Transcriptomic and Metabolomic Analysis Reveal Possible Molecular Mechanisms Regulating Tea Plant Growth Elicited by Chitosan Oligosaccharide
Abstract
1. Introduction
2. Results
2.1. Yield Measurement of Tea Leaves
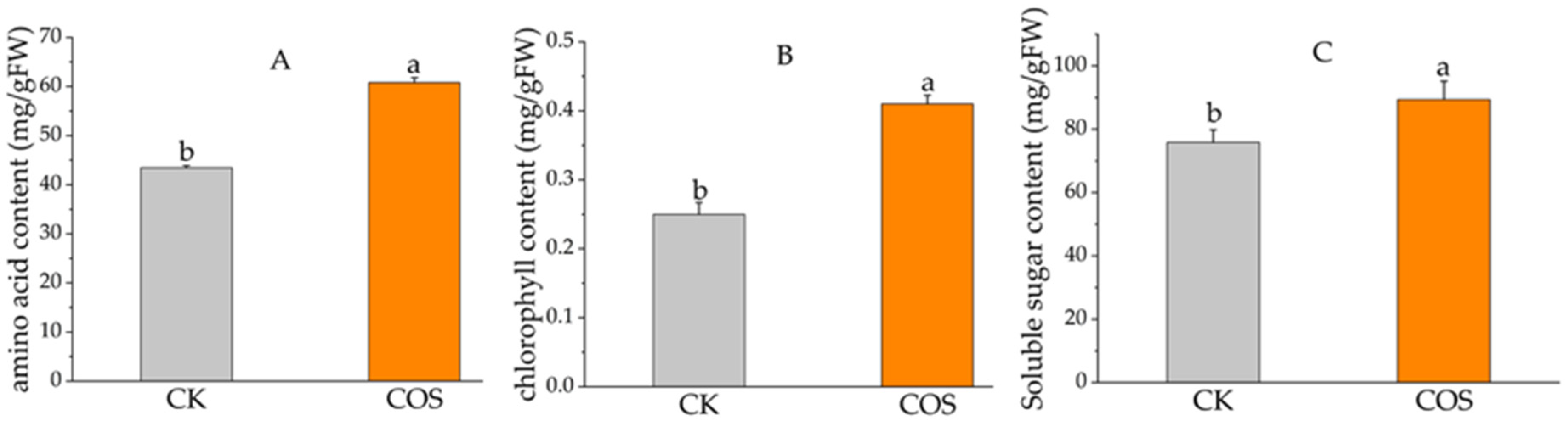
2.2. Changes of Chlorophyll, Soluble Sugar, and Amino Acid Content in Tea Leaves
2.3. Transcriptomic Analysis
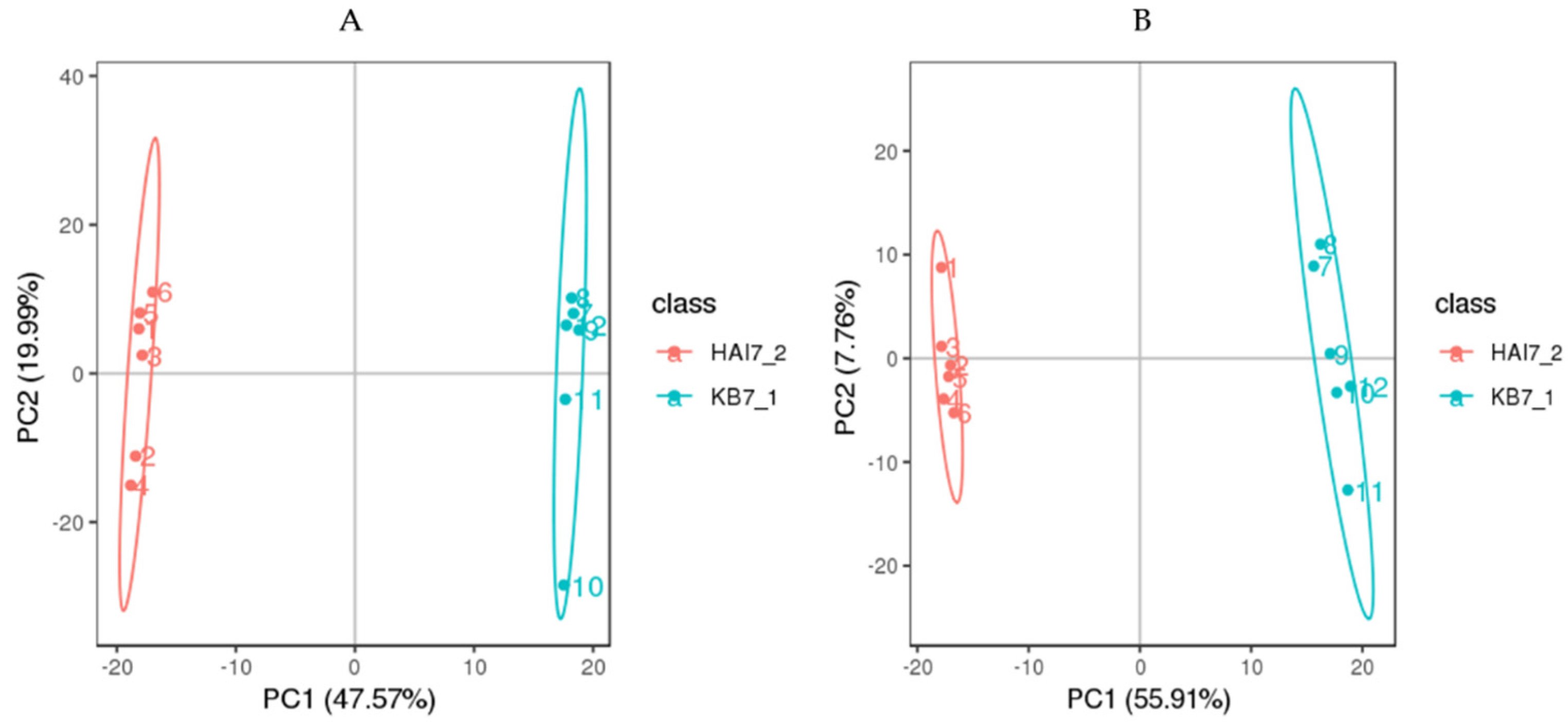
2.3.1. Transcriptome Sequencing and Assembly
2.3.2. Differentially Expressed Gene (DEG) Analysis
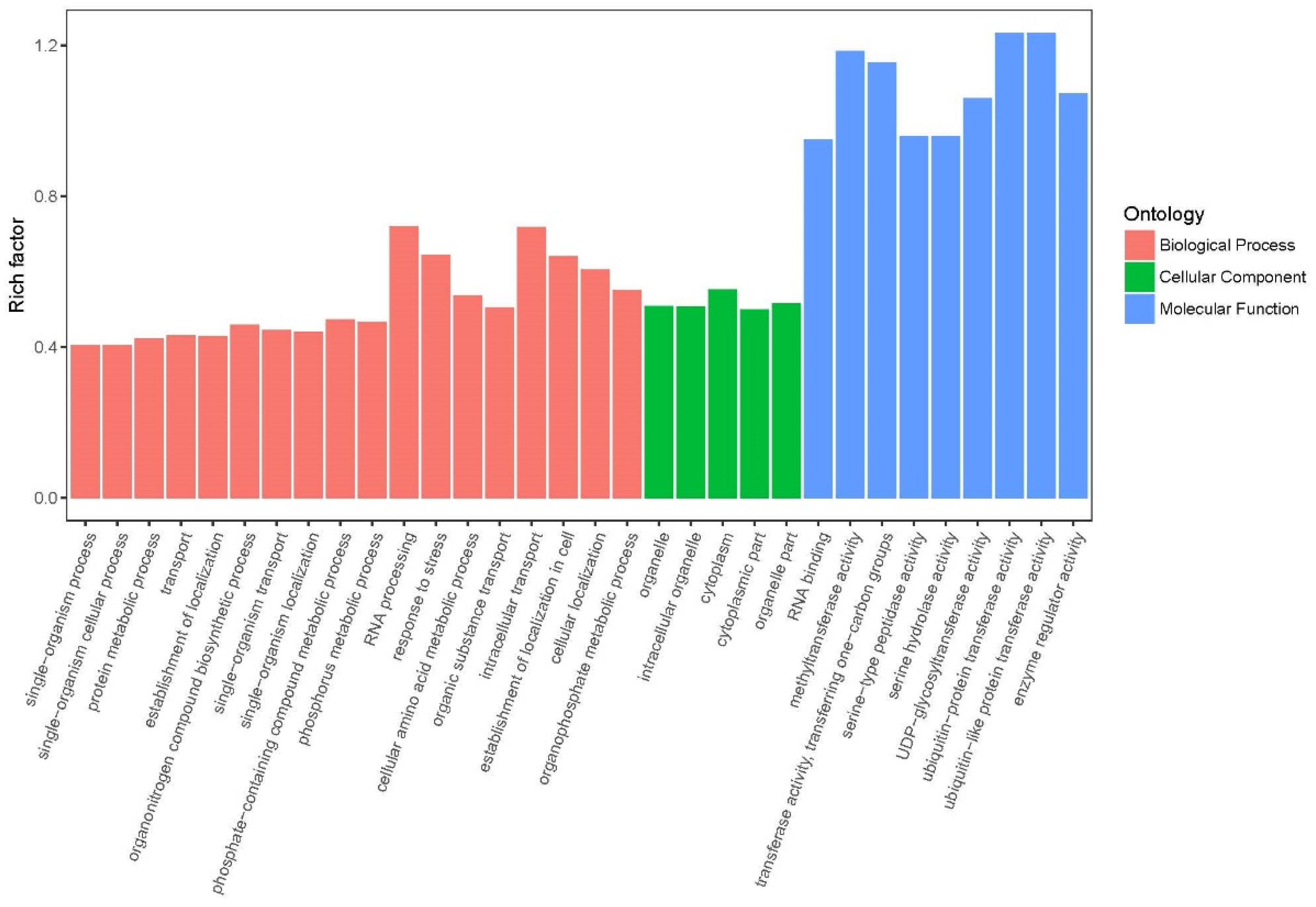
2.3.3. Gene Ontology (GO) Annotation
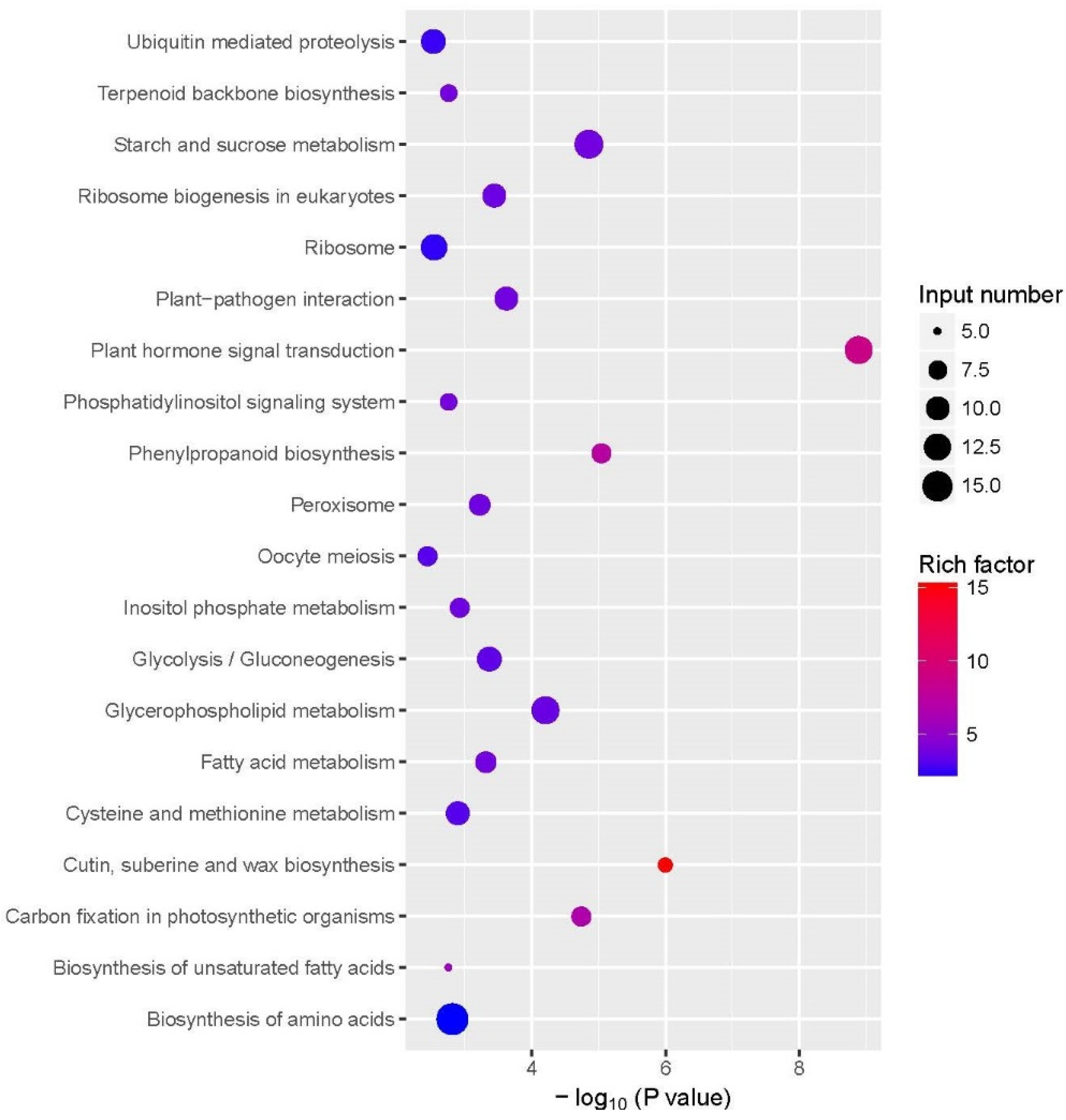
2.3.4. Kyoto Encyclopedia of Genes and Genomes (KEGG) Annotation
2.3.5. Validation of qRT-PCR (Quantitative Real-Time Polymerase Chain Reaction)
2.4. Metabolome Data Analysis
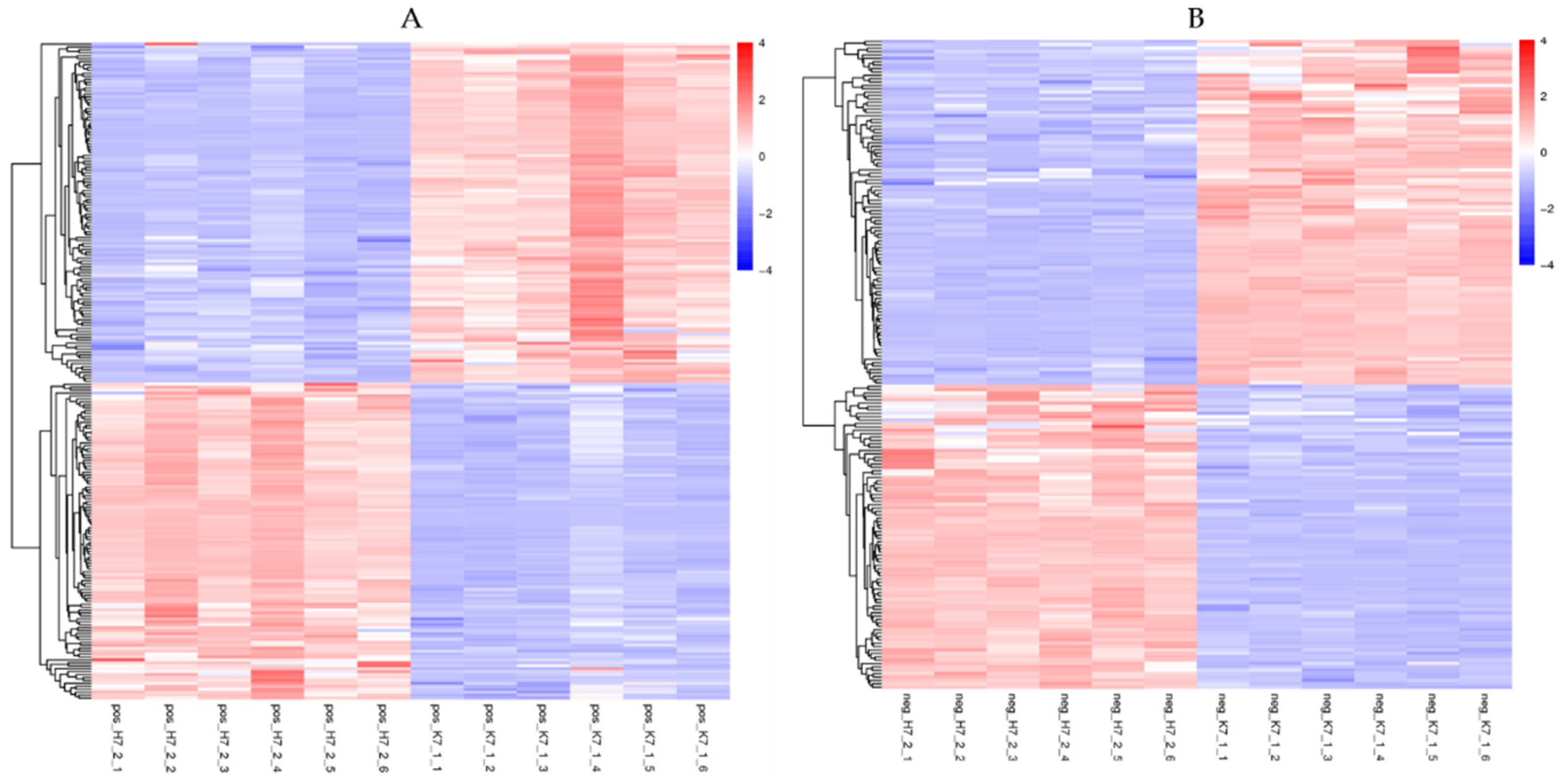
2.4.1. Effect of COS on Differential Changes in Tea Metabolites
2.4.2. KEGG Analysis of DMs Response to COS Treatment
2.5. Combination of the Metabolic Profiling and Transcriptomic Analysis
3. Discussion
4. Materials and Methods
4.1. Field Experiment, Sample Collection and Yield Estimate
4.2. Determination of Chlorophyll, Soluble Sugar and Amino Acid Content
4.3. Transcriptomic Profiling
4.4. Metabolite Profiling
4.4.1. Extraction and Detection of Metabolites
4.4.2. Data Processing and Statistics
4.5. Validation of qRT-PCR
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, Z.J.; Ma, H.Y.; Zhuang, J. ITRAQ-based proteomics monitors the withering dynamics in postharvest leaves of tea plant (Camellia sinensis). Mol. Genet. Genom. 2017, 293, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Jayasinghe, S.L.; Kumar, L. Potential impact of the current and future climate on the yeld, quality, and climate suitability for tea [Camellia sinensis (L.) O. Kuntze]: A systematic review. Agronomy 2021, 11, 619. [Google Scholar] [CrossRef]
- Kumar, A.; Seema. Monitoring of fluoride contamination and correlation with physicochemical parameters of surface soil and ground water near tea-garden of Thakurganj block of Kishanganj, Bihar, India. Am. J. Environ. Eng. 2016, 6, 38–51. [Google Scholar]
- Zhou, Y. Risk regulation in the application of the pesticide residue standards—A case study of Chinese tea. Sch. J. Agric. Vet. Sci. 2016, 3, 389–394. [Google Scholar]
- Mourato, S.; Ozdemiroglu, E.; Foster, V. Evaluating health and environmental impacts of pesticide use: Implications for the design of ecolabels and pesticide taxes. Environ. Sci. Technol. 2000, 34, 1456–1461. [Google Scholar] [CrossRef]
- Chen, H.; Hao, Z.X.; Wang, Q.H.; Jiang, Y.; Pan, R.; Wang, C.; Liu, X.; Lu, C.Y. Occurrence and risk assessment of organophosphorus pesticide residues in Chinese tea. Hum. Ecol. Risk Assess. Int. J. 2016, 22, 28–38. [Google Scholar] [CrossRef]
- Minhas, P.S.; Rane, J.; Pasala, R.K. Plant Bioregulators: A Stress Mitigation Strategy for Resilient Agriculture. In Abiotic Stress Management for Resilient Agriculture; Minhas, P., Rane, J., Pasala, R., Eds.; Springer: Singapore, 2017; pp. 235–259. [Google Scholar]
- Mukarram, M.; Khan, M.; Choudhary, S.; Zehra, A.; Aftab, T. Natural Polysaccharides: Novel Plant Growth Regulators. In Plant Growth Regulators; Aftab, T., Hakeem, K.R., Eds.; Springer: Aligarh, India, 2021; pp. 335–354. [Google Scholar]
- Mzibra, A.; Aasfar, A.; El Arroussi, H.; Khouloud, M.; Dhiba, D.; Kadmiri, I.M.; Bamouh, A. Polysaccharides extracted from moroccan seaweed: A promising source of tomato plant growth promoters. J. Appl. Phycol. 2018, 30, 2953–2962. [Google Scholar] [CrossRef]
- Mondal, M.; Malek, M.A.; Puteh, A.B.; Ismail, M.R.; Naher, L. Effect of foliar application of chitosan on growth and yield inokra. Aust. J. Crop Sci. 2012, 6, 918–921. [Google Scholar]
- Xia, W.; Liu, P.; Zhang, J.; Chen, J. Biological activities of chitosan and chitooligosaccharides. Food Hydrocoll. 2011, 25, 170–179. [Google Scholar] [CrossRef]
- Badawy, M.; Rabea, E.I. A biopolymer chitosan and its derivatives as promising antimicrobial agents against plant pathogens and their applications in crop protection. Int. J. Carbohydr. Chem. 2011, 2011, 29. [Google Scholar] [CrossRef]
- Manjunatha, G.; Roopa, K.S.; Prashanth, G.N.; Shekar Shetty, H. Chitosan enhances disease resistance in pearl millet against downy mildew caused by Sclero-spora graminicola and defence-related enzyme activation. Pest Manag. Sci. Former. Pestic. Sci. 2008, 64, 1250–1257. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, D.; Hemantaranjan, A.; Singh, B.; Bhanu, A.N. A future perspective in crop protection: Chitosan and its oligosaccharides. Adv. Plants Agric. Res. 2014, 1, 6. [Google Scholar]
- Wink, M. Plant breeding: Importance of plant secondary metabolites for protection against pathogens and herbivores. Theor. Appl. Genet. 1988, 75, 225–233. [Google Scholar] [CrossRef]
- Chibu, H.; Shibayama, H.; Arima, S. Effects of chitosan application on the shoot growth of rice and soybean. Jpn. J. Crop Sci. 2002, 71, 212–219. [Google Scholar] [CrossRef][Green Version]
- Rabea, E.I.; Badawy, M.E.; Steurbaut, W.; Stevens, C.V. In vitro assessment of N-(benzyl) chitosan derivatives against some plant pathogenic bacteria and fungi. Eur. Polym. J. 2009, 45, 237–245. [Google Scholar] [CrossRef]
- Sathiyabama, M.; Akila, G.; Charles, R.E. Chitosan-induced defence responses in tomato plants against early blight disease caused by Alternaria solani (Ellis and Martin) Sorauer. Arch. Pflanzenschutz 2014, 47, 1963–1973. [Google Scholar] [CrossRef]
- Islam, M.M.; Kabir, M.H.; Mamun, A.; Islam, M.; Das, P. Studies on yield and yield attributes in tomato and chilli using foliar application of oligo-chitosan. GSC Biol. Pharm. Sci. 2018, 3, 20–28. [Google Scholar] [CrossRef]
- Ibraheim, S.; Mohsen, A. Effect of chitosan and nitrogen rates on growth and productivity of summer squash plants. Middle East 2015, 4, 673–681. [Google Scholar]
- Sheikha, S.; Al-Malki, F.M. Growth and chlorophyll responses of bean plants to the chitosan applications. Eur. J. Sci. Res. 2011, 50, 124–134. [Google Scholar]
- Falcón-Rodríguez, A.B.; Costales, D.; Gónzalez-Pea, D.; Morales, D.; Cabrera, J.C. Chitosans of different molecular weight enhance potato (Solanum tuberosum L.) yield in a field trial. Span. J. Agric. Res. 2017, 15, e0902. [Google Scholar] [CrossRef]
- Wang, M.; Chen, Y.; Zhang, R.; Wang, W.; Zhao, X.; Du, Y.; Yin, H. Effects of chitosan oligosaccharides on the yield components and production quality of different wheat cultivars (Triticum aestivum L.) in Northwest China. Field Crops Res. 2015, 172, 11–20. [Google Scholar] [CrossRef]
- Tham, L.X.; Nagasawa, N.; Matsuhashi, S.; Kume, T. Effect of radiation-degraded chitosan on plants stressed with vanadium. Radiat. Phys. Chem. 2001, 61, 171–175. [Google Scholar] [CrossRef]
- Yan, L.; Wei, L.J.; Wang, Q.; Li, H.; Zhao, X.; Hou, H.; Du, Y. Effects of oligochitosan on photosynthetic parameter of Brassica napus L. leaves. Chin. Agric. Sci. Bull. 2010, 26, 132–136. [Google Scholar]
- Zhang, X.; Li, K.; Xing, R.; Liu, S.; Li, P. Metabolite profiling of wheat seedlings induced by chitosan: Revelation of the enhanced carbon and nitrogen metabolism. Front. Plant Sci. 2017, 8, 2017. [Google Scholar] [CrossRef] [PubMed]
- Ou, L.; Zhang, Q.; Ji, D.; Li, Y.; Zhou, X.; Jin, L. Physiological, transcriptomic investigation on the tea plant growth and yield motivation by chitosan oligosaccharides. Horticulturae 2022, 8, 68. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Ou, L.; Ji, D.; Liu, T.; Lan, R.; Li, X.; Jin, L. Response to the cold stress signaling of the tea plant (Camellia sinensis) elicited by chitosan oligosaccharide. Agronomy 2020, 10, 915. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhang, H.; Wang, T.; Chen, S.; Dai, S. Proteomics-based investigation of salt-responsive mechanisms in plant roots. J. Proteom. 2013, 82, 230–253. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, L.; Zeng, K. Efficacy of Pichia membranaefaciens combined with chitosan against Colletotrichum gloeosporioides in citrus fruits and possible modes of action. Biol. Control 2016, 96, 39–47. [Google Scholar] [CrossRef]
- Winkler, A.J.; Dominguez-Nunez, J.A.; Aranaz, I.; Poza-Carrion, C.; Ramonell, K.; Somerville, S.; Berrocal-Lobo, M. Short-chain chitin oligomers: Promoters of plant growth. Mar. Drugs 2017, 15, 40. [Google Scholar] [CrossRef]
- Wang, H.; Guo, X.; Li, Q.; Lu, Y.; Yan, S. Integrated transcriptomic and metabolic framework for carbon metabolism and plant hormones regulation in Vigna radiata during post-germination seedling growth. Sci. Rep. 2020, 10, 3745. [Google Scholar] [CrossRef]
- An, L.; Ma, J.; Wang, H.; Li, F.; Qin, D.; Wu, J.; Zhu, G.; Zhang, J.; Yuan, Y.; Zhou, L.; et al. NMR-based global metabolomics approach to decipher the metabolic effects of three plant growth regulators on strawberry maturation. Food Chem. 2018, 269, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Suo, J.; Liu, S.; Xu, J.; Yang, T.; Xiang, N.; Wu, Y.; Lu, B.; Qin, R.; Liu, H.; et al. Combined transcriptomic and metabolomic analysis reveals the potential mechanism of seed germination and young seedling growth in Tamarix hispida. BMC Genom. 2022, 23, 109. [Google Scholar] [CrossRef] [PubMed]
- Dzung, N.A.; Khanh, V.; Dzung, T.T. Research on impact of chitosan oligomers on biophysical characteristics, growth, development and drought resistance of coffee. Carbohydr. Polym. 2011, 84, 751–755. [Google Scholar] [CrossRef]
- Dalio, R.; Pinheiro, H.P.; Sodek, L.; Haddad, C. The effect of 24-epibrassinolide and clotrimazole on the adaptation of Cajanus cajan (L.) Millsp. to salinity. Acta Physiol. Plant. 2011, 33, 1887–1896. [Google Scholar] [CrossRef]
- Balal, R.M.; Shahid, M.A.; Javaid, M.M.; Iqbal, Z.; Liu, G.D.; Zotarelli, L.; Khan, N. Chitosan alleviates phytotoxicity caused by boron through augmented polyamine metabolism and antioxidant activities and reduced boron concentration in Cucumis sativus L. Acta Physiol. Plant. 2017, 39, 31. [Google Scholar] [CrossRef]
- Dar, T.A.; Uddin, M.; Khan, M.M.A.; Ali, A.; Mir, S.R.; Varshney, L. Effect of Co-60 gamma irradiated chitosan and phosphorus fertilizer on growth, yield and trigonelline content of Trigonella foenum-graecum L. J. Radiat. Res. Appl. Sci. 2015, 8, 446–458. [Google Scholar] [CrossRef]
- Uthairatanakij, A.; Silva, J.; Obsuwan, K. Chitosan for improving orchid production and quality. Orchid. Sci. Biotechnol. 2007, 1, 1–5. [Google Scholar]
- Ahmed, A.; Nesiem, A.E.; Allam, H.A.; El-Wakil, A.F. Effect of pre-harvest chitosan foliar application on growth, yield and chemical composition of Washington navel orange trees grown in two different regions. Afr. J. Biochem. Res. 2016, 10, 59–69. [Google Scholar] [CrossRef]
- Studer, A.J.; Gandin, A.; Kolbe, A.R.; Wang, L.; Cousins, A.B.; Brutnell, T.P. A limited role for carbonic anhydrase in C4 photosynthesis as revealed by a ca1ca2 double mutant in maize. Plant Physiol. 2014, 165, 608. [Google Scholar] [CrossRef]
- Rueda-López, M.; Cañas, R.A.; Canales, J.; Cánovas, F.M.; Ávila, C. The overexpression of the pine transcription factor PpDof5 in Arabidopsis leads to increased lignin content and affects carbon and nitrogen metabolism. Physiol. Plant. 2015, 155, 369–383. [Google Scholar] [CrossRef]
- Masahiro, T.; Miki, N.; Yukinori, Y.; Shigeru, S. Carbon metabolism in the Calvin cycle. Plant Tissue Cult. Lett. 2005, 22, 355–360. [Google Scholar]
- Zhang, X.; Li, K.; Xing, R.; Liu, S.; Chen, X.; Yang, H.; Li, P. miRNA and mRNA expression profiles reveal insight into chitosan-mediated regulation of plant growth. J. Agric. Food Chem. 2018, 66, 3810–3822. [Google Scholar] [CrossRef] [PubMed]
- Akimoto-Tomiyama, C.; Sakata, K.; Yazaki, J.; Nakamura, K.; Fujii, F.; Shimbo, K.; Yamamoto, K.; Sasaki, T.; Kishimoto, N.; Kikuchi, S. Rice gene expression in response to N-acetylchitooligosaccharide elicitor: Comprehensive analysis by DNA microarray with randomly selected ESTs. Plant Mol. Biol. 2003, 52, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, J.X.; Terce-Laforgue, T.; Armengaud, P.; Clement, G.; Renou, J.P.; Pelletier, S.; Catterou, M.; Azzopardi, M.; Gibon, Y.; Lea, P.J. Characterization of a NADH-dependent glutamate dehydrogenase mutant of arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell 2012, 24, 4044–4065. [Google Scholar] [CrossRef] [PubMed]
- Elboutachfaiti, R.; Molinié, R.; Mathiron, D.; Maillot, Y.; Fontaine, J.X.; Pilard, S.; Quéro, A.; Brasselet, C.; Dols-Lafargue, M.; Delattre, C.; et al. Secondary metabolism rearrangements in Linum usitatissimum L. after biostimulation of roots with COS oligosaccharides from fungal cell wall. Molecules 2022, 27, 2372. [Google Scholar] [CrossRef]
- Chamnanmanoontham, N.; Pongprayoon, W.; Pichayangkura, R.; Roytrakul, S.; Chadchawan, S. Chitosan enhances rice seedling growth via gene expression network between nucleus and chloroplast. Plant Growth Regul. 2015, 75, 101–114. [Google Scholar] [CrossRef]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2013, 25, 402–408. [Google Scholar] [CrossRef]




| Yield Indicators | CK | COS Treatment (with Different Times of Dilution) | ||
|---|---|---|---|---|
| 3.2 g/667 m2 (1:500) | 2.0 g/667 m2 (1:800) | 1.6 g/667 m2 (1:1000) | ||
| Bud density (bud/m2) | 1028 ± 48b | 1223 ± 60a | 1288 ± 38a | 1269 ± 64a |
| 100-bud weight (g/100 bud) | 6.76 ± 0.04b | 7.68 ± 0.07a | 7.77 ± 0.04a | 7.70 ± 0.07a |
| Actual output (g/m2) | 29.35 ± 0.10b | 33.96 ± 0.04a | 36.33 ± 0.06a | 34.66 ± 0.08a |
| Samples | Raw Reads | Clean Reads | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|
| TreT7_1(COS) | 84011624 | 78935806 | 100 | 99.4 | 46.81 |
| TreT7_2(COS) | 106371756 | 99995940 | 100 | 99.3 | 46.73 |
| TreT7_3(COS) | 70432706 | 66250686 | 100 | 99.3 | 46.73 |
| ConT7_1(CK) | 81114666 | 75623100 | 100 | 99.3 | 46.94 |
| ConT7_2(CK) | 96205136 | 89960092 | 100 | 99.3 | 46.74 |
| ConT7_3(CK) | 76155870 | 70900816 | 100 | 99.3 | 46.8 |
| Compared Samples | Total Metabolites | Total DMs | Upregulated DMs | Down Regulated DMs |
|---|---|---|---|---|
| COS vs. control pos | 748 | 224 | 108 | 116 |
| COS vs. control neg | 591 | 192 | 90 | 102 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, D.; Ou, L.; Ren, X.; Yang, X.; Tan, Y.; Zhou, X.; Jin, L. Transcriptomic and Metabolomic Analysis Reveal Possible Molecular Mechanisms Regulating Tea Plant Growth Elicited by Chitosan Oligosaccharide. Int. J. Mol. Sci. 2022, 23, 5469. https://doi.org/10.3390/ijms23105469
Ji D, Ou L, Ren X, Yang X, Tan Y, Zhou X, Jin L. Transcriptomic and Metabolomic Analysis Reveal Possible Molecular Mechanisms Regulating Tea Plant Growth Elicited by Chitosan Oligosaccharide. International Journal of Molecular Sciences. 2022; 23(10):5469. https://doi.org/10.3390/ijms23105469
Chicago/Turabian StyleJi, Dezhong, Lina Ou, Xiaoli Ren, Xiuju Yang, Yanni Tan, Xia Zhou, and Linhong Jin. 2022. "Transcriptomic and Metabolomic Analysis Reveal Possible Molecular Mechanisms Regulating Tea Plant Growth Elicited by Chitosan Oligosaccharide" International Journal of Molecular Sciences 23, no. 10: 5469. https://doi.org/10.3390/ijms23105469
APA StyleJi, D., Ou, L., Ren, X., Yang, X., Tan, Y., Zhou, X., & Jin, L. (2022). Transcriptomic and Metabolomic Analysis Reveal Possible Molecular Mechanisms Regulating Tea Plant Growth Elicited by Chitosan Oligosaccharide. International Journal of Molecular Sciences, 23(10), 5469. https://doi.org/10.3390/ijms23105469






