Lactobacillus brevis BGZLS10-17 and Lb. plantarum BGPKM22 Exhibit Anti-Inflammatory Effect by Attenuation of NF-κB and MAPK Signaling in Human Bronchial Epithelial Cells
Abstract
1. Introduction
2. Results
2.1. Safety Assessment of LAB Strains
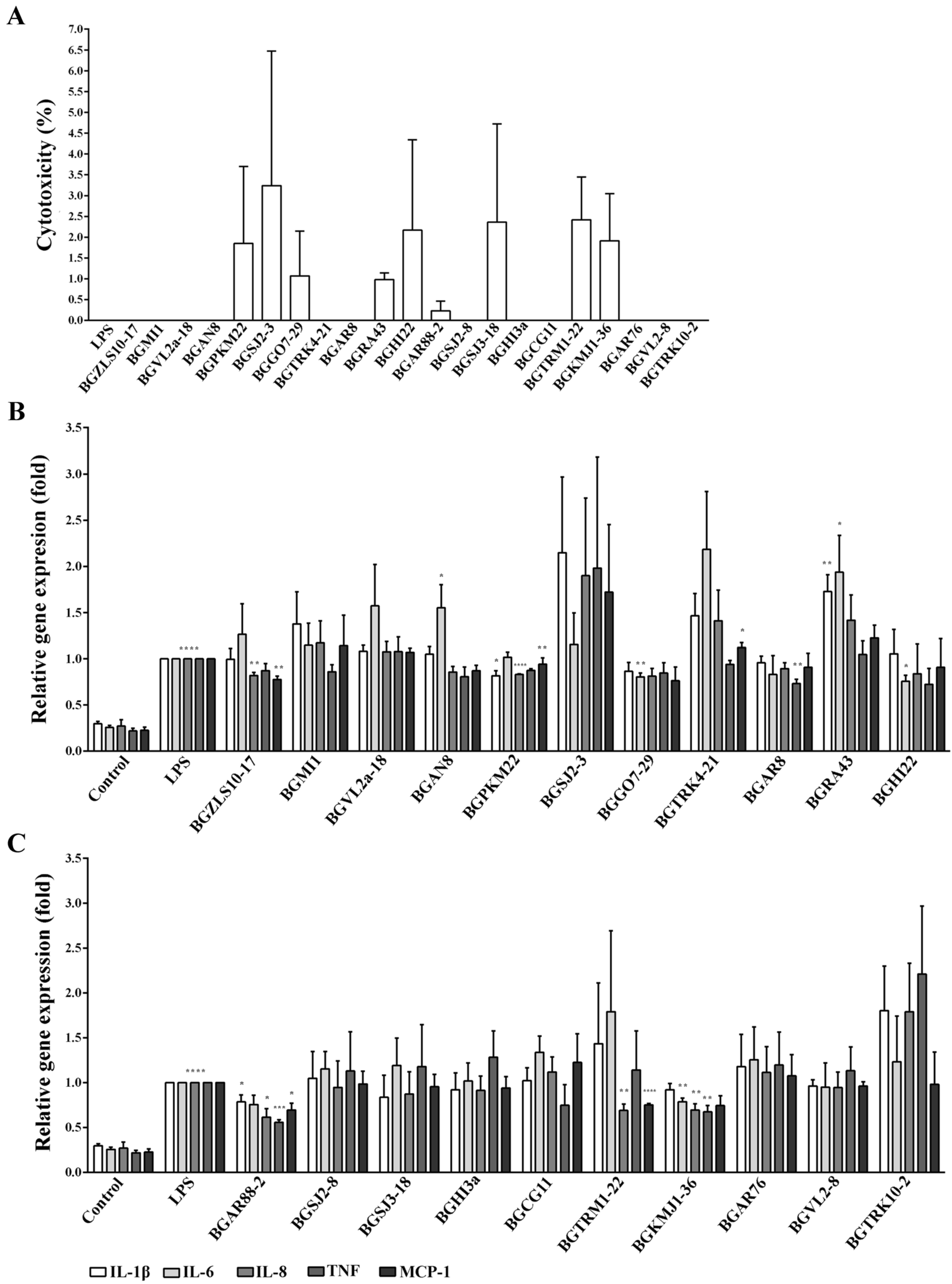
2.2. The Cytotoxicity
2.3. Immunomodulatory Effects of 21 LAB Strains on the Expression of Pro-Inflammatory Genes
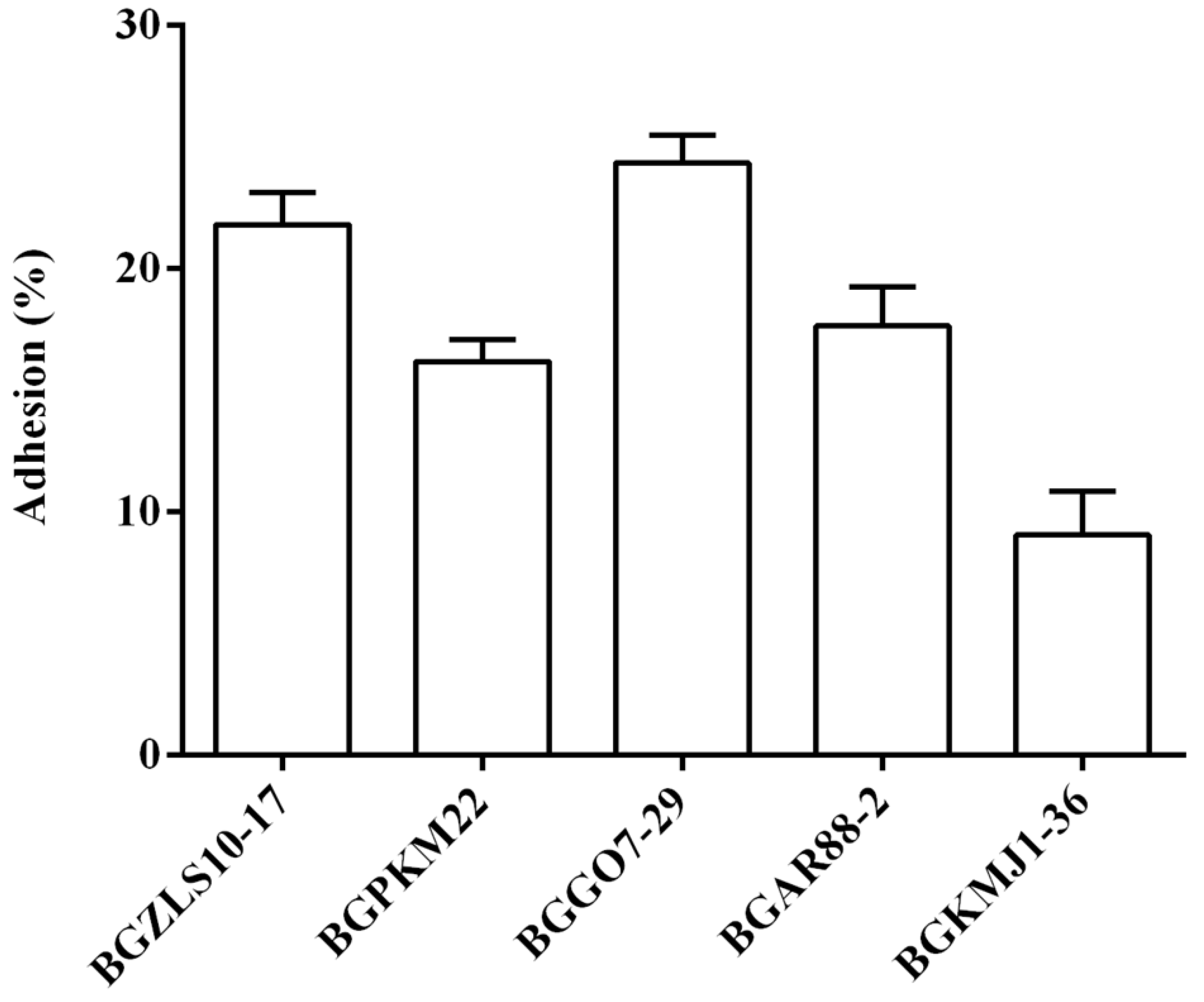
2.4. Adhesion of Selected LAB Strains to BEAS-2B Cells
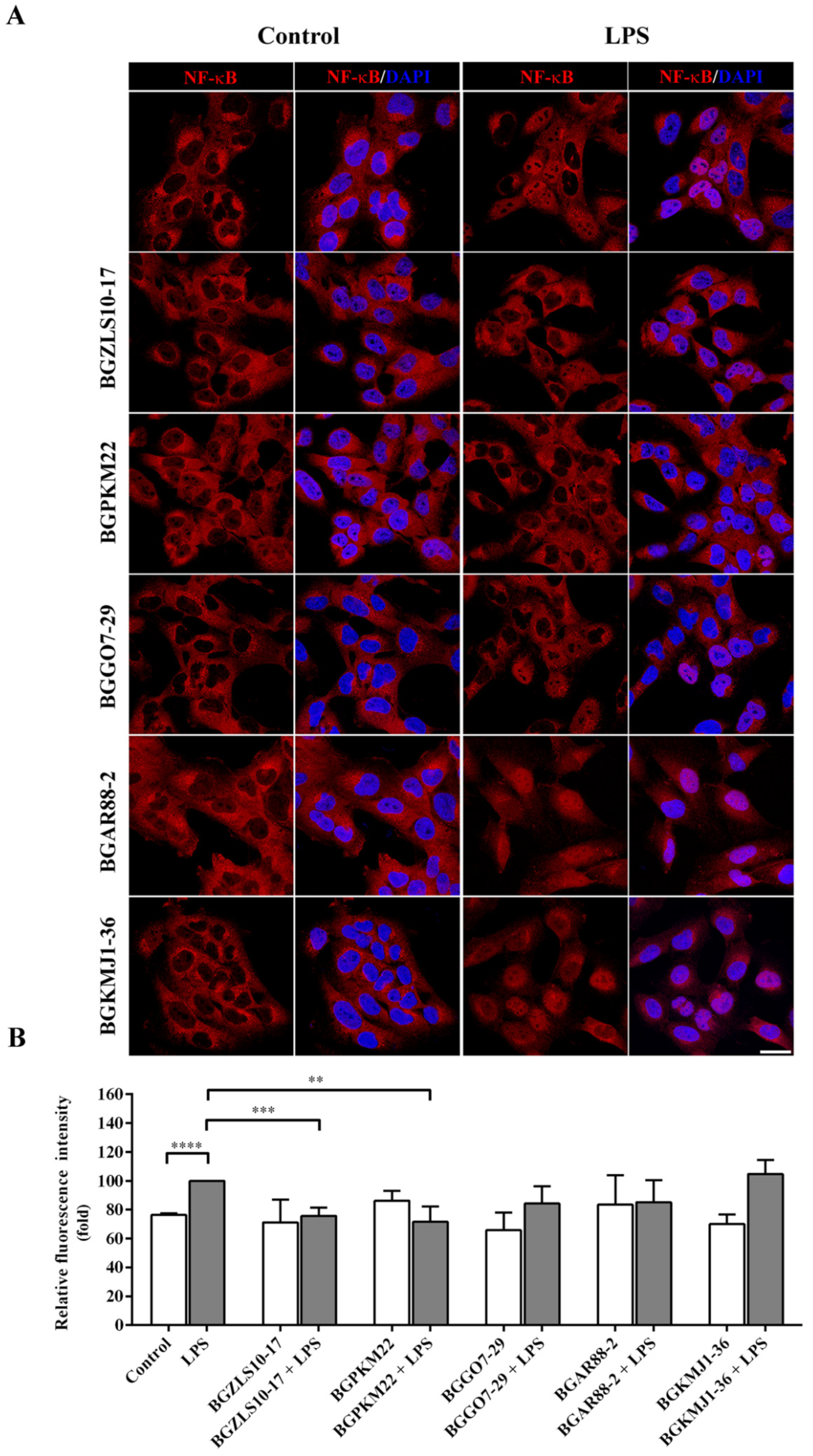
2.5. LAB Strains BGZLS10-17 and BGPKM22 Attenuated LPS-Induced Nuclear Translocation of NF-κB
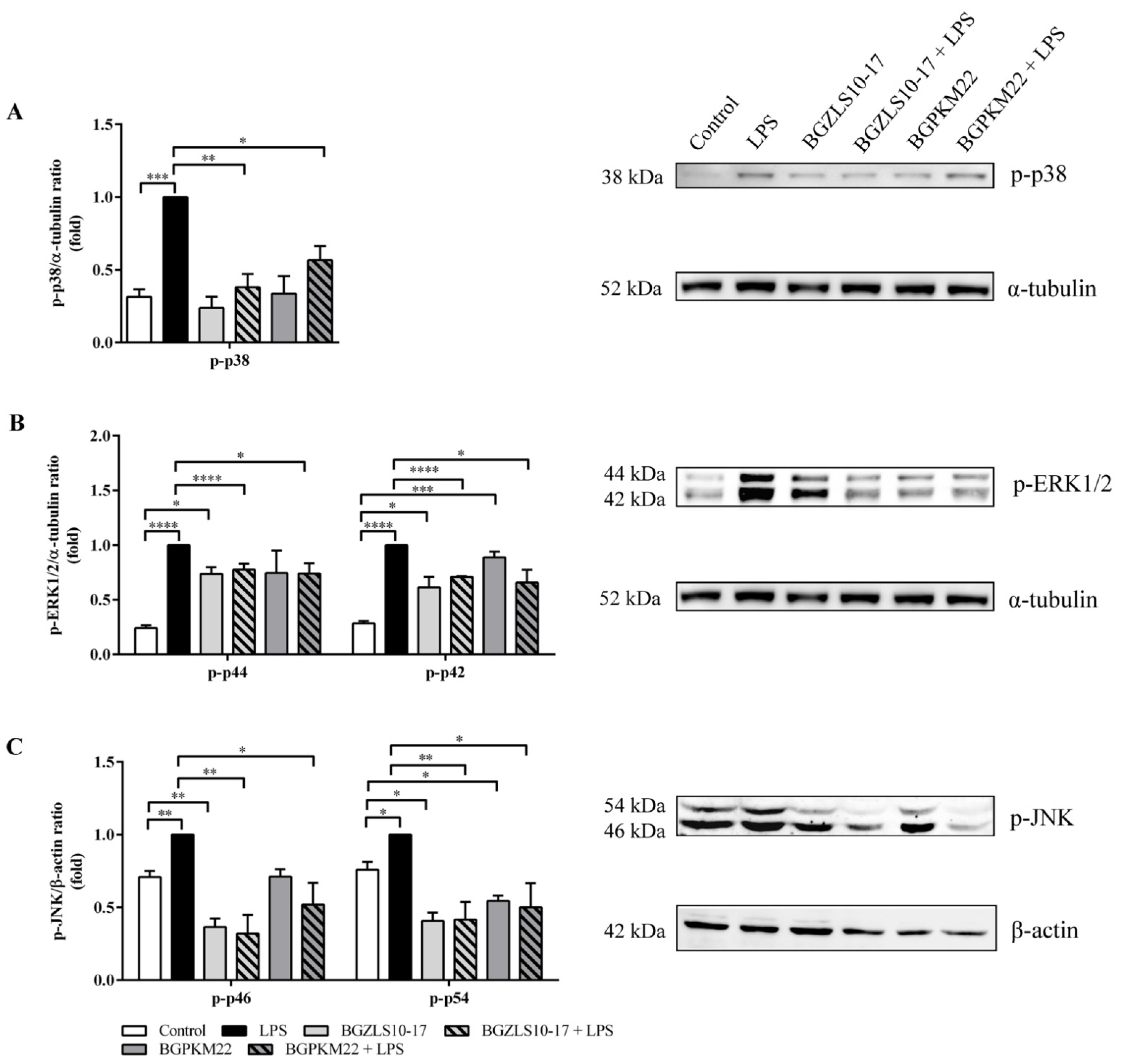
2.6. LAB Strains BGZLS10-17 and BGPKM22 Attenuate LPS-Induced Activation of p38, ERK1/2, and JNK MAPK Signaling
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Media, and Growth Conditions
4.2. Antibiotic Susceptibility
4.3. Hemolytic and Gelatinase Activity Assays
4.4. Cell Culture
4.5. Lactate Dehydrogenase (LDH) Release Assay
4.6. Quantitative Real Time-PCR (qRT-PCR) Assay
4.7. Adhesion Assay
4.8. Immunocytochemistry
4.9. Image Acquisition and Analysis
4.10. Western Blotting
4.11. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Gao, W.; Li, L.; Wang, Y.; Zhang, S.; Adcock, I.M.; Barnes, P.J.; Mao, H.; Xin, Y. Bronchial epithelial cells: The key effector cells in the pathogenesis of chronic obstructive pulmonary disease? Respirology 2015, 20, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Whitsett, J.A.; Alenghat, T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 2015, 16, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Fabbrizzi, A.; Amedei, A.; Lavorini, F.; Renda, T.; Fontana, G. The lung microbiome: Clinical and therapeutic implications. Inter. Emerg. Med. 2019, 14, 1241–1250. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, E.; Escribano-Vazquez, U.; Descamps, D.; Cherbuy, C.; Langella, P.; Riffault, S.; Aude, R.; Muriel, T. Paradigms of lung microbiota functions in health and disease, particularly, in asthma. Front. Physiol. 2018, 9, 1168. [Google Scholar] [CrossRef] [PubMed]
- Pattaroni, C.; Watzenboeck, M.L.; Schneidegger, S.; Kieser, S.; Wong, N.C.; Bernasconi, E.; Pernot, J.; Mercier, L.; Knapp, S.; Nicod, L.P.; et al. Early-life formation of the microbial and immunological environment of the human airways. Cell Host Microbe 2018, 24, 857–865.e4. [Google Scholar] [CrossRef] [PubMed]
- Frati, F.; Salvatori, C.; Incorvaia, C.; Bellucci, A.; Di-Cara, G.; Marcucci, F.; Esposito, S. The role of the microbiome in asthma: The Gut-Lung Axis. Int. J. Mol. Sci. 2018, 20, 123. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. Kinases as novel therapeutic targets in asthma and chronic obstructive pulmonary disease. Pharmacol. Rev. 2016, 68, 788–815. [Google Scholar] [CrossRef]
- Llewellyn, A.; Foey, A. Probiotic modulation of innate cell pathogen sensing and signaling events. Nutrients 2017, 9, 1156. [Google Scholar] [CrossRef]
- Barnes, P.J. Inflammatory mechanisms in patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2016, 138, 16–27. [Google Scholar] [CrossRef]
- Yang, X.; Li, H.; Ma, Q.; Zhang, Q.; Wang, C. Neutrophilic asthma is associated with increased airway bacterial burden and disordered community composition. Biomed. Res. Int. 2018, 2018, 9230234. [Google Scholar] [CrossRef]
- Roesch, E.A.; Nichols, D.P.; Chmiel, J.F. Inflammation in cystic fibrosis: An update. Pediatr. Pulmonol. 2018, 53, S30–S50. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Chang, A.B.; Bell, S.C. Anti-inflammatory therapies in bronchiectasis. Eur. Respir. Monogr. 2011, 52, 223–238. [Google Scholar] [CrossRef]
- Desai, O.; Winkler, J.; Minasyan, M.; Herzog, E.L. The role of immune and inflammatory cells in idiopathic pulmonary fibrosis. Front. Med. (Lausanne) 2018, 5, 43. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.W.; Chen, J.; Ruan, Y.C.; Zhou, T.; Chen, Y.; Chen, Y.; Tsang, L.L.; Chan, H.C.; Peng, Y.Z. CFTR-regulated MAPK/NF-κB signaling in pulmonary inflammation in thermal inhalation injury. Sci. Rep. 2015, 5, 15946. [Google Scholar] [CrossRef]
- Dong, J.; Ma, Q. In Vivo activation and pro-fibrotic function of NF-κB in fibroblastic cells during pulmonary inflammation and fibrosis induced by carbon nanotubes. Front. Pharmacol. 2019, 10, 1140. [Google Scholar] [CrossRef]
- Li, K.; Chen, Z.; Huang, Y.; Zhang, R.; Luan, X.; Lei, T.; Chen, L. Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety. Respir. Res. 2019, 20, 272. [Google Scholar] [CrossRef]
- Ubags, N.D.J.; Marsland, B.J. Mechanistic insight into the function of the microbiome in lung diseases. Eur. Respir. J. 2017, 11, 50. [Google Scholar] [CrossRef]
- O’Dwyer, D.N.; Dickson, R.P.; Moore, B.B. The lung microbiome, immunity, and the pathogenesis of chronic lung disease. J. Immunol. 2016, 196, 4839–4847. [Google Scholar] [CrossRef]
- Mammen, M.J.; Sethi, S. COPD and the microbiome. Respirology 2016, 21, 590–599. [Google Scholar] [CrossRef]
- Dicker, A.J.; Chalmers, J.D. Microbial dysbiosis in bronchiectasis and cystic fibrosis. Arch. Bronconeumol. 2017, 53, 471–472. [Google Scholar] [CrossRef][Green Version]
- Toraldo, D.M.; Conte, L. Influence of the lung microbiota dysbiosis in chronic obstructive pulmonary disease exacerbations: The controversial use of corticosteroid and antibiotic treatments and the role of eosinophils as a disease marker. J. Clin. Med. Res. 2019, 11, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Oh, N.S.; Joung, J.Y.; Lee, J.Y.; Kim, Y. Probiotic and anti-inflammatory potential of Lactobacillus rhamnosus 4B15 and Lactobacillus gasseri 4M13 isolated from infant feces. PLoS ONE 2018, 13, e0192021. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Yamazaki, T.; Ohshio, K.; Sugamata, M.; Yoshikawa, M.; Kanauchi, O.; Morita, Y. A specific strain of lactic acid bacteria, Lactobacillus paracasei, inhibits inflammasome activation in vitro and prevents inflammation-related disorders. J. Immunol. 2020, 205, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Rajilić-Stojanović, M.; Dimitrijević, S.; and Golić, N. Lactic acid bacteria in the gut. In Lactic acid Bacteria, 5th ed.; Chapter, 24, Vinderola, G., Ouwehand, A., Salminen, S., von Wright, A., Eds.; CRC Press: Boca Raton, FL, USA, 2019; p. 383. [Google Scholar]
- Terzić-Vidojević, A.; Veljović, K.; Tolinački, M.; Živković, M.; Lukić, J.; Lozo, J.; Fira, Đ.; Jovčić, B.; Strahinić, I.; Begović, J.; et al. Diversity of non-starter lactic acid bacteria in autochthonous dairy products from Western Balkan Countries Technological and probiotic properties. Food Res. Int. 2020, 136, 109494. [Google Scholar] [CrossRef] [PubMed]
- Popović, N.; Brdarić, E.; Ðokić, J.; Dinić, M.; Veljović, K.; Golić, N.; Terzić-Vidojević, A. Yogurt produced by novel natural starter cultures improves gut epithelial barrier in vitro. Microorganisms 2020, 8, 1586. [Google Scholar] [CrossRef] [PubMed]
- Dinić, M.; Herholz, M.; Kačarević, U.; Radojević, D.; Novović, K.; Đokić, J.; Trifunović, A.; Golić, N. Host-commensal interaction promotes health and lifespan in Caenorhabditis elegans through the activation of HLH-30/TFEB-mediated autophagy. Aging 2021, 13, 8040–8054. [Google Scholar] [CrossRef]
- Dinić, M.; Jakovljević, S.; Đokić, J.; Popović, N.; Radojević, D.; Strahinić, I.; Golić, N. Probiotic-mediated p38 MAPK immune signaling prolongs thesurvival of Caenorhabditis elegans exposed to pathogenic bacteria. Sci. Rep. 2021, 11, 21258. [Google Scholar] [CrossRef]
- Veljović, K.; Terzić-Vidojević, A.; Vukasinović, M.; Strahinić, I.; Begović, J.; Lozo, J.; Ostojic, M.; Topisirovic, L. Preliminary characterization of lactic acid bacteria isolated from Zlatar cheese. J. Appl. Microbiol. 2007, 103, 2142–2152. [Google Scholar] [CrossRef]
- Sokovic-Bajić, S.; Đokić, J.; Dinić, M.; Veljović, K.; Golić, N.; Mihajlović, S.; Tolinacki, M. GABA-producing natural dairy isolate from artisanal Zlatar cheese attenuates gut inflammation and strengthens gut epithelial barrier in vitro. Front. Microbiol. 2019, 10, 527. [Google Scholar] [CrossRef]
- Terzic-Vidojevic, A.; Tolinacki, M.; Nikolic, M.; Veljovic, K.; Jovanovic, S.; Macej, O.; Topisirovic, L. Vlasina raw goat’s milk cheese: Evaluation and selection of autochthonous lactic acid bacteria as starter cultures. Food Technol. Biotechnol. 2013, 51, 253–264. [Google Scholar]
- Golić, N.; Čadež, N.; Terzić-Vidojević, A.; Šuranská, H.; Beganović, J.; Lozo, J.; Kos, B.; Sušković, J.; Raspor, P.; Topisirović, L. Evaluation of lactic acid bacteria and yeast diversity in traditional white pickled and fresh soft cheeses from the mountain regions of Serbia and lowland regions of Croatia. Int. J. Food Microbiol. 2013, 166, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Terzić-Vidojević, A.; Mihajlović, S.; Uzelac, G.; Veljović, K.; Tolinački, M.; Nikolić, M.; Topisirovic, L.; Kojic, M. Characterization of lactic acid bacteria isolated from artisanal Travnik young cheeses, sweet creams and sweet kajmaks over four seasons. Food Microbiol. 2014, 39, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, M.; Terzic-Vidojevic, A.; Jovcic, B.; Begovic, J.; Golic, N.; Topisirovic, L. Characterization of lactic acid bacteria isolated from Bukuljac, a homemade goat’s milk cheese. Int. J. Food Microbiol. 2008, 122, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Strahinic, I.; Lozo, J.; Terzic-Vidojevic, A.; Fira, Đ.; Kojic, M.; Golic, N.; Begovic, J. Technological and probiotic potential of BGRA43 a natural isolate of Lactobacillus helveticus. Front. Microbiol. 2013, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Veljović, K.; Dinić, M.; Lukić, J.; Mihajlović, S.; Tolinački, M.; Živković, M.; Begović, J.; Mrvaljević, I.; Golić, N.; Terzić-Vidojević, A. Promotion of early gut colonization by probiotic intervention on microbiota diversity in pregnant sows. Front. Microbiol. 2017, 8, 2028. [Google Scholar] [CrossRef] [PubMed]
- Golić, N.; Veljović, K.; Popović, N.; Djokić, J.; Strahinić, I.; Mrvaljević, I.; Terzić-Vidojević, A. In vitro and in vivo antagonistic activity of new probiotic culture against Clostridium difficile and Clostridium perfringens. BMC Microbiol. 2017, 17, 108. [Google Scholar] [CrossRef]
- Lozo, J.; Jovcic, B.; Kojic, M.; Dalgalarrondo, M.; Chobert, J.M.; Haertlé, T.; Topisirovic, L. Molecular characterization of a novel bacteriocin and an unusually large aggregation factor of Lactobacillus paracasei subsp. paracasei BGSJ2-8, a natural isolate from home-made cheese. Curr. Microbiol. 2007, 55, 266–271. [Google Scholar] [CrossRef]
- Strahinic, I.; Kojic, M.; Tolinacki, M.; Fira, D.; Topisirovic, L. The presence of prtP proteinase gene in natural isolate Lactobacillus plantarum BGSJ3-18. Lett. Appl. Microbiol. 2010, 50, 43–49. [Google Scholar] [CrossRef]
- EFSA Biohaz Panel (EFSA Panel on Biological Hazards); Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Escámez, P.S.F.; Girones, R.; Herman, L.; Koutsoumanis, K.; et al. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 8: Suitability of taxonomic units notified to EFSA until March 2018. EFSA J. 2018, 16, e05315. [Google Scholar] [CrossRef]
- GBD Chronic Respiratory Disease Collaborators. Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir. Med. 2020, 8, 585–596. [Google Scholar] [CrossRef]
- Barnes, P.J. New anti-inflammatory targets for chronic obstructive pulmonary disease. Nat. Rev. Drug Discov. 2013, 12, 543–559. [Google Scholar] [CrossRef]
- Cantin, A.M.; Hartl, D.; Konstan, M.W.; Chmiel, J.F. Inflammation in cystic fibrosis lung disease: Pathogenesis and therapy. J. Cyst. Fibros. 2015, 14, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Funke, M.; Geiser, T. Idiopathic pulmonary fibrosis: The turning point is now! Swiss Med. Wkly. 2015, 145, w14139. [Google Scholar] [CrossRef]
- Gross, N.J.; Barnes, P.J. New therapies for asthma and chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2017, 195, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Mitri, C.; Xu, Z.; Bardin, P.; Corvol, H.; Touqui, L.; Tabary, O. Novel anti-inflammatory approaches for cystic fibrosis lung disease: Identification of molecular targets and design of innovative therapies. Front. Pharmacol. 2020, 11, 1096. [Google Scholar] [CrossRef] [PubMed]
- Mancabelli, L.; Milani, C.; Fontana, F.; Lugli, G.A.; Tarracchini, C.; Turroni, F.; van Sinderen, D.; Ventura, M. Mapping bacterial diversity and metabolic functionality of the human respiratory tract microbiome. J. Oral Microbiol. 2022, 14, 2051336. [Google Scholar] [CrossRef]
- Gill, N.; Wlodarska, M.; Finlay, B.B. The future of mucosal immunology: Studying an integrated system-wide organ. Nat. Immunol. 2010, 11, 558–560. [Google Scholar] [CrossRef]
- Kulas, J.; Mirkov, I.; Tucovic, D.; Zolotarevski, L.; Glamoclija, J.; Veljovic, K.; Tolinacki, M.; Golic, N.; Kataranovski, M. Pulmonary Aspergillus fumigatus infection in rats affects gastrointestinal homeostasis. Immunobiology 2019, 224, 116–123. [Google Scholar] [CrossRef]
- Pulvirenti, G.; Parisi, G.F.; Giallongo, A.; Papale, M.; Manti, S.; Savasta, S.; Licari, A.; Marseglia, G.L.; Leonardi, S. Lower airway microbiota. Front. Pediatr. 2019, 7, 393. [Google Scholar] [CrossRef]
- Villena, J.; Kitazawa, H. Modulation of intestinal TLR4-inflammatory signaling pathways by probiotic microorganisms: Lessons learned from Lactobacillus jensenii TL2937. Front. Immunol. 2014, 4, 512. [Google Scholar] [CrossRef]
- Wu, B.G.; Segal, L.N. Chapter 7: Lung Microbiota and its Impact on the Mucosal Immune Phenotype. Microbiol. Spectr. 2017, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Yadava, K.; Pattaroni, C.; Sichelstiel, A.K.; Trompette, A.; Gollwitzer, E.S.; Salami, O.; von Garnier, C.; Nicod, L.P.; Marsland, B.J. Microbiota Promotes Chronic Pulmonary Inflammation by Enhancing IL-17A and Autoantibodies. Am. J. Respir. Crit. Care Med. 2016, 193, 975–987. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.C.; Cho, W.K.; Oh, J.H.; Im, G.Y.; Jeong, Y.H.; Yang, M.C.; Ma, J.Y. Fermentation by Lactobacillus enhances anti-inflammatory effect of Oyaksungisan on LPS- stimulated RAW 264.7 mouse macrophage cells. BMC Complement. Altern. Med. 2012, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Indira, M.; Venkateswarulu, T.C.; Peele, A.K.; Bobby, N.; Krupanidhi, S. Bioactive molecules of probiotic bacteria and their mechanism of action: A review. Biotech 2019, 9, 306. [Google Scholar] [CrossRef] [PubMed]
- Hemarajata, P.; Versalovic, J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterol. 2013, 6, 39–51. [Google Scholar] [CrossRef]
- Muñoz-Provencio, D.; Llopis, M.; Antolín, M.; de Torres, I.; Guarner, F.; Pérez-Martínez, G.; Monedero, V. Adhesion properties of Lactobacillus casei strains to resected intestinal fragments and components of the extracellular matrix. Arch. Microbiol. 2009, 191, 153–161. [Google Scholar] [CrossRef]
- Valenzuela, A.S.; Omar, N.B.; Abriouel, H.; López, R.L.; Veljovic, K.; Canamero, M.M.; Kojic, M.; Topisirovic, L.; Gálvez, A. Virulence factors, antibiotic resistance, and bacteriocins in enterococci from artisan foods of animal origin. Food Control 2009, 20, 381–385. [Google Scholar] [CrossRef]
- Lopes, M.F.; Simões, A.P.; Tenreiro, R.; Marques, J.J.F.; Crespo, M.T.B. Activity and expression of a virulence factor, gelatinase, in dairy enterococci. Int. J. Food Microbiol. 2006, 112, 208–214. [Google Scholar] [CrossRef]
- Mushtaq, N.; Ezzati, M.; Hall, L.; Dickson, I.; Kirwan, M.; Ken, M.Y.; Mudway, I.S.; Grigg, J. Adhesion of Streptococcus pneumoniae to human airway epithelial cells exposed to urban particulate matter. J. Allergy Clin. Immunol. 2011, 127, 1236–1242. [Google Scholar] [CrossRef]
- Available online: www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ (accessed on 22 April 2022).
| Bacterial Strain | Source | References |
|---|---|---|
| Lactobacillus brevis BGZLS10-17 | 10-day-old semi-hard cheese | [29,30] |
| Lactobacillus plantarum BGMI1 | White cheese | IMGGE collection |
| Lactobacillus plantarum BGVL2a-18 | 15-day-old raw goat milk cheese | [31] |
| Lactobacillus plantarum BGAN8 | Artisanal soft cheese | IMGGE collection |
| Lactobacillus plantarum BGPKM22 | Artisanal sour milk | IMGGE collection |
| Lactobacillus plantarum BGSJ2-3 | White cow cheese | IMGGE collection |
| Lactobacillus plantarum BGGO7-29 | 60-day-old white pickled cheese | [32] |
| Lactococcus lactis subsp. lactis BGTRK4-21 | Sweet kajmak | [33] |
| Lactococcus lactis subsp. lactis BGAR8 | 5-day-old goat cheese | [34] |
| Lactobacillus helveticus BGRA43 | Human intestinum | [35,36,37] |
| Lactobacillus rhamnosus BGHI22 | Human intestinum, neonatus | IMGGE collection |
| Lactobacillus paracasei BGAR88-2 | 5-day-old goat cheese | [34] |
| Lactobacillus paracasei BGSJ2-8 | White cow cheese | [38] |
| Lactobacillus rhamnosus BGSJ3-18 | White cow cheese | [39] |
| Lactobacillus brevis BGHI3a | Human intestinum, neonatus | IMGGE collection |
| Lactobacillus paraplantarum BGCG11 | Old full-fat cheese | IMGGE collection |
| Lactococcus lactis subsp. cremoris BGTRM1-22 | Sweet cream | [33] |
| Streptococcus thermophilus BGKMJ1-36 | Artisanal sour milk | [26] |
| Lactobacillus paracasei BGAR76 | 5-day-old goat cheese | [34] |
| Lactococcus lactis subsp. lactis BGVL2-8 | 5-day-old raw goat milk cheese | [31] |
| Lactococcus lactis subsp. lactis biovar. diacetylactis BGTRK10-2 | Sweet kajmak | [33] |
| Antibiotic with the MIC (ug/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Strains | Amp | Van | Gen | Kan | Str | Ery | Cln | Tet | Chl |
| Lb. plantarum | |||||||||
| BGMI1 | <1 | n.r. | 8 | 32 | n.r. | <1 | 2 | 16 | 4 |
| BGVL2a-18 | 1 | n.r. | 4 | 16 | n.r. | <1 | 2 | 16 | 4 |
| BGAN8 | 1 | n.r. | 8 | 16 | n.r. | <1 | 2 | 16 | 4 |
| BGPKM22 | <1 | n.r. | 4 | 16 | n.r. | <1 | 2 | 16 | 4 |
| BGSJ2-3 | 1 | n.r. | 4 | 32 | n.r. | <1 | 2 | 16 | 4 |
| BGGO7-29 | 1 | n.r. | 8 | 16 | n.r. | <1 | 2 | 16 | 4 |
| BGCG11 | <1 | n.r. | 4 | 8 | n.r. | <1 | 1 | 8 | 1 |
| Lb. brevis | |||||||||
| BGZLS10-17 | 1 | n.r. | 4 | 32 | 32 | <1 | 2 | 4 | 2 |
| BGHI3a | 1 | n.r. | 4 | 32 | 16 | <1 | 2 | 4 | 2 |
| Lb. helveticus | |||||||||
| BGRA43 | <1 | 1 | 4 | 4 | 4 | <1 | 1 | 2 | 1 |
| Lb. rhamnosus | |||||||||
| BGHI22 | 2 | n.r. | 8 | 32 | 16 | <1 | 2 | 2 | 2 |
| BGSJ3-18 | 2 | n.r. | 8 | 16 | 16 | <1 | 2 | 2 | 2 |
| Lb. casei | |||||||||
| BGSJ2-8 | 2 | n.r. | 16 | 16 | 32 | <1 | 2 | 2 | 2 |
| BGAR76 | 1 | n.r. | 8 | 32 | 32 | <1 | 2 | 2 | 2 |
| BG88-2 | 1 | n.r. | 8 | 32 | 32 | <1 | 2 | 2 | 2 |
| St. thermophilus | |||||||||
| BGKMJ1-36 | <1 | <0.5 | 16 | n.r. | 32 | 1 | 0.5 | 2 | 1 |
| Lc. lactis | |||||||||
| BGTRK4-21 | <1 | 2 | 16 | 32 | 16 | <1 | <1 | 2 | 4 |
| BGAR8 | <1 | 2 | 16 | 32 | 16 | <1 | <1 | 2 | 4 |
| BGAR76 | 1 | 2 | 16 | 32 | 16 | <1 | <1 | 2 | 2 |
| BGVL2-8 | <1 | 2 | 8 | 32 | 8 | <1 | <1 | 2 | 2 |
| BGTRK10-2 | <1 | 2 | 16 | 32 | 16 | <1 | <1 | 2 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stankovic, M.; Veljovic, K.; Popovic, N.; Kojic, S.; Dunjic Manevski, S.; Radojkovic, D.; Golic, N. Lactobacillus brevis BGZLS10-17 and Lb. plantarum BGPKM22 Exhibit Anti-Inflammatory Effect by Attenuation of NF-κB and MAPK Signaling in Human Bronchial Epithelial Cells. Int. J. Mol. Sci. 2022, 23, 5547. https://doi.org/10.3390/ijms23105547
Stankovic M, Veljovic K, Popovic N, Kojic S, Dunjic Manevski S, Radojkovic D, Golic N. Lactobacillus brevis BGZLS10-17 and Lb. plantarum BGPKM22 Exhibit Anti-Inflammatory Effect by Attenuation of NF-κB and MAPK Signaling in Human Bronchial Epithelial Cells. International Journal of Molecular Sciences. 2022; 23(10):5547. https://doi.org/10.3390/ijms23105547
Chicago/Turabian StyleStankovic, Marija, Katarina Veljovic, Nikola Popovic, Snezana Kojic, Sofija Dunjic Manevski, Dragica Radojkovic, and Natasa Golic. 2022. "Lactobacillus brevis BGZLS10-17 and Lb. plantarum BGPKM22 Exhibit Anti-Inflammatory Effect by Attenuation of NF-κB and MAPK Signaling in Human Bronchial Epithelial Cells" International Journal of Molecular Sciences 23, no. 10: 5547. https://doi.org/10.3390/ijms23105547
APA StyleStankovic, M., Veljovic, K., Popovic, N., Kojic, S., Dunjic Manevski, S., Radojkovic, D., & Golic, N. (2022). Lactobacillus brevis BGZLS10-17 and Lb. plantarum BGPKM22 Exhibit Anti-Inflammatory Effect by Attenuation of NF-κB and MAPK Signaling in Human Bronchial Epithelial Cells. International Journal of Molecular Sciences, 23(10), 5547. https://doi.org/10.3390/ijms23105547








