Heterologous Expression of Dehydration-Inducible MfbHLH145 of Myrothamnus flabellifoli Enhanced Drought and Salt Tolerance in Arabidopsis
Abstract
1. Introduction
2. Results and Discussions
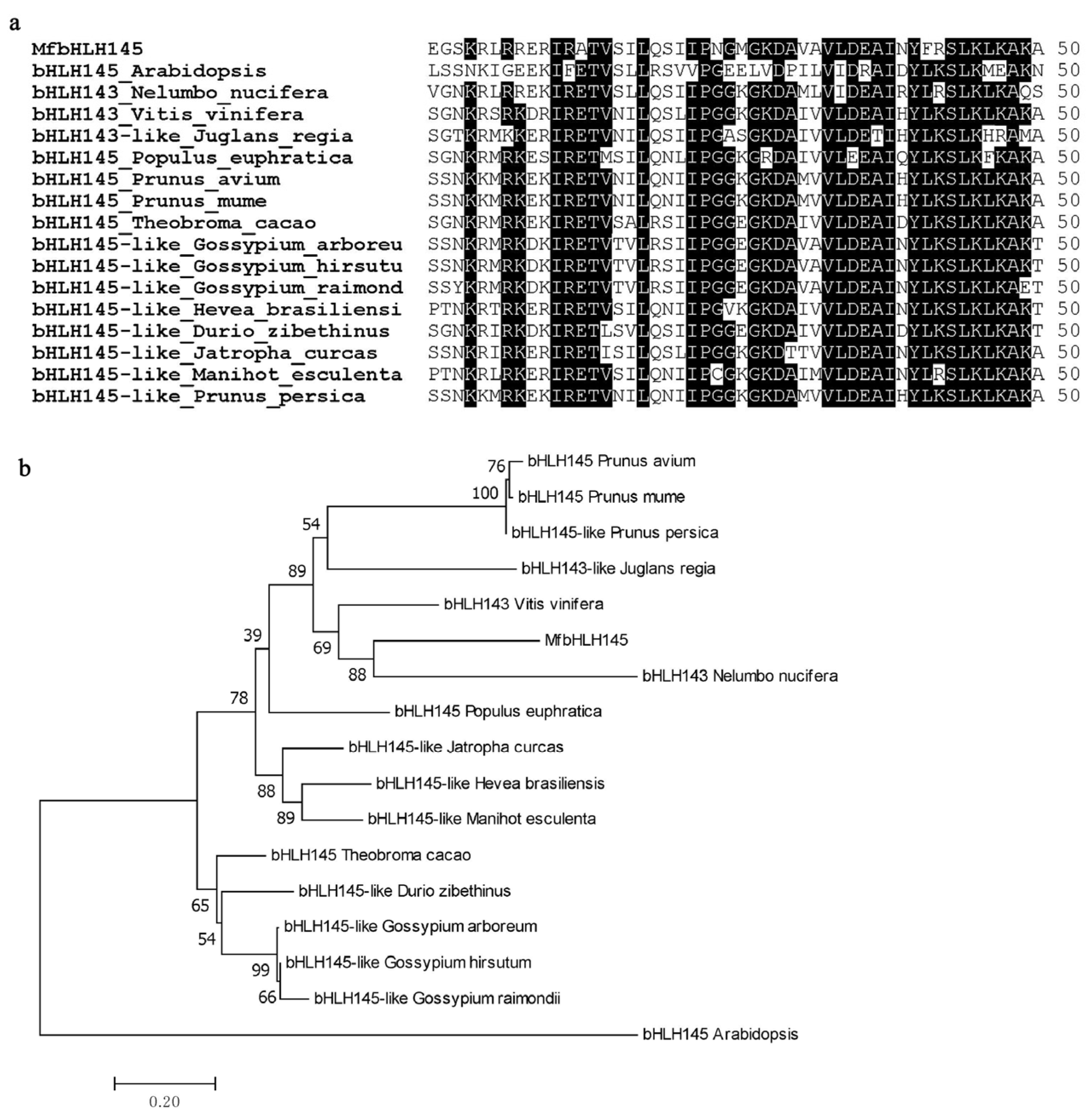
2.1. Isolation and Sequence Analysis of MfbHLH145
2.2. MfbHLH145 Is Localized in the Nucleus
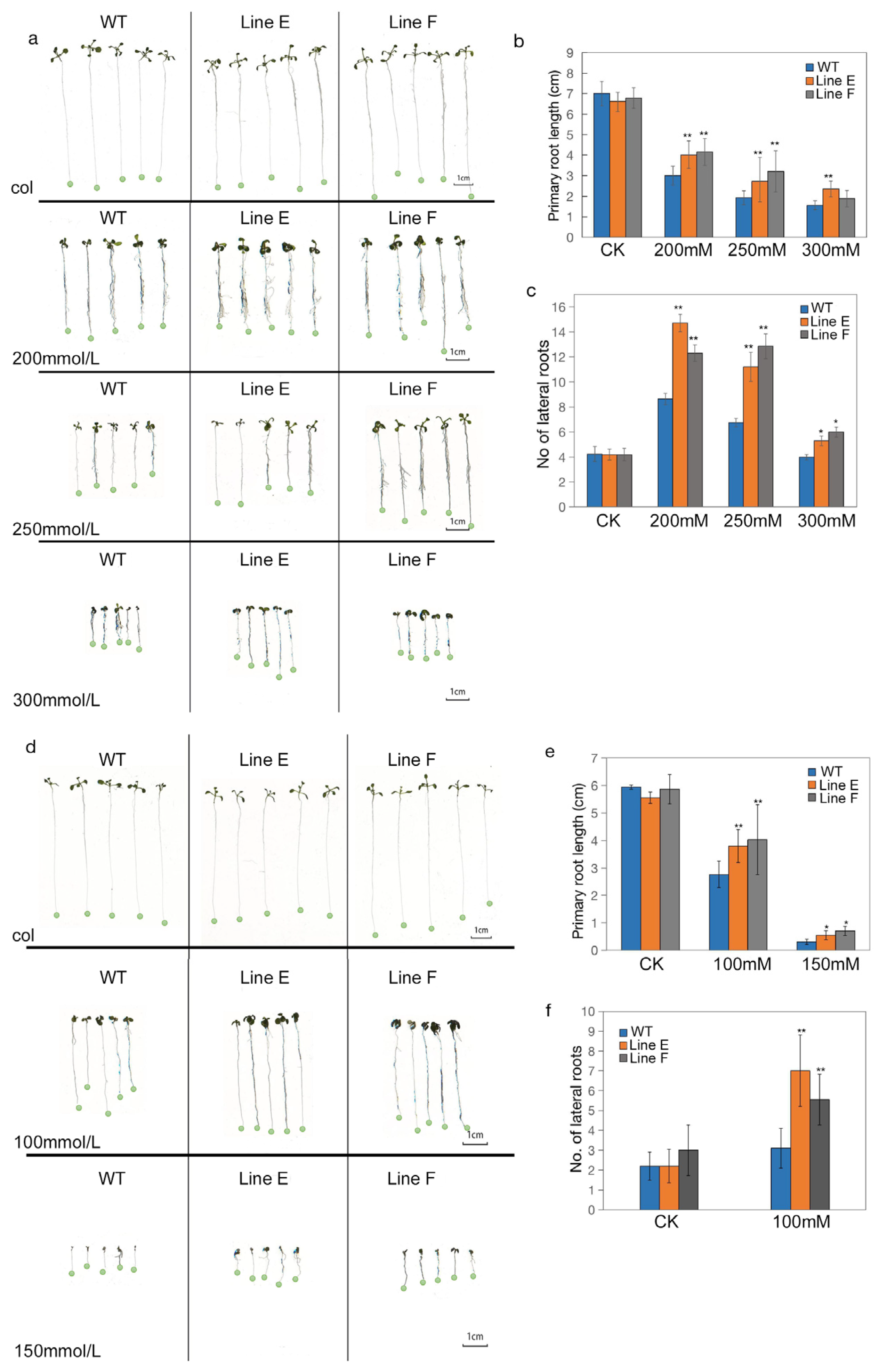
2.3. Overexpression of MfbHLH145 in Arabidopsis Enhanced Tolerance to Drought and Salt
2.4. MfbHLH145 Enhanced Leaf Water Retention Capacity under Drought and Salt Stresses
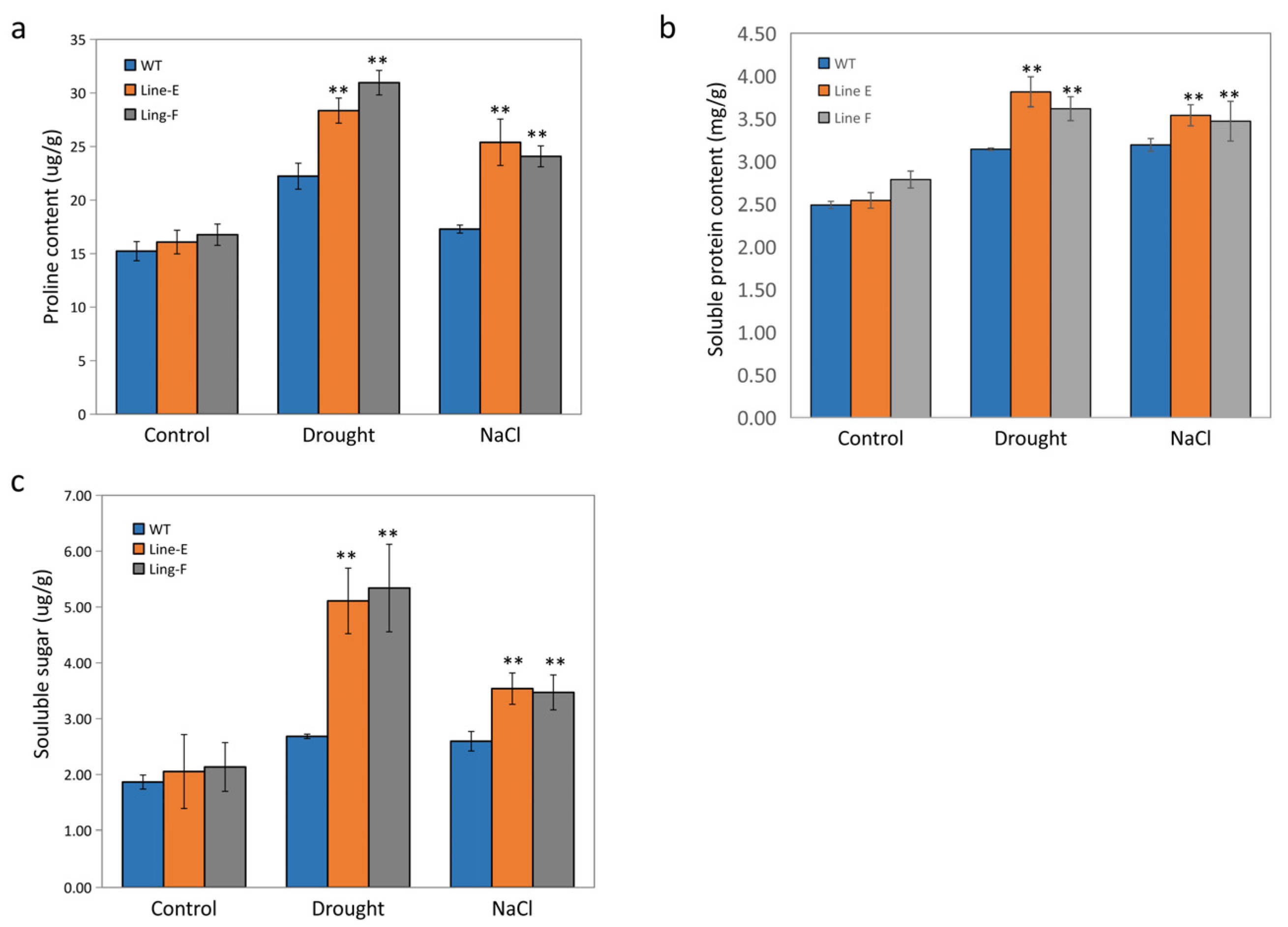
2.5. MfbHLH145 Increased Osmolytes Accumulation under Drought and Salt Stresses
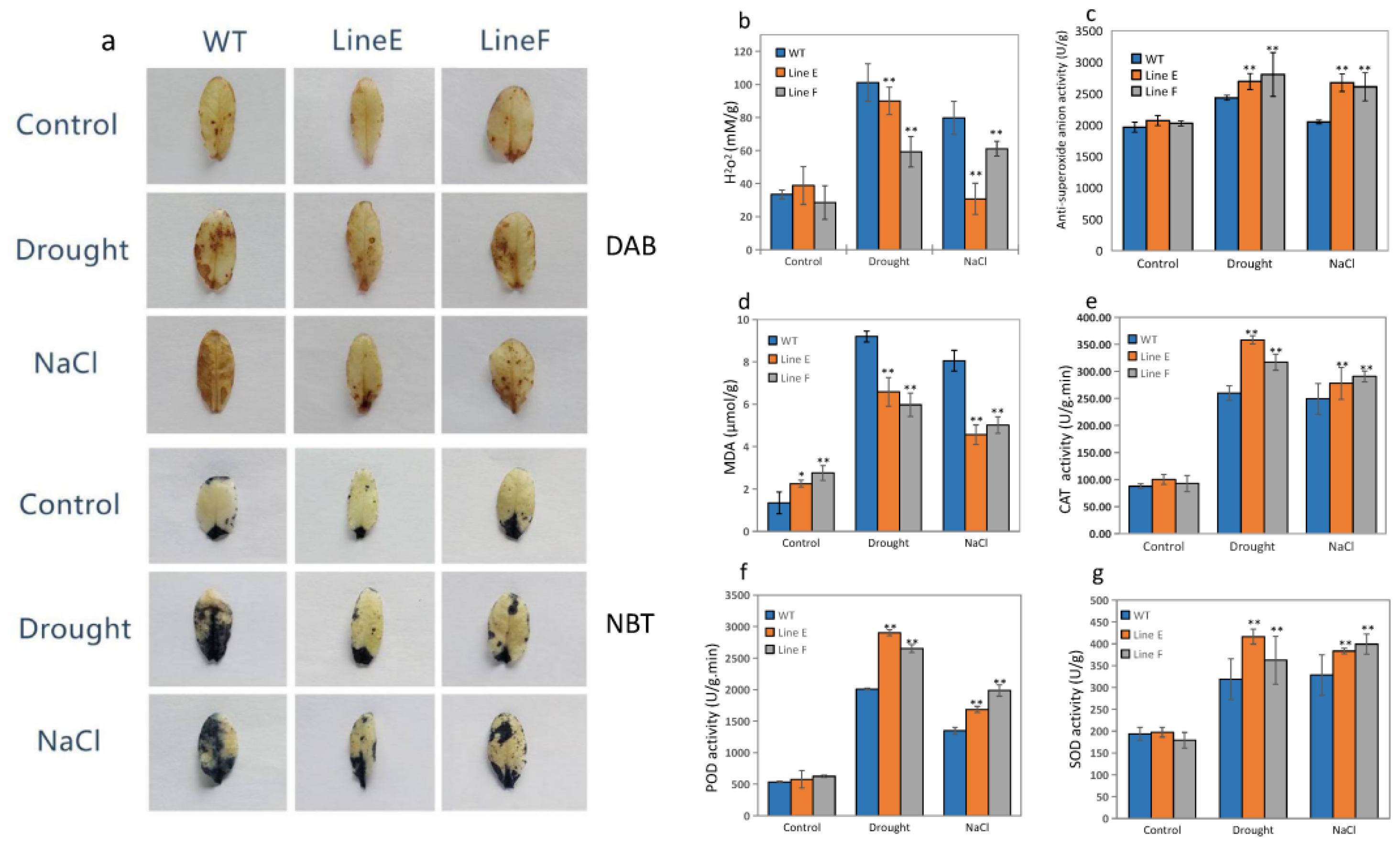
2.6. MfbHLH145 Decreased Stress-Induced Oxidative Damage through Increasing Antioxidant Enzyme Activities
3. Conclusions
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Gene Cloning and Sequence Analysis
4.3. Subcellular Localization of MfbHLH145
4.4. Generation of Transgenic Plants
4.5. Drought and Salt Treatment
4.6. Assays of Water Loss and Stomatal Aperture Closure
4.7. Measurement of Biochemistry Parameters Related to Stress Responses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shao, H.B.; Chu, L.Y.; Jaleel, C.A.; Manivannan, P.; Panneerselvam, R.; Shao, M.A. Understanding water deficit stress-induced changes in the basic metabolism of higher plants-biotechnologically and sustainably improving agriculture and the ecoenvironment in arid regions of the globe. Crit. Rev. Biotechnol. 2009, 29, 131–151. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yang, B.M.; Deyholos, M.K. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol. Genet. Genomics. 2009, 282, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Atchley, W.R.; Fitch, W.M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–5176. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.J.; Atchley, W.R. Phylogenetic analysis of plant basic helix-loop-helix proteins. J. Mol. Evol. 2003, l56, 742–750. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helixtranscription factor family. Plant Cell. 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Li, X.X.; Duan, X.P.; Jiang, H.X.; Sun, Y.J.; Tang, Y.P.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-wide analysis of basic/helix-loop-helixtranscription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef]
- Bernhardt, C.; Lee, M.M.; Gonzalez, A.; Zhang, F.; Lloyd, A.; Schiefelbein, J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 2003, 130, 6431–6439. [Google Scholar] [CrossRef]
- Karas, B.; Amyot, L.; Johansen, C.; Sato, S.; Tabata, S.; Kawaguchi, M.; Szczyglowski, K. Conservation of Lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol. 2009, 151, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Monte, E.; Tepperman, J.M.; Al-Sady, B.; Kaczorowski, K.A.; Alonso, J.M.; Ecker, J.R.; Li, X.; Zhang, Y.; Quail, P.H. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 2004, 101, 16091–16098. [Google Scholar] [CrossRef] [PubMed]
- Godiard, L.; Lepage, A.; Moreau, S.; Laporte, D.; Verdenaud, M.; Timmers, T.; Gamas, P. MtbHLH1, a bHLH transcription factor involved in Medicago truncatula nodule vascular patterning and nodule to plant metabolic exchanges. New Phytol. 2011, 191, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Huq, E.; Quail, P.H. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002, 21, 2441–2450. [Google Scholar] [CrossRef]
- Yuan, Y.; Wu, H.; Wang, N.; Li, J.; Zhao, W.; Du, J.; Wang, D.; Ling, H.-Q. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008, 18, 385–396. [Google Scholar] [CrossRef]
- Wu, H.; Chen, C.; Du, J.; Liu, H.; Cui, Y.; Zhang, Y.; He, Y.; Wang, Y.; Chu, C.; Feng, Z.; et al. Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 2012, 158, 790–800. [Google Scholar] [CrossRef]
- Sivitz, A.B.; Hermand, V.; Curie, C.; Vert, G. Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS ONE 2012, 7, e44843. [Google Scholar] [CrossRef]
- Liu, W.; Tai, H.; Li, S.; Gao, W.; Zhao, M.; Xie, C.; Li, W.X. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef]
- Meng, C.M.; Zhang, T.Z.; Guo, W.Z. Molecular Cloning and Characterization of a Novel Gossypium hirsutum L. bHLH Gene in Response to ABA and Drought Stresses. Plant Mol. Biol. Rep. 2009, 27, p381–p387. [Google Scholar] [CrossRef]
- Ottow, E.A.; Polle, A.; Brosche, M.; Kangasjärvi, J.; Dibrov, P.; Zörb, C. Molecular characterization of PeNhaD1: The first member of the NhaD Na+/H+ antiporter family of plant origin. Plant Mol. Biol. 2005, 58, 75–88. [Google Scholar] [CrossRef]
- Tang, S.; Liang, H.; Yan, D.; Zhao, Y.; Han, X.; Carlson, J.E.; Xia, X.; Yin, W. Populus euphratica: The transcriptomic response to drought stress. Plant Mol. Biol. 2013, 8, 539–557. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhu, H.; Chen, D.; Li, Z.; Peng, R.; Yao, Q. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tiss. Organ. Cult. 2016, 125, 387–398. [Google Scholar] [CrossRef]
- Seo, J.-S.; Joo, J.; Kim, M.-J.; Kim, Y.-K.; Nahm, B.H.; Song, S.I.; Cheong, J.-J.; Lee, J.S.; Kim, J.-K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.R.; Yao, S.F.; Hao, L.; Zhao, Y.Y.; Lu, W.J.; Xiao, K. Wheat bHLH-type transcription factor gene TabHLH1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-associated pathway. Plant Cell Rep. 2016, 35, 2309–2323. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.P.; Nguema-Ona, E.; Chevalier, L.; Lindsey, G.G.; Brandt, W.F.; Lerouge, P.; Farrant, J.M.; Driouich, A. Response of the leaf cell wall to desiccation in the resurrection plant Myrothamnus flabellifolius. Plant Physiol. 2006, 141, 651–662. [Google Scholar] [CrossRef]
- Moore, J.P.; Lindsey, G.G.; Farrant, J.M.; Brandt, W.F. An overview of the biology of the desiccation-tolerant resurrection plant Myrothamnus flabellifolia. Ann. Bot. 2007, 99, 211. [Google Scholar] [CrossRef]
- Ma, C.; Wang, H.; Macnish, A.J.; Estrada-Melo, A.C.; Lin, J.; Chang, Y.; Reid, M.S.; Jiang, C.-Z. Transcriptomic analysis reveals numerous diverse protein kinases and transcription factors involved in desiccation tolerance in the resurrection plant Myrothamnus flabellifolia. Hortic. Res. 2015, 2, 15034. [Google Scholar] [CrossRef]
- Xu, D.; Marquis, K.; Pei, J.; Fu, S.C.; Cağatay, T.; Grishin, N.V.; Chook, M.Y. LocNES: A computational tool for locating classical NESs in CRM1 cargo proteins. Bioinformatics 2015, 31, 1357–1365. [Google Scholar] [CrossRef]
- Li, Z.X.; Liu, C.; Zhang, Y.; Wang, B.M.; Ran, Q.J.; Zhang, J.R. The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis. J. Exp. Bot. 2019, 70, 5471–5486. [Google Scholar] [CrossRef]
- Ko, S.-S.; Li, M.-J.; Ho, Y.-C.; Yu, C.-P.; Yang, T.-T.; Lin, Y.-J.; Hsing, H.-C.; Chen, T.-K.; Jhong, C.-M.; Li, W.-H.; et al. Rice transcription factor GAMYB modulates bHLH142 and is homeostatically regulated by TDR during anther tapetal and pollen development. J. Exp. Bot. 2021, 72, 4888–4903. [Google Scholar] [CrossRef]
- Bello, B.K.; Hou, Y.; Zhao, J.; Jiao, G.; Wu, Y.; Li, Z.; Wang, Y.; Tong, X.; Wang, W.; Yuan, W.; et al. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant Biotechnol. J. 2019, 17, 1222–1235. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Besseau, S.; Törönen, P.; Sipari, N.; Kollist, H.; Holm, L.; Palva, T. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013, 200, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Patanè, C.; Scordia, D.; Testa, G.; Cosentino, S.L. Physiological screening for drought tolerance in Mediterranean long-storage tomato. Plant Sci. 2016, 249, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef]
- Zhao, Q.; Fan, Z.; Qiu, L.; Che, Q.; Wang, T.; Li, Y.; Wang, Y. MdbHLH130, an Apple bHLH Transcription Factor, Confers Water Stress Resistance by Regulating Stomatal Closure and ROS Homeostasis in Transgenic Tobacco. Front. Plant Sci. 2020, 11, 543696. [Google Scholar] [CrossRef]
- Manavalan, L.P.; Guttikonda, S.K.; Tran, L.S.; Nguyen, H.T. Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 2009, 50, 1260–1276. [Google Scholar] [CrossRef]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, H.; Ali, J.; Li, Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in Rice. PLoS ONE 2014, 9, e92913. [Google Scholar] [CrossRef]
- Huang, Z.; He, J.; Xia, D.; Zhong, X.-J.; Li, X.; Sun, L.-X.; Cai, S.-Z. Evaluation of physiological responses and tolerance to low-temperature stress of four Iceland poppy (Papaver nudicaule) varieties. J. Plant Interac. 2016, 11, 1117–1123. [Google Scholar] [CrossRef][Green Version]
- Qiu, J.R.; Huang, Z.; Xiang, X.Y.; Xu, W.X.; Wang, J.T.; Chen, J.; Song, L.; Xiao, Y.; Li, X.; Ma, J.; et al. MfbHLH38, a Myrothamnus flabellifolia bHLH transcription factor, confers tolerance to drought and salinity stresses in Arabidopsis. BMC Plant Biol. 2020, 20, 542. [Google Scholar] [CrossRef]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–72. [Google Scholar] [CrossRef]
- Xiong, L.; Zhu, J.K. Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 2002, 25, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Jin, S.-H.; Yang, L.; Song, L.; Wang, Y.-H.; Jian, L.-L.; Jiang, C.-Z. Heterologous Expression of Dehydration-Inducible MfbHLH145 of Myrothamnus flabellifoli Enhanced Drought and Salt Tolerance in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 5546. https://doi.org/10.3390/ijms23105546
Huang Z, Jin S-H, Yang L, Song L, Wang Y-H, Jian L-L, Jiang C-Z. Heterologous Expression of Dehydration-Inducible MfbHLH145 of Myrothamnus flabellifoli Enhanced Drought and Salt Tolerance in Arabidopsis. International Journal of Molecular Sciences. 2022; 23(10):5546. https://doi.org/10.3390/ijms23105546
Chicago/Turabian StyleHuang, Zhuo, Si-Han Jin, Li Yang, Li Song, Yuan-Hong Wang, Lin-Li Jian, and Cai-Zhong Jiang. 2022. "Heterologous Expression of Dehydration-Inducible MfbHLH145 of Myrothamnus flabellifoli Enhanced Drought and Salt Tolerance in Arabidopsis" International Journal of Molecular Sciences 23, no. 10: 5546. https://doi.org/10.3390/ijms23105546
APA StyleHuang, Z., Jin, S.-H., Yang, L., Song, L., Wang, Y.-H., Jian, L.-L., & Jiang, C.-Z. (2022). Heterologous Expression of Dehydration-Inducible MfbHLH145 of Myrothamnus flabellifoli Enhanced Drought and Salt Tolerance in Arabidopsis. International Journal of Molecular Sciences, 23(10), 5546. https://doi.org/10.3390/ijms23105546





