23(10):5375. https://doi.org/10.3390/ijms23105375
Chicago/Turabian StyleMarzol, Eliana, Cecilia Borassi, Mariana Carignani Sardoy, Philippe Ranocha, Ariel A. Aptekmann, Mauro Bringas, Janice Pennington, Julio Paez-Valencia, Javier Martínez Pacheco, Diana R. Rodríguez-Garcia, and et al. 2022. "Class III Peroxidases PRX01, PRX44, and PRX73 Control Root Hair Growth in Arabidopsis thaliana" International Journal of Molecular Sciences 23, no. 10: 5375. https://doi.org/10.3390/ijms23105375
APA StyleMarzol, E., Borassi, C., Carignani Sardoy, M., Ranocha, P., Aptekmann, A. A., Bringas, M., Pennington, J., Paez-Valencia, J., Martínez Pacheco, J., Rodríguez-Garcia, D. R., Rondón Guerrero, Y. d. C., Peralta, J. M., Fleming, M., Mishler-Elmore, J. W., Mangano, S., Blanco-Herrera, F., Bedinger, P. A., Dunand, C., Capece, L., ... Estevez, J. M. (2022). Class III Peroxidases PRX01, PRX44, and PRX73 Control Root Hair Growth in Arabidopsis thaliana. International Journal of Molecular Sciences, 23(10), 5375. https://doi.org/10.3390/ijms23105375






