Lb1G04202, an Uncharacterized Protein from Recretohalophyte Limonium bicolor, Is Important in Salt Tolerance
Abstract
:1. Introduction
2. Results
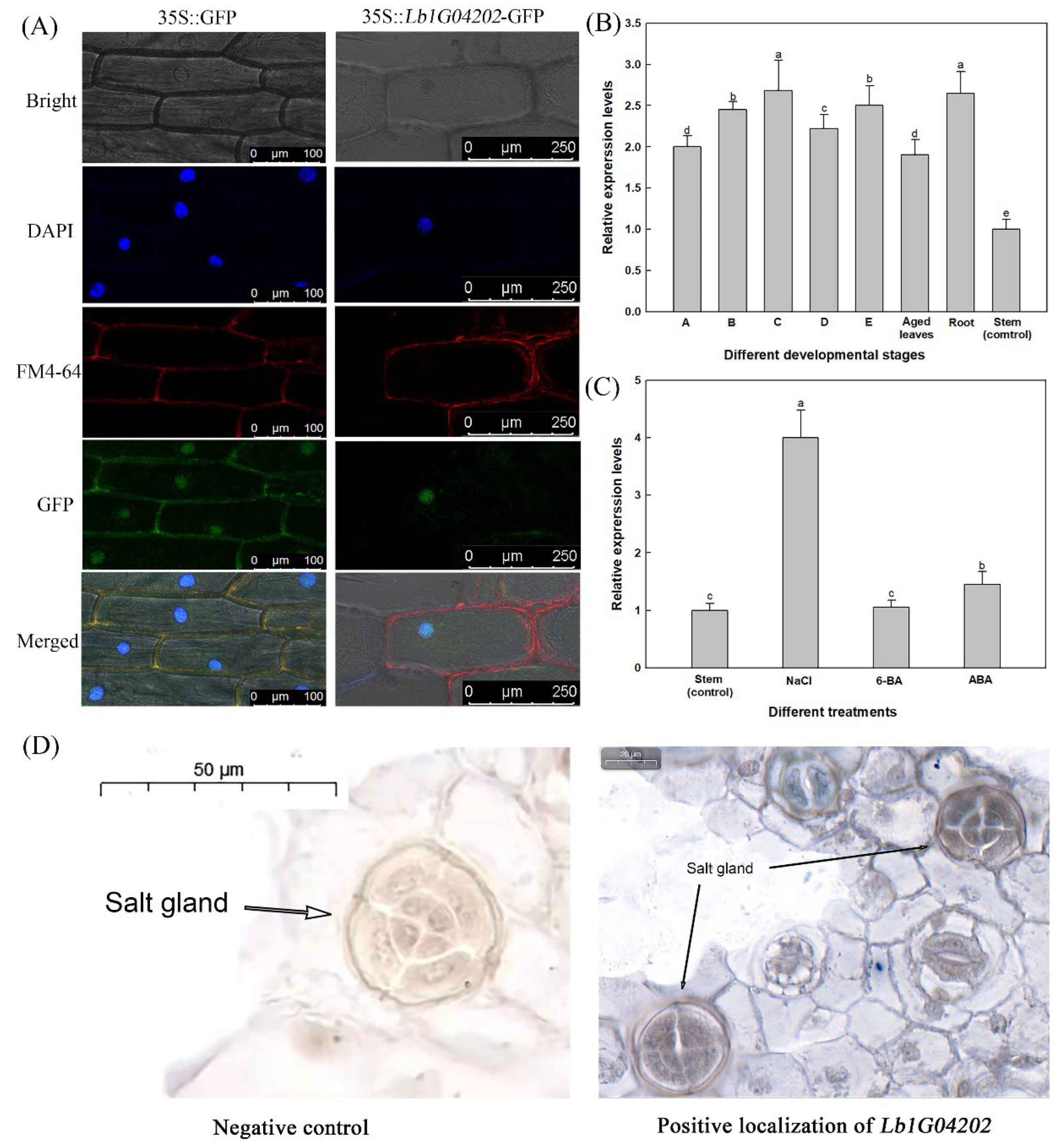
2.1. Lb1G04202 Encodes an Uncharacterized Protein, Is Expressed in Salt Glands and Is Highly Expressed after Salt Treatment
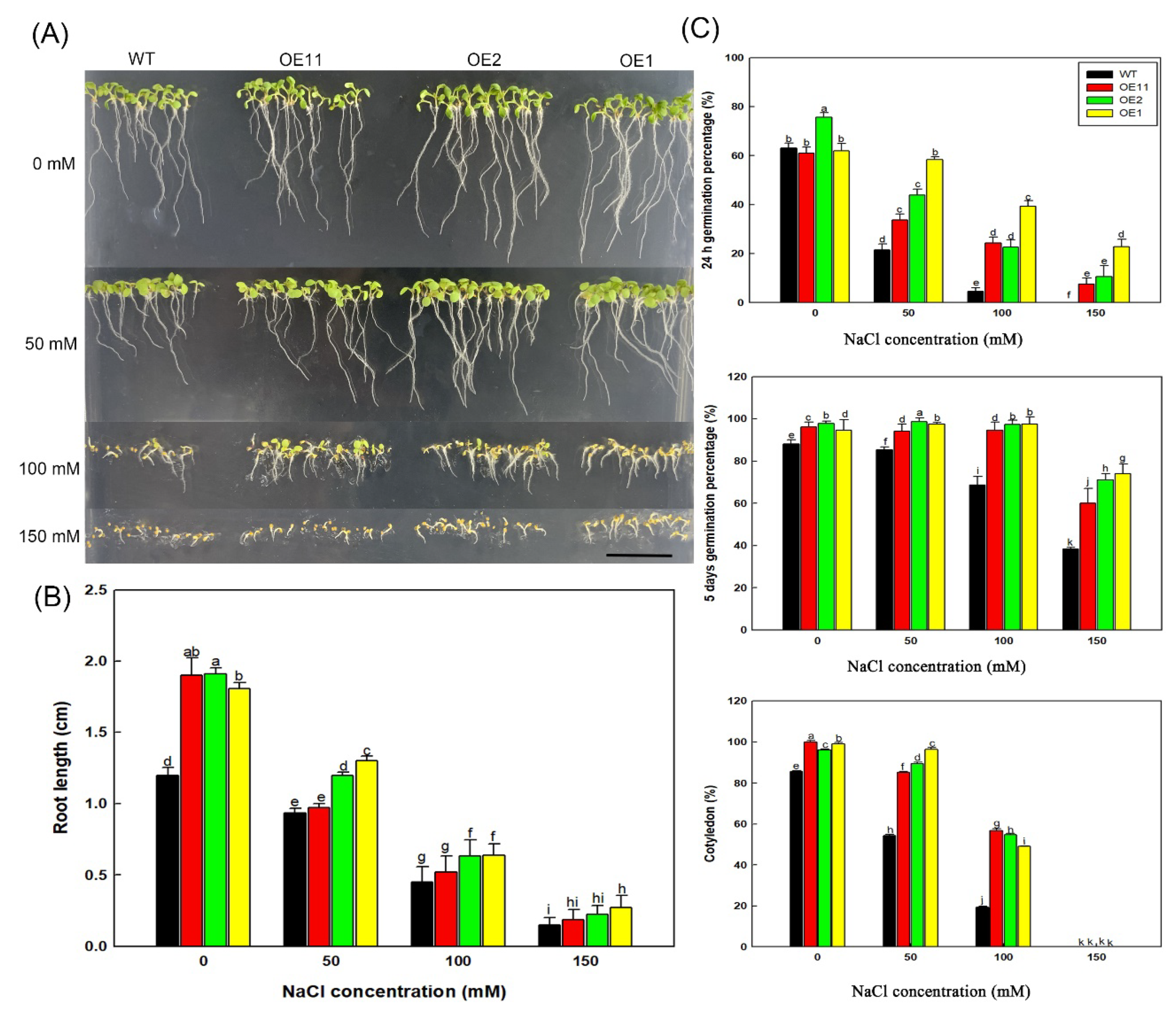
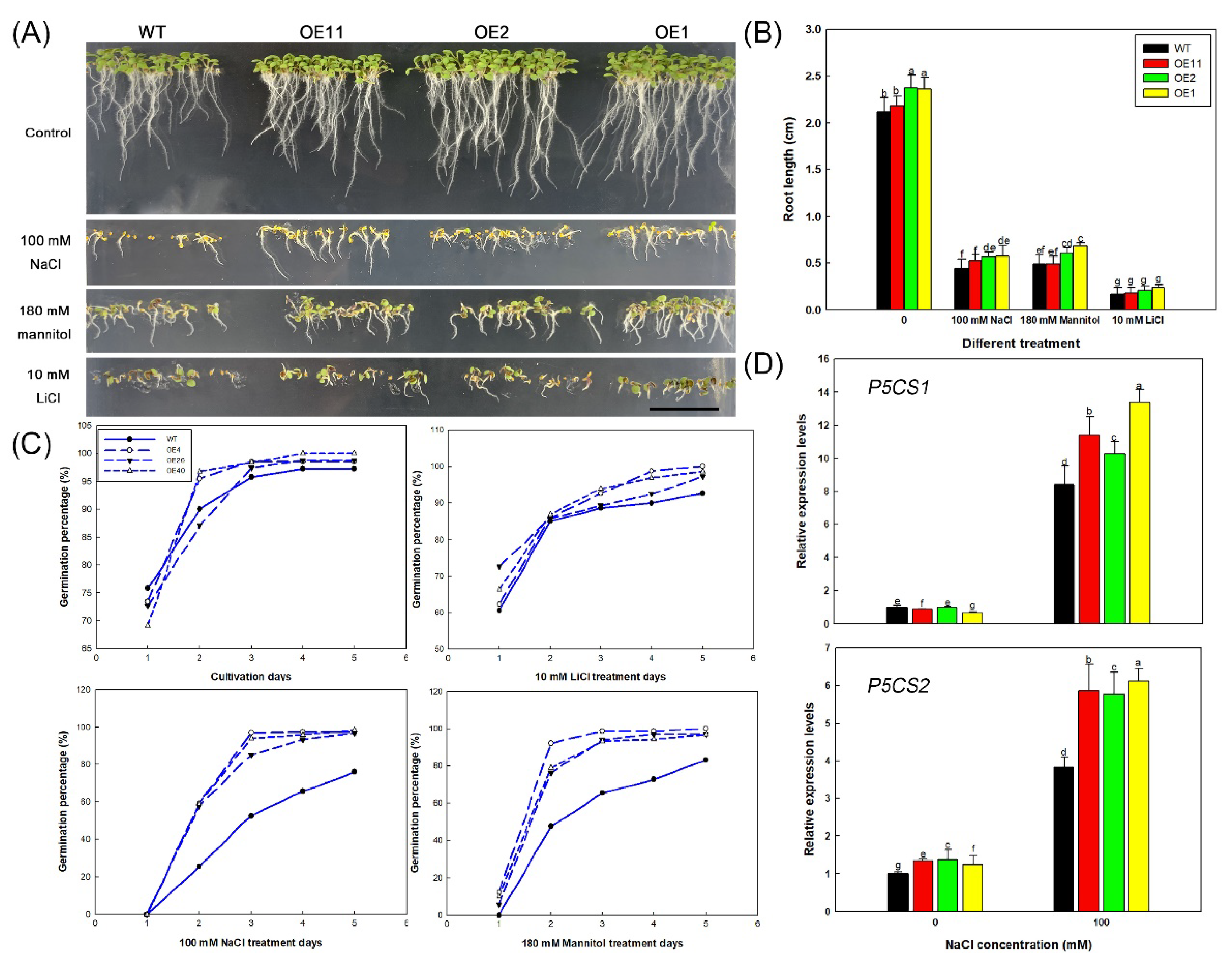
2.2. Heterologous Expression of Lb1G04202 Increases Arabidopsis Salt Resistance during Germination
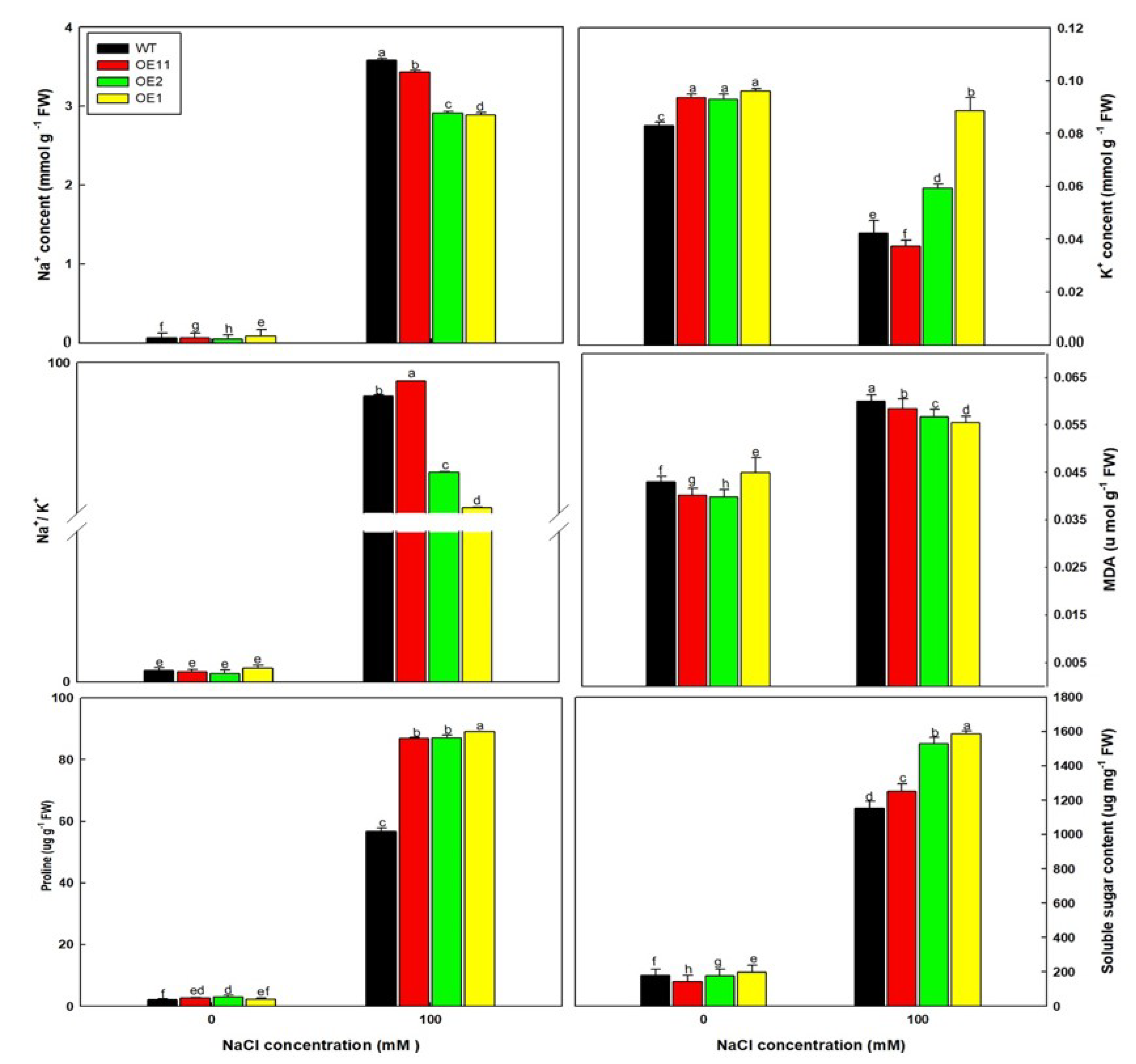
2.3. Lb1G04202 Reduces NaCl Damage at the Seedling Stage
2.4. Lb1G04202 Enhances Salt Tolerance by Alleviating Osmotic Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
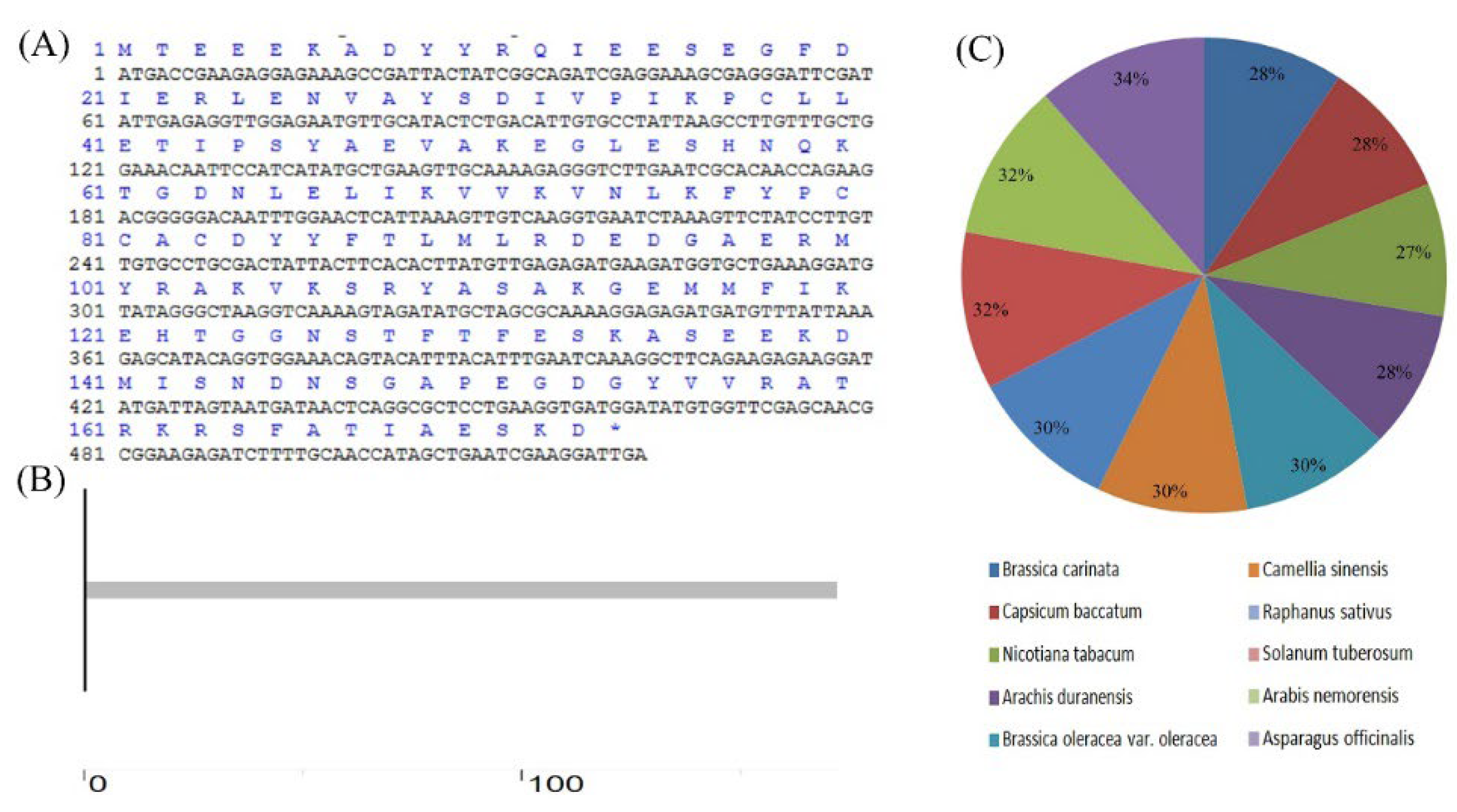
4.2. Cloning and Bioinformatic Analysis of Lb1G04202
4.3. Localization of Lb1G04202
4.4. Analysis of the Expression Pattern of Lb1G04202 during the Development of L. bicolor and under Different Treatments
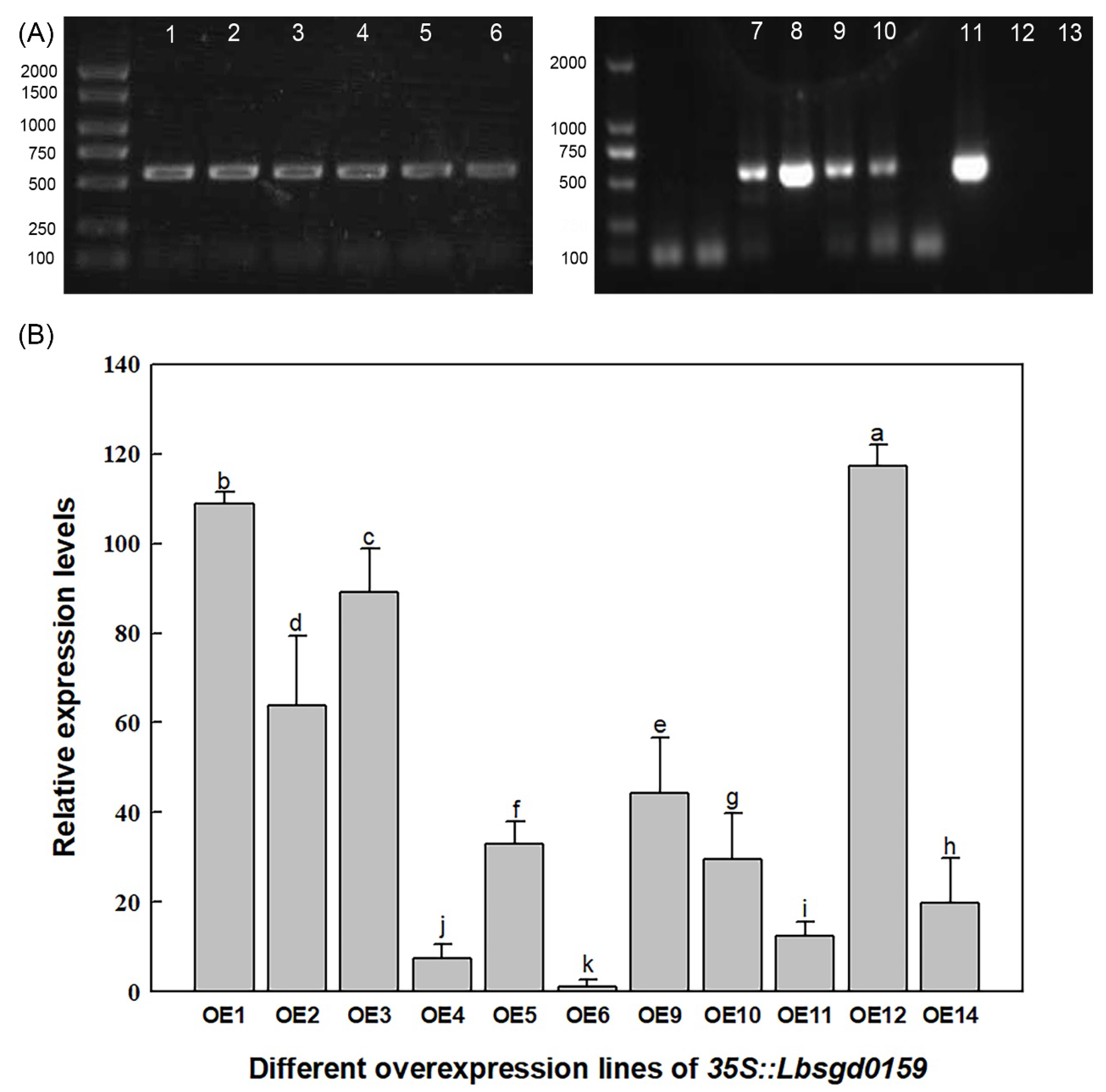
4.5. Heterologous Expression in Arabidopsis Lines
4.6. Determination of Salt Tolerance Index during Germination
4.7. Measurement of Physiological Indexes under Salt Treatment at Seedling Stages
4.8. Performance of Transgenic Lines under Single Ion and Osmotic Stress
4.9. Expression Analysis of Osmotic Stress-Related Marker Genes in Transgenic Arabidopsis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| L. bicolor | Limonium bicolor |
| TTG1 | TRANSPARENT TESTA GLABRA1 |
| SAD2 | SUPER SENSITIVE TO ABA AND DROUGHT2 |
| TRY | TRIPTYCHON |
| CPC | CAPRICE |
| AtSOS1 | SALT OVERLY SENSITIVE 1 |
| AtSOS3 | SALT OVERLY SENSITIVE 3 |
| AtP5CS1 | DELTA1-PYRROLINE-5-CARBOXYLATE SYNTHASE 1 |
| AtP5CS2 | DELTA1-PYRROLINE-5-CARBOXYLATE SYNTHASE 2 |
| Col-0 | Columbia-0 |
| qPCR | quantitative PCR |
| CDS | coding sequence |
| WT | wild-type |
References
- Hou, D.; Bolan, N.S.; Tsang, D.C.W.; Kirkham, M.B.; O’Connor, D. Sustainable soil use and management: An interdisciplinary and systematic approach. Sci. Total Environ. 2020, 729, 138961. [Google Scholar] [CrossRef] [PubMed]
- Perri, S.; Suweis, S.; Holmes, A.; Marpu, P.R.; Entekhabi, D.; Molini, A. River basin salinization as a form of aridity. Proc. Natl. Acad. Sci. USA 2020, 117, 17635–17642. [Google Scholar] [CrossRef] [PubMed]
- Singh, A. Soil salinization management for sustainable development: A review. J. Environ. Manag. 2021, 277, 111383. [Google Scholar] [CrossRef] [PubMed]
- Litalien, A.; Zeeb, B. Curing the earth: A review of anthropogenic soil salinization and plant-based strategies for sustainable mitigation. Sci. Total Environ. 2020, 698, 134235. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Hayashi, N.; Kobayashi, M.; Aoki, N.; Miyao, A.; Mitsuhara, I.; Ichikawa, H.; Komatsu, S.; Hirochika, H.; Kikuchi, S.; et al. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 2012, 69, 26–36. [Google Scholar] [CrossRef]
- Song, J.; Wang, B. Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 2015, 115, 541–553. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Zhang, J.J.S.; Review, T. Research Advances in Physiological Ecology Adaptation of Plant Salt-tolerance. Sci. Technol. Rev. 2011, 29, 76–79. [Google Scholar]
- Yuan, F.; Guo, J.; Shabala, S.; Wang, B. Reproductive Physiology of Halophytes: Current Standing. Front. Plant Sci. 2018, 9, 1954. [Google Scholar] [CrossRef]
- Guo, J.; Du, M.; Lu, C.; Wang, B. NaCl improves reproduction by enhancing starch accumulation in the ovules of the euhalophyte Suaeda salsa. BMC Plant Biol. 2020, 20, 262. [Google Scholar] [CrossRef] [PubMed]
- Leng, B.Y.; Yuan, F.; Dong, X.X.; Wang, B.S. Salt gland distribution in Limonium bicolor at the individual level. Earth Environ. Sci. 2018, 113, 012202. [Google Scholar] [CrossRef]
- Böhm, J.; Messerer, M.; Müller, H.; Scholz-Starke, J.; Gradogna, A.; Scherzer, S.; Maierhofer, T.; Bazihizina, N.; Zhang, H.; Stigloher, C.J.C.B. Understanding the Molecular Basis of Salt Sequestration in Epidermal Bladder Cells of Chenopodium quinoa. Curr. Biol. 2018, 28, 3075–3085.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, J.; Liu, J.; Na, Z.; Sa, R.; Jiang, L. Technology, Effect of Salt Stress on Antioxidant Enzymes, Soluble Sugar and Yield of Oat. J. Integr. Agric. 2013, 12, 1441–1449. [Google Scholar] [CrossRef]
- Luo, D.; Zhou, Q.; Wu, Y.; Chai, X.; Liu, W.; Wang, Y.; Yang, Q.; Wang, Z.; Liu, Z. Full-length transcript sequencing and comparative transcriptomic analysis to evaluate the contribution of osmotic and ionic stress components towards salinity tolerance in the roots of cultivated alfalfa (Medicago sativa L.). BMC Plant Biol. 2019, 19, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, P.; Wang, Z. Effects of different types of halophytes on the concentration of cadmium in coastal saline soil. Acta Ecol. Sin. 2017, 37, 4656–4662. [Google Scholar]
- Yuan, F.; Leng, B.Y.; Wang, B.S. Research Progress in Salt Secretion of Salt Glands in Plants. Plant Physiol. J. 2015, 7, 1531–1537. [Google Scholar]
- Yuan, F.; Lyu, M.J.; Leng, B.Y.; Zhu, X.G.; Wang, B.S. The transcriptome of NaCl-treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol. Biol. 2016, 91, 241–256. [Google Scholar] [CrossRef]
- Semenova, G.A.; Fomina, I.R.; Biel, K.Y. Structural features of the salt glands of the leaf of Distichlis spicata ‘Yensen 4a’ (Poaceae). Protoplasma 2010, 240, 75–82. [Google Scholar] [CrossRef]
- Arisz, W.; Camphuis, I.; Heikens, H.; Tooren, V. The secretion of the salt glands of Limonium latifolium Ktze. Acta Bot. Neerl. 1955, 4, 322–338. [Google Scholar] [CrossRef]
- Levering, C.A.; Thomson, W.W. The ultrastructure of the salt gland of Spartina foliosa. Planta 1971, 97, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, H.; Lüttge, U. Die Salzdrüsen von Limonium vulgare. Planta 1967, 70, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Jelte, R.; Ingrid, R.; Taede, S. A light and electron-microscopical study on the structure and function of the salt gland of Glaux maritima L. New Phytol. 1977, 79, 665–671. [Google Scholar]
- Morris, L.; Yun, K.; Rutter, A.; Zeeb, B.A. Characterization of Excreted Salt from the Recretohalophytes Distichlis spicata and Spartina pectinata. J. Environ. Qual. 2019, 48, 1775–1780. [Google Scholar] [CrossRef]
- Feng, Z.; Sun, Q.; Deng, Y. Study on pathway and characteristics of ion secretion of salt glands of Limonium bicolor. Acta Physiol. Plant. 2014, 36, 2729–2741. [Google Scholar] [CrossRef]
- Lu, C.; Feng, Z.; Yuan, F. The SNARE protein LbSYP61 participates in salt secretion in Limonium bicolor. Environ. Exp. Bot. 2020, 176, 104076. [Google Scholar] [CrossRef]
- Deng, Y.; Feng, Z.; Yuan, F.; Guo, J.; Suo, S.; Wang, B. Identification and functional analysis of the autofluorescent substance in Limonium bicolor salt glands. Plant Physiol. Biochem. 2015, 97, 20–27. [Google Scholar] [CrossRef]
- Ding, F.; Chen, M.; Sui, N. Ca2+ significantly enhanced development and salt-secretion rate of salt glands of Limonium bicolor under NaCl treatment. S. Afr. J. Bot. 2010, 76, 95–101. [Google Scholar] [CrossRef] [Green Version]
- Yuan, F.; Chen, M.; Yang, J. A system for the transformation and regeneration of the recretohalophyte Limonium bicolor. Vitr. Cell. Dev. Biol.-Plant 2014, 50, 610–617. [Google Scholar] [CrossRef]
- Xu, Y.; Jiao, X.; Wang, X.; Zhang, H.; Wang, B.; Yuan, F. Importin-beta From the Recretohalophyte Limonium bicolor Enhances Salt Tolerance in Arabidopsis thaliana by Reducing Root Hair Development and Abscisic Acid Sensitivity. Front. Plant Sci. 2020, 11, 582459. [Google Scholar] [CrossRef]
- Yuan, F.; Leng, B.; Zhang, H.; Wang, X.; Han, G.; Wang, B. A WD40-Repeat Protein from the Recretohalophyte Limonium bicolor Enhances Trichome Formation and Salt Tolerance in Arabidopsis. Front. Plant Sci. 2019, 10, 1456. [Google Scholar] [CrossRef]
- Leng, B.; Wang, X.; Yuan, F.; Zhang, H.; Lu, C.; Chen, M.; Wang, B. Heterologous expression of the Limonium bicolor MYB transcription factor LbTRY in Arabidopsis thaliana increases salt sensitivity by modifying root hair development and osmotic homeostasis. Plant Sci. 2021, 302, 110704. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, Y.; Xu, Y.; Wang, B.; Yuan, F. A novel gene LbHLH from the halophyte Limonium bicolor enhances salt tolerance via reducing root hair development and enhancing osmotic resistance. BMC Plant Biol. 2021, 21, 284. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Li, J.; Qiu, S.; Qi, F.; Su, H.; Bu, Q.; Jiang, R.; Tang, K.; Zhang, L.; Chen, W. The transcription factors TLR1 and TLR2 negatively regulate trichome density and artemisinin levels in Artemisia annua. J. Integr. Plant Biol. 2022, 35355415. [Google Scholar] [CrossRef] [PubMed]
- Abea, S.; Sadob, A.; Tanakac, K.; Kisugic, T. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. USA 2014, 111, 18084–18089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Wang, C.; Han, X.; Tang, S.; Liu, S.; Xia, X.; Yin, W. A novel bHLH transcription factor PebHLH35 from Populus euphratica confers drought tolerance through regulating stomatal development, photosynthesis and growth in Arabidopsis. Biochem. Biophys. Res. Commun. 2014, 450, 453–458. [Google Scholar] [CrossRef]
- Imamura, T.; Yasui, Y.; Koga, H.; Takagi, H.; Abe, A.; Nishizawa, K.; Mizuno, N.; Ohki, S.; Mizukoshi, H.; Mori, M. A novel WD40-repeat protein involved in formation of epidermal bladder cells in the halophyte quinoa. Commun. Biol. 2020, 3, 513. [Google Scholar] [CrossRef]
- Wang, D.; Liu, H.; Wang, H.; Zhang, P.; Shi, C. A novel sucrose transporter gene IbSUT4 involves in plant growth and response to abiotic stress through the ABF-dependent ABA signaling pathway in Sweetpotato. BMC Plant Biol. 2020, 20, 157. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhu, C.; Jin, L.; Xiao, A.; Duan, J.; Ma, L. A novel gene of Kalanchoe daigremontiana confers plant drought resistance. Sci. Rep. 2018, 8, 2547. [Google Scholar] [CrossRef] [Green Version]
- Yuan, F.; Lyu, M.J.; Leng, B.Y.; Zheng, G.Y.; Feng, Z.T.; Li, P.H.; Zhu, X.G.; Wang, B.S. Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 2015, 38, 1637–1657. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Q.; Zhang, Y.; Cui, J.; Chen, G.; Xie, B.; Wu, C.; Liu, H. Synergistic and antagonistic effects of salinity and pH on germination in switchgrass (Panicum virgatum L.). PLoS ONE 2014, 9, e85282. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.F.; Song, J.; Fan, H.; Zhou, S.; Zhao, M. Growth response to ionic and osmotic stress of NaCl in salt-tolerant and salt-sensitive maize. J. Integr. Plant Biol. 2010, 52, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Dix, P.J.; Mclysaght, U.A.; Plunkett, A. Salt stress: Resistance mechanisms and in vitro selection procedures. Plant Tissue Cult. Its Agric. Appl. 1986, 469–478. [Google Scholar]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Q.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.-K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.-D.; Wang, P.; Bao, Z.; Ma, Q.; Duan, L.-J.; Bao, A.-K.; Zhang, J.-L.; Wang, S.-M. SOS1, HKT1;5, and NHX1 Synergistically Modulate Na+ Homeostasis in the Halophytic Grass Puccinellia tenuiflora. Front. Plant Sci. 2017, 8, 576. [Google Scholar] [CrossRef] [Green Version]
- Yuan, F.; Chen, M.; Leng, B.Y.; Wang, B.S. An efficient autofluorescence method for screening Limonium bicolor mutants for abnormal salt gland density and salt secretion. S. Afr. J. Bot Afr. J. Bot. 2013, 88, 110–117. [Google Scholar] [CrossRef]
- Sui, N.; Tian, S.; Wang, W.; Wang, M.; Fan, H. Overexpression of Glycerol-3-Phosphate Acyltransferase from Suaeda salsa Improves Salt Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1337. [Google Scholar] [CrossRef]
- Hu, Y.M.; Su, Q.; Zu, Y.; Liu, J.W. Coexpression of Genes Related to Glycine Betaine Biosynthesis from Salicornia europaea Increase Salt Tolerance of Transgenic Tobacco. Chin. Agric. Sci. Bull. 2010, 26, 55–59. [Google Scholar]
- Mach, J. Fear Not the Unknown: OPENER as a Study in Shedding Light on Genes with Unknown Function. Plant Cell 2019, 31, 1420. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhu, J. A calcium sensor homolog required for plant salt tolerance. Science 1998, 280, 1943–1945. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Barrena, M.J.; Fujii, H.; Angulo, I.; Martinez-Ripoll, M.; Zhu, J.K.; Albert, A. The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol. Cell 2007, 26, 427–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabro, G.; Kovács, I.; Pavet, V.; Szabados, L.; Alvarez, M.E. Proline Accumulation and AtP5CS2 Gene Activation Are Induced by Plant-Pathogen Incompatible Interactions in Arabidopsis. Mol. Plant Microbe Interact. 2004, 17, 343–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nounjan, N.; Theerakulpisut, P. Effects of exogenous proline and trehalose on physiological responses in rice seedlings during salt-stress and after recovery. Plant Soil Environ. 2012, 58, 309–315. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.J.; Ma, X.L.; Wan, P.; Liu, L.Y. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Muchate, N.S.; Nikalje, G.C.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. Plant Salt Stress: Adaptive Responses, Tolerance Mechanism and Bioengineering for Salt Tolerance. Bot. Rev. 2016, 82, 371–406. [Google Scholar] [CrossRef]
- Han, G.; Yuan, F.; Guo, J.; Zhang, Y.; Sui, N.; Wang, B. AtSIZ1 improves salt tolerance by maintaining ionic homeostasis and osmotic balance in Arabidopsis. Plant Sci. 2019, 285, 55–67. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Wang, B.; Yuan, F. Lb1G04202, an Uncharacterized Protein from Recretohalophyte Limonium bicolor, Is Important in Salt Tolerance. Int. J. Mol. Sci. 2022, 23, 5401. https://doi.org/10.3390/ijms23105401
Wang X, Wang B, Yuan F. Lb1G04202, an Uncharacterized Protein from Recretohalophyte Limonium bicolor, Is Important in Salt Tolerance. International Journal of Molecular Sciences. 2022; 23(10):5401. https://doi.org/10.3390/ijms23105401
Chicago/Turabian StyleWang, Xi, Baoshan Wang, and Fang Yuan. 2022. "Lb1G04202, an Uncharacterized Protein from Recretohalophyte Limonium bicolor, Is Important in Salt Tolerance" International Journal of Molecular Sciences 23, no. 10: 5401. https://doi.org/10.3390/ijms23105401
APA StyleWang, X., Wang, B., & Yuan, F. (2022). Lb1G04202, an Uncharacterized Protein from Recretohalophyte Limonium bicolor, Is Important in Salt Tolerance. International Journal of Molecular Sciences, 23(10), 5401. https://doi.org/10.3390/ijms23105401





