Exploring BODIPY-Based Sensor for Imaging of Intracellular Microviscosity in Human Breast Cancer Cells
Abstract
:1. Introduction
2. Results and Discussion
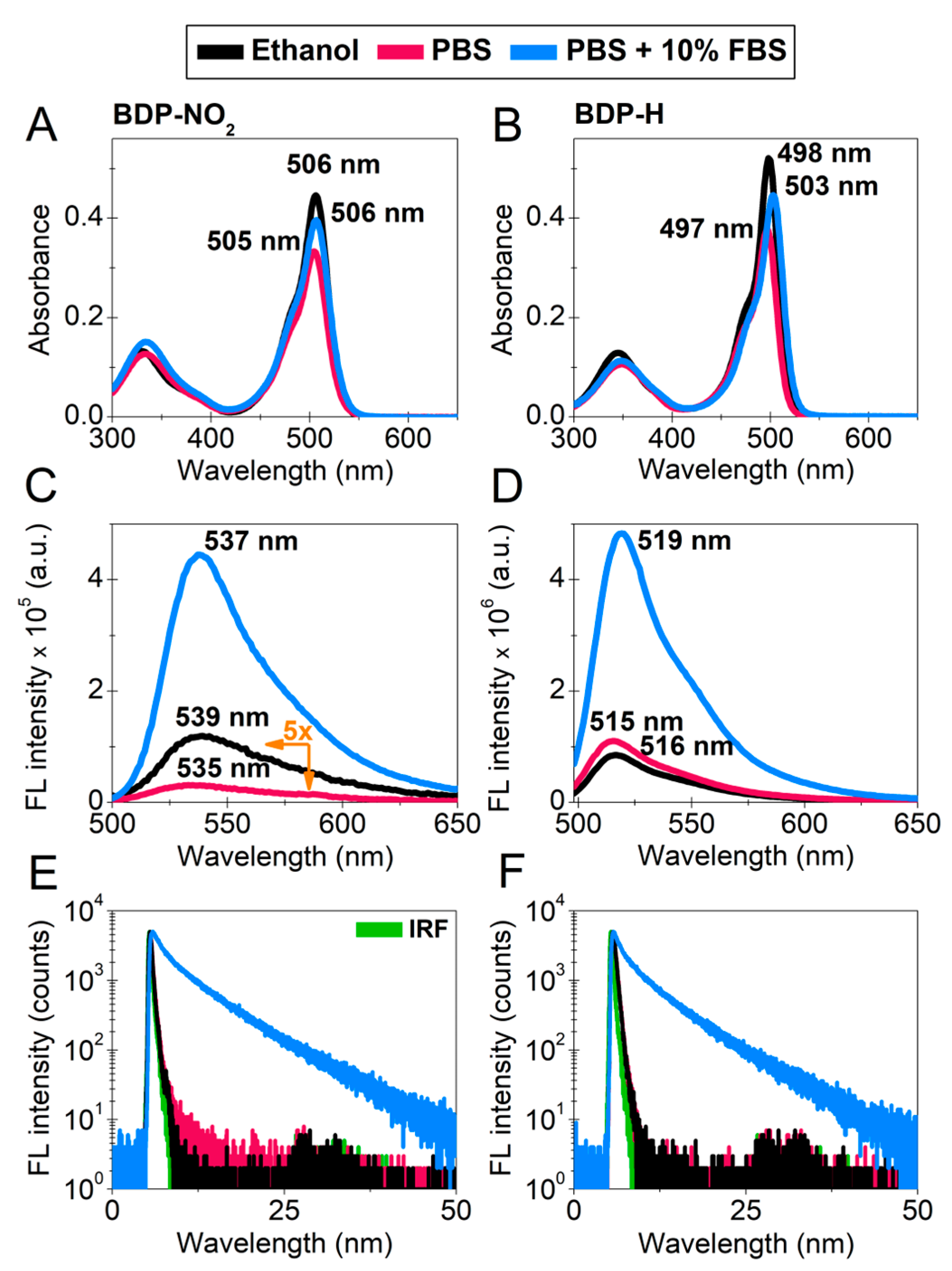
2.1. Photophysical Characterization
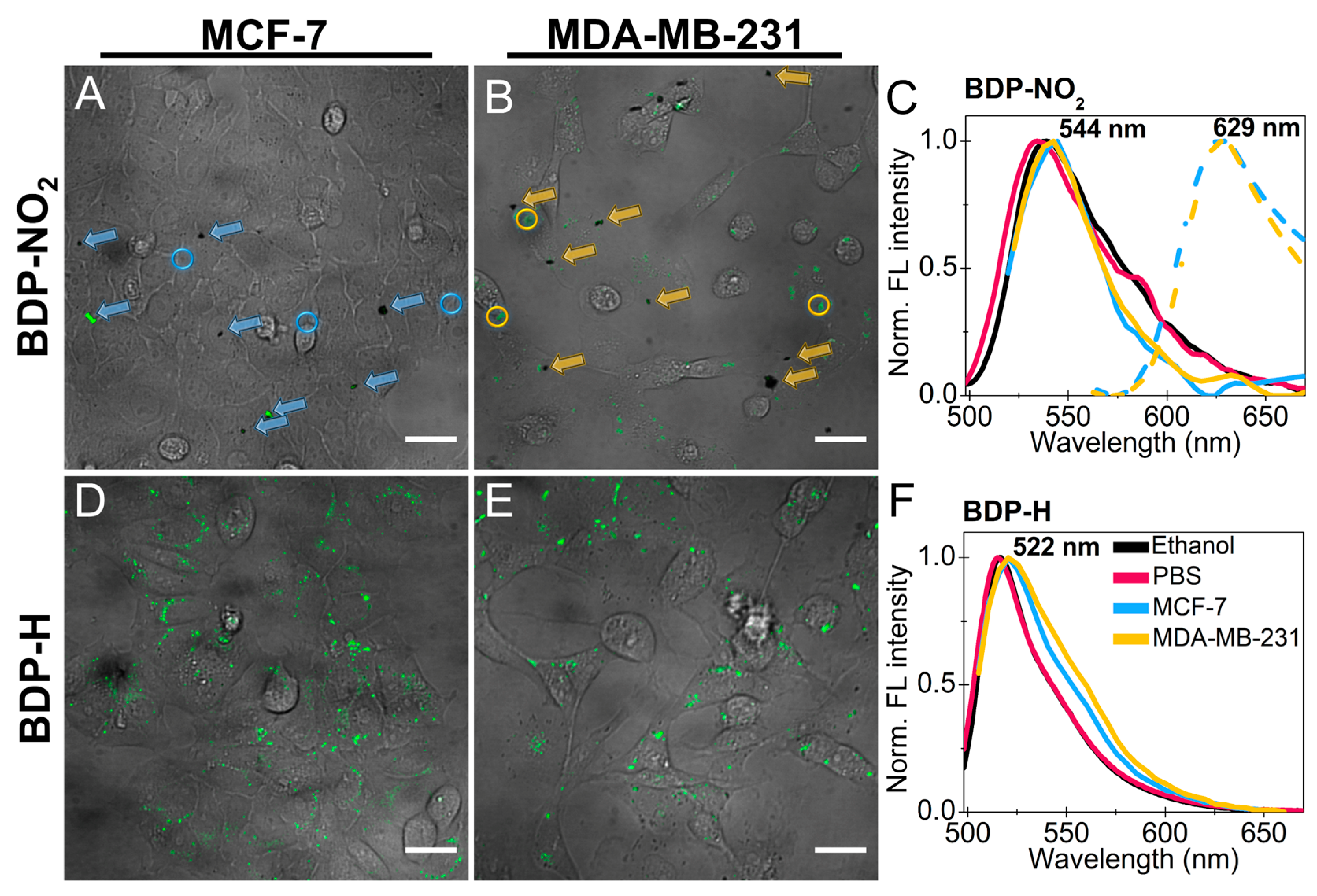
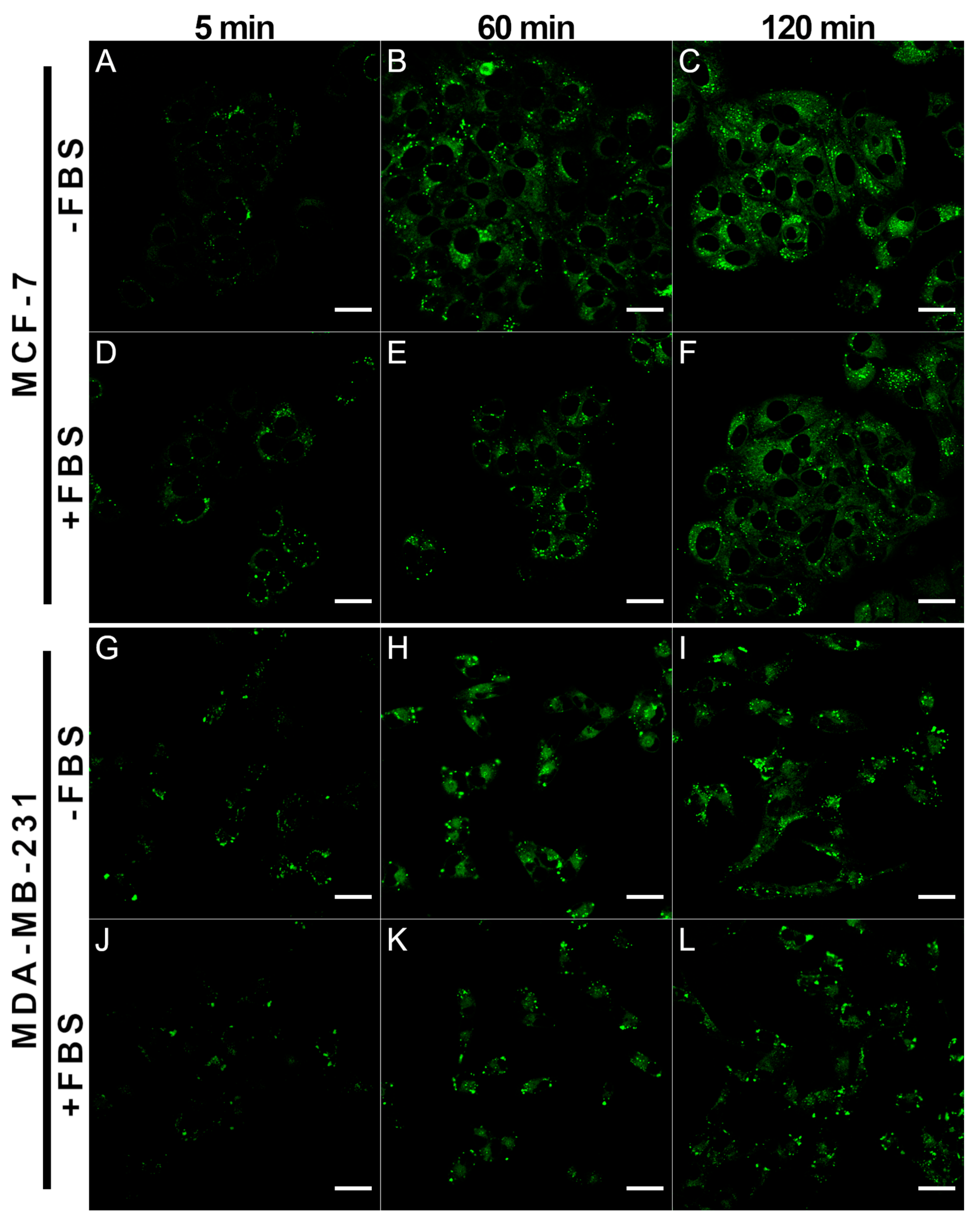
2.2. Accumulation of BDP-NO2 and BDP-H in Breast Cancer Cells
2.3. Localization of BDP-H in Breast Cancer Cells
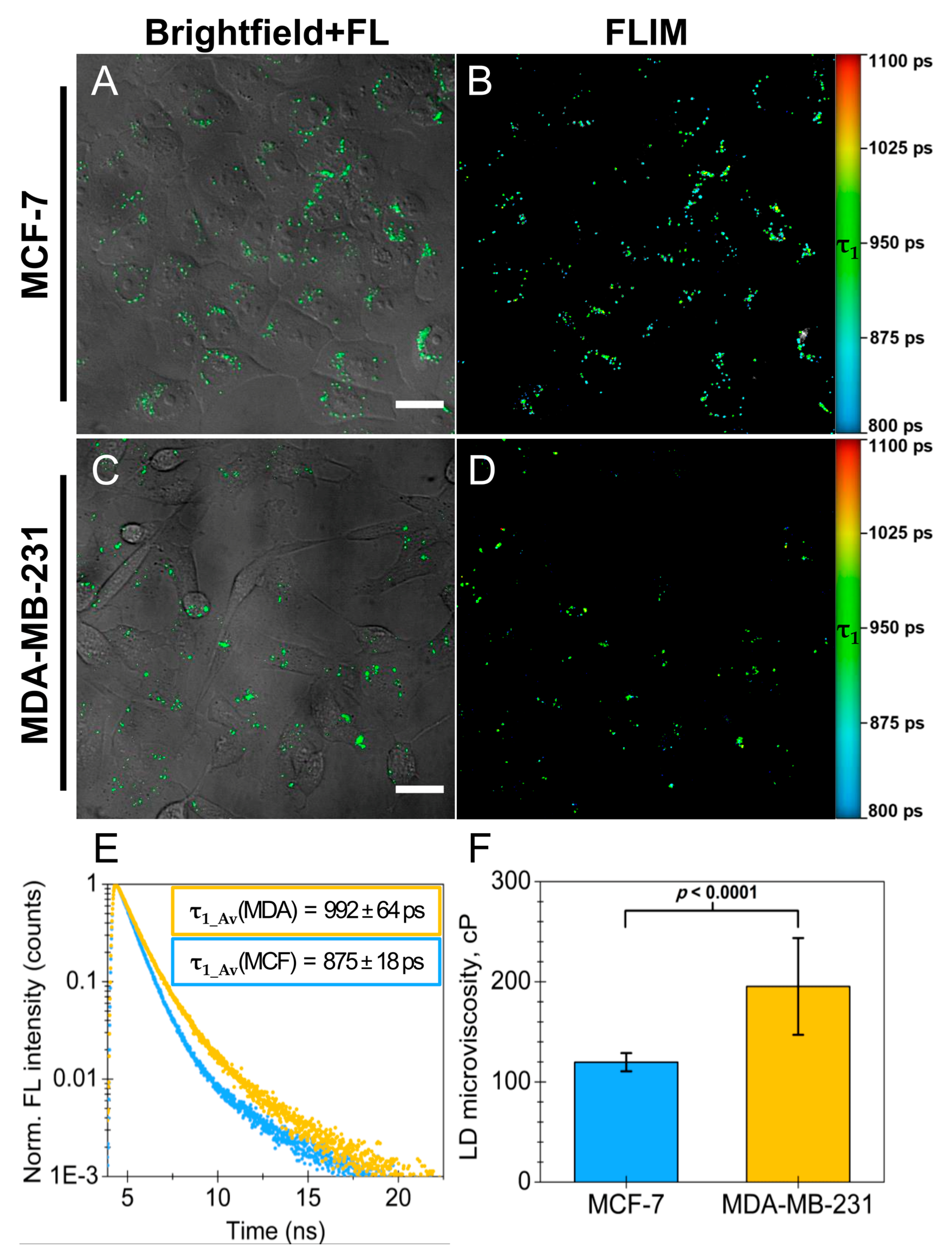
2.4. Utilizing BDP-H and FLIM to Measure Lipid Droplet Microviscosity in Cancer Cells
3. Material and Methods
3.1. Molecular Rotors
3.2. Commercial Probes Used for Colocalization Experiments
3.3. Spectral Analysis
3.4. Analysis of Spectral Measurements
3.5. Cell Culturing
3.6. Treatment of Cells with Molecular Rotors and Commercial Probes
3.7. Cellular Microscopy
3.8. Colocalization Analysis
3.9. Fluorescence Lifetime Imaging Microscopy
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luby-Phelps, K. Cytoarchitecture and Physical Properties of Cytoplasm: Volume, Viscosity, Diffusion, Intracellular Surface Area. In International Review of Cytology; Microcompartmentation and Phase Separation in Cytoplasm; Walter, H., Brooks, D.E., Srere, P.A., Eds.; Academic Press: Cambridge, MA, USA, 1999; Volume 192, pp. 189–221. [Google Scholar]
- Kuimova, M.K. Mapping Viscosity in Cells Using Molecular Rotors. Phys. Chem. Chem. Phys. 2012, 14, 12671–12686. [Google Scholar] [CrossRef] [PubMed]
- Kuimova, M.K.; Botchway, S.W.; Parker, A.W.; Balaz, M.; Collins, H.A.; Anderson, H.L.; Suhling, K.; Ogilby, P.R. Imaging Intracellular Viscosity of a Single Cell during Photoinduced Cell Death. Nat. Chem. 2009, 1, 69–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadiv, O.; Shinitzky, M.; Manu, H.; Hecht, D.; Roberts, C.T.; LeRoith, D.; Zick, Y. Elevated Protein Tyrosine Phosphatase Activity and Increased Membrane Viscosity Are Associated with Impaired Activation of the Insulin Receptor Kinase in Old Rats. Biochem. J. 1994, 298, 443–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deliconstantinos, G.; Villiotou, V.; Stavrides, J.C. Modulation of Particulate Nitric Oxide Synthase Activity and Peroxynitrite Synthesis in Cholesterol Enriched Endothelial Cell Membranes. Biochem. Pharmacol. 1995, 49, 1589–1600. [Google Scholar] [CrossRef]
- Zubenko, G.S.; Kopp, U.; Seto, T.; Firestone, L.L. Platelet Membrane Fluidity Individuals at Risk for Alzheimer’s Disease: A Comparison of Results from Fluorescence Spectroscopy and Electron Spin Resonance Spectroscopy. Psychopharmacology 1999, 145, 175–180. [Google Scholar] [CrossRef]
- Halpern, H.J.; Chandramouli, G.V.R.; Barth, E.D.; Yu, C.; Peric, M.; Grdina, D.J.; Teicher, B.A. Diminished Aqueous Microviscosity of Tumors in Murine Models Measured with in Vivo Radiofrequency Electron Paramagnetic Resonance1. Cancer Res. 1999, 59, 5836–5841. [Google Scholar]
- Shimolina, L.E.; Izquierdo, M.A.; López-Duarte, I.; Bull, J.A.; Shirmanova, M.V.; Klapshina, L.G.; Zagaynova, E.V.; Kuimova, M.K. Imaging Tumor Microscopic Viscosity in Vivo Using Molecular Rotors. Sci. Rep. 2017, 7, 41097. [Google Scholar] [CrossRef] [Green Version]
- Haidekker, M.A.; Theodorakis, E.A. Molecular Rotors—Fluorescent Biosensors for Viscosity and Flow. Org. Biomol. Chem. 2007, 5, 1669–1678. [Google Scholar] [CrossRef]
- Kuimova, M.K.; Yahioglu, G.; Levitt, J.A.; Suhling, K. Molecular Rotor Measures Viscosity of Live Cells via Fluorescence Lifetime Imaging. J. Am. Chem. Soc. 2008, 130, 6672–6673. [Google Scholar] [CrossRef] [Green Version]
- Chambers, J.E.; Kubánková, M.; Huber, R.G.; López-Duarte, I.; Avezov, E.; Bond, P.J.; Marciniak, S.J.; Kuimova, M.K. An Optical Technique for Mapping Microviscosity Dynamics in Cellular Organelles. ACS Nano 2018, 12, 4398–4407. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Chi, W.; Qiao, Q.; Kokate, S.V.; Cabrera, E.P.; Xu, Z.; Liu, X.; Chang, Y.-T. Molecular Mechanism of Viscosity Sensitivity in BODIPY Rotors and Application to Motion-Based Fluorescent Sensors. ACS Sens. 2020, 5, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Förster, T.; Hoffmann, G. Die Viskositätsabhängigkeit Der Fluoreszenzquantenausbeuten Einiger Farbstoffsysteme. Z. Phys. Chem. 1971, 75, 63–76. [Google Scholar] [CrossRef]
- Vyšniauskas, A.; López-Duarte, I.; Duchemin, N.; Vu, T.T.; Wu, Y.; Budynina, E.M.; Volkova, Y.A.; Cabrera, E.P.; Ramírez-Ornelas, D.E.; Kuimova, M.K. Exploring Viscosity, Polarity and Temperature Sensitivity of BODIPY-Based Molecular Rotors. Phys. Chem. Chem. Phys. 2017, 19, 25252–25259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suhling, K.; French, P.M.W.; Phillips, D. Time-Resolved Fluorescence Microscopy. Photochem. Photobiol. Sci. 2005, 4, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Suhling, K.; Levitt, J.A.; Chung, P.-H.; Kuimova, M.K.; Yahioglu, G. Fluorescence Lifetime Imaging of Molecular Rotors in Living Cells. J. Vis. Exp. 2012, 60, e2925. [Google Scholar] [CrossRef]
- Loudet, A.; Burgess, K. BODIPY Dyes and Their Derivatives: Syntheses and Spectroscopic Properties. Chem. Rev. 2007, 107, 4891–4932. [Google Scholar] [CrossRef]
- Boens, N.; Leen, V.; Dehaen, W. Fluorescent Indicators Based on BODIPY. Chem. Soc. Rev. 2012, 41, 1130–1172. [Google Scholar] [CrossRef]
- Kowada, T.; Maeda, H.; Kikuchi, K. BODIPY-Based Probes for the Fluorescence Imaging of Biomolecules in Living Cells. Chem. Soc. Rev. 2015, 44, 4953–4972. [Google Scholar] [CrossRef]
- Lee, J.-S.; Kim, H.K.; Feng, S.; Vendrell, M.; Chang, Y.-T. Accelerating Fluorescent Sensor Discovery: Unbiased Screening of a Diversity-Oriented BODIPY Library. Chem. Commun. 2011, 47, 2339–2341. [Google Scholar] [CrossRef]
- Toliautas, S.; Dodonova, J.; Žvirblis, A.; Čiplys, I.; Polita, A.; Devižis, A.; Tumkevičius, S.; Šulskus, J.; Vyšniauskas, A. Enhancing the Viscosity-Sensitive Range of a BODIPY Molecular Rotor by Two Orders of Magnitude. Chem. Eur. J. 2019, 25, 10342–10349. [Google Scholar] [CrossRef]
- Michels, L.; Gorelova, V.; Harnvanichvech, Y.; Borst, J.W.; Albada, B.; Weijers, D.; Sprakel, J. Complete Microviscosity Maps of Living Plant Cells and Tissues with a Toolbox of Targeting Mechanoprobes. Proc. Natl. Acad. Sci. USA 2020, 117, 18110–18118. [Google Scholar] [CrossRef] [PubMed]
- Subiros-Funosas, R.; Lam Ho, V.C.; Barth, N.D.; Mendive-Tapia, L.; Pappalardo, M.; Barril, X.; Ma, R.; Zhang, C.-B.; Qian, B.-Z.; Sintes, M.; et al. Fluorogenic Trp(RedBODIPY) Cyclopeptide Targeting Keratin 1 for Imaging of Aggressive Carcinomas. Chem. Sci. 2020, 11, 1368–1374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maleckaitė, K.; Narkevičius, D.; Žilėnaitė, R.; Dodonova-Vaitkūnienė, J.; Toliautas, S.; Tumkevičius, S.; Vyšniauskas, A. Give or Take: Effects of Electron-Accepting/-Withdrawing Groups in Red-Fluorescent BODIPY Molecular Rotors. Molecules 2022, 27, 23. [Google Scholar] [CrossRef] [PubMed]
- Reese, A.E.; Lochenie, C.; Geddis, A.; Machado, L.A.; de Souza, M.C.; Marques, F.F.C.; de Simone, C.A.; Gouvêa, M.M.; Pedrosa, L.F.; da Silva Júnior, E.N.; et al. Rational Design and Synthesis of Large Stokes Shift 2,6-Sulphur-Disubstituted BODIPYs for Cell Imaging. Chemosensors 2022, 10, 19. [Google Scholar] [CrossRef]
- Li, F.; Yang, S.I.; Ciringh, Y.; Seth, J.; Martin, C.H.; Singh, D.L.; Kim, D.; Birge, R.R.; Bocian, D.F.; Holten, D.; et al. Design, Synthesis, and Photodynamics of Light-Harvesting Arrays Comprised of a Porphyrin and One, Two, or Eight Boron-Dipyrrin Accessory Pigments. J. Am. Chem. Soc. 1998, 120, 10001–10017. [Google Scholar] [CrossRef]
- Benniston, A.C.; Clift, S.; Harriman, A. Intramolecular Charge-Transfer Interactions in a Julolidine–Bodipy Molecular Assembly as Revealed via 13C NMR Chemical Shifts. J. Mol. Struct. 2011, 985, 346–354. [Google Scholar] [CrossRef]
- Polita, A.; Toliautas, S.; Žvirblis, R.; Vyšniauskas, A. The Effect of Solvent Polarity and Macromolecular Crowding on the Viscosity Sensitivity of a Molecular Rotor BODIPY-C10. Phys. Chem. Chem. Phys. 2020, 22, 8296–8303. [Google Scholar] [CrossRef]
- Descalzo, A.B.; Ashokkumar, P.; Shen, Z.; Rurack, K. On the Aggregation Behaviour and Spectroscopic Properties of Alkylated and Annelated Boron-Dipyrromethene (BODIPY) Dyes in Aqueous Solution. ChemPhotoChem 2020, 4, 120–131. [Google Scholar] [CrossRef] [Green Version]
- Levitt, J.A.; Kuimova, M.K.; Yahioglu, G.; Chung, P.-H.; Suhling, K.; Phillips, D. Membrane-Bound Molecular Rotors Measure Viscosity in Live Cells via Fluorescence Lifetime Imaging. J. Phys. Chem. C 2009, 113, 11634–11642. [Google Scholar] [CrossRef] [Green Version]
- López-Duarte, I.; Vu, T.T.; Izquierdo, M.A.; Bull, J.A.; Kuimova, M.K. A Molecular Rotor for Measuring Viscosity in Plasma Membranes of Live Cells. Chem. Commun. 2014, 50, 5282–5284. [Google Scholar] [CrossRef]
- Shirmanova, M.V.; Lukina, M.M.; Shimolina, L.E.; Kuimova, M.K.; Dudenkova, V.V.; Shcheslavskiy, V.I.; Shcheslavskiy, V.I.; Zagaynova, E.V. Probing Energy Metabolism and Microviscosity in Cancer Using FLIM. In Proceedings of the Clinical and Preclinical Optical Diagnostics (2017), Munich, Germany, 25–29 June 2017; Optical Society of America: Washington, DC, USA, 2017; p. 1041102. [Google Scholar]
- Shimolina, L.E.; Gulin, A.A.; Paez-Perez, M.; López-Duarte, I.; Druzhkova, I.N.; Lukina, M.M.; Gubina, M.V.; Brooks, N.J.; Zagaynova, E.V.; Kuimova, M.K.; et al. Mapping Cisplatin-Induced Viscosity Alterations in Cancer Cells Using Molecular Rotor and Fluorescence Lifetime Imaging Microscopy. J. Biomed. Opt. 2020, 25, 126004. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular Portraits of Human Breast Tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Holliday, D.L.; Speirs, V. Choosing the Right Cell Line for Breast Cancer Research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yersal, O.; Barutca, S. Biological Subtypes of Breast Cancer: Prognostic and Therapeutic Implications. World J. Clin. Oncol. 2014, 5, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Comşa, Ş.; Cîmpean, A.M.; Raica, M. The Story of MCF-7 Breast Cancer Cell Line: 40 Years of Experience in Research. Anticancer Res. 2015, 35, 3147–3154. [Google Scholar]
- Anders, C.K.; Carey, L.A. Biology, Metastatic Patterns, and Treatment of Patients with Triple-Negative Breast Cancer. Clin. Breast Cancer 2009, 9, S73–S81. [Google Scholar] [CrossRef]
- Gjerdrum, C.; Tiron, C.; Høiby, T.; Stefansson, I.; Haugen, H.; Sandal, T.; Collett, K.; Li, S.; McCormack, E.; Gjertsen, B.T.; et al. Axl Is an Essential Epithelial-to-Mesenchymal Transition-Induced Regulator of Breast Cancer Metastasis and Patient Survival. Proc. Natl. Acad. Sci. USA 2010, 107, 1124–1129. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M. Fetal Bovine Serum. Mater. Methods 2013, 3, 175. [Google Scholar] [CrossRef]
- Wu, Y.; Štefl, M.; Olzyńska, A.; Hof, M.; Yahioglu, G.; Yip, P.; Casey, D.R.; Ces, O.; Humpolíčková, J.; Kuimova, M.K. Molecular Rheometry: Direct Determination of Viscosity in Lo and Ld Lipid Phases via Fluorescence Lifetime Imaging. Phys. Chem. Chem. Phys. 2013, 15, 14986–14993. [Google Scholar] [CrossRef] [Green Version]
- Karolin, J.; Johansson, L.B.-A.; Strandberg, L.; Ny, T. Fluorescence and Absorption Spectroscopic Properties of Dipyrrometheneboron Difluoride (BODIPY) Derivatives in Liquids, Lipid Membranes, and Proteins. J. Am. Chem. Soc. 1994, 116, 7801–7806. [Google Scholar] [CrossRef]
- Maleckaite, K.; Dodonova, J.; Toliautas, S.; Zilenaite, R.; Jurgutis, D.; Karabanovas, V.; Tumkevicius, S.; Vysniauskas, A. Designing a Red-Emitting Viscosity Sensitive BODIPY Fluorophore for Intracellular Viscosity Imaging. Chem. Eur. J. 2021, 27, 16768–16775. [Google Scholar] [CrossRef] [PubMed]
- DiDonato, D.; Brasaemle, D.L. Fixation Methods for the Study of Lipid Droplets by Immunofluorescence Microscopy. J. Histochem. Cytochem. 2003, 51, 773–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jüngst, C.; Klein, M.; Zumbusch, A. Long-Term Live Cell Microscopy Studies of Lipid Droplet Fusion Dynamics in Adipocytes. J. Lipid Res. 2013, 54, 3419–3429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, S.; Parton, R.G. Lipid Droplets: A Unified View of a Dynamic Organelle. Nat. Rev. Mol. Cell Biol. 2006, 7, 373–378. [Google Scholar] [CrossRef]
- Fujimoto, T.; Ohsaki, Y.; Cheng, J.; Suzuki, M.; Shinohara, Y. Lipid Droplets: A Classic Organelle with New Outfits. Histochem. Cell Biol. 2008, 130, 263–279. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, T.; Parton, R.G. Not Just Fat: The Structure and Function of the Lipid Droplet. Cold Spring Harb. Perspect. Biol. 2011, 3, a004838. [Google Scholar] [CrossRef] [Green Version]
- Blanchette-Mackie, E.J.; Dwyer, N.K.; Barber, T.; Coxey, R.A.; Takeda, T.; Rondinone, C.M.; Theodorakis, J.L.; Greenberg, A.S.; Londos, C. Perilipin Is Located on the Surface Layer of Intracellular Lipid Droplets in Adipocytes. J. Lipid Res. 1995, 36, 1211–1226. [Google Scholar] [CrossRef]
- Tauchi-Sato, K.; Ozeki, S.; Houjou, T.; Taguchi, R.; Fujimoto, T. The Surface of Lipid Droplets Is a Phospholipid Monolayer with a Unique Fatty Acid Composition. J. Biol. Chem. 2002, 277, 44507–44512. [Google Scholar] [CrossRef] [Green Version]
- Walther, T.C.; Farese, R.V. The Life of Lipid Droplets. Biochim. Biophys. Acta 2009, 1791, 459–466. [Google Scholar] [CrossRef]
- Jacquier, N.; Choudhary, V.; Mari, M.; Toulmay, A.; Reggiori, F.; Schneiter, R. Lipid Droplets Are Functionally Connected to the Endoplasmic Reticulum in Saccharomyces Cerevisiae. J. Cell Sci. 2011, 124, 2424–2437. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, V.; Schneiter, R. A Unique Junctional Interface at Contact Sites Between the Endoplasmic Reticulum and Lipid Droplets. Front. Cell Dev. Biol. 2021, 9, 703. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Plouffe, B.D.; Belov, A.M.; Ray, S.; Wang, X.; Murthy, S.K.; Karger, B.L.; Ivanov, A.R. An Integrated Platform for Isolation, Processing, and Mass Spectrometry-Based Proteomic Profiling of Rare Cells in Whole Blood. Mol. Cell Proteom. 2015, 14, 1672–1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.; Chattoraj, S.; Mondal, T.; Bhattacharyya, K. Dynamics in Cytoplasm, Nucleus, and Lipid Droplet of a Live CHO Cell: Time-Resolved Confocal Microscopy. Langmuir 2013, 29, 7975–7982. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Condello, S.; Thomes-Pepin, J.; Ma, X.; Xia, Y.; Hurley, T.D.; Matei, D.; Cheng, J.-X. Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell Stem Cell 2017, 20, 303–314.e5. [Google Scholar] [CrossRef] [Green Version]
- Cruz, A.L.S.; Barreto, E.D.A.; Fazolini, N.P.B.; Viola, J.P.B.; Bozza, P.T. Lipid Droplets: Platforms with Multiple Functions in Cancer Hallmarks. Cell Death Dis. 2020, 11, 105. [Google Scholar] [CrossRef] [Green Version]
- Tosi, M.R.; Tugnoli, V. Cholesteryl Esters in Malignancy. Clin. Chim. Acta 2005, 359, 27–45. [Google Scholar] [CrossRef]
- Antalis, C.J.; Uchida, A.; Buhman, K.K.; Siddiqui, R.A. Migration of MDA-MB-231 Breast Cancer Cells Depends on the Availability of Exogenous Lipids and Cholesterol Esterification. Clin. Exp. Metastasis 2011, 28, 733–741. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; López-Vilaró, L.; Nasarre, L.; Perez-Olabarria, M.; Vázquez, T.; Escuin, D.; Badimon, L.; Barnadas, A.; Lerma, E.; Llorente-Cortés, V. Intratumor Cholesteryl Ester Accumulation Is Associated with Human Breast Cancer Proliferation and Aggressive Potential: A Molecular and Clinicopathological Study. BMC Cancer 2015, 15, 460. [Google Scholar] [CrossRef] [Green Version]
- Antalis, C.J.; Arnold, T.; Rasool, T.; Lee, B.; Buhman, K.K.; Siddiqui, R.A. High ACAT1 Expression in Estrogen Receptor Negative Basal-like Breast Cancer Cells Is Associated with LDL-Induced Proliferation. Breast Cancer Res. Treat. 2010, 122, 661–670. [Google Scholar] [CrossRef]
- Yue, S.; Li, J.; Lee, S.-Y.; Lee, H.J.; Shao, T.; Song, B.; Cheng, L.; Masterson, T.A.; Liu, X.; Ratliff, T.L.; et al. Cholesteryl Ester Accumulation Induced by PTEN Loss and PI3K/AKT Activation Underlies Human Prostate Cancer Aggressiveness. Cell Metab. 2014, 19, 393–406. [Google Scholar] [CrossRef] [Green Version]
- Nipper, M.E.; Majd, S.; Mayer, M.; Lee, J.C.-M.; Theodorakis, E.A.; Haidekker, M.A. Characterization of Changes in the Viscosity of Lipid Membranes with the Molecular Rotor FCVJ. Biochim. Biophys. Acta (BBA)-Biomembr. 2008, 1778, 1148–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Róg, T.; Pasenkiewicz-Gierula, M.; Vattulainen, I.; Karttunen, M. Ordering Effects of Cholesterol and Its Analogues. Biochim. Biophys. Acta (BBA)-Biomembr. 2009, 1788, 97–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jurgutis, D.; Jarockyte, G.; Poderys, V.; Dodonova-Vaitkuniene, J.; Tumkevicius, S.; Vysniauskas, A.; Rotomskis, R.; Karabanovas, V. Exploring BODIPY-Based Sensor for Imaging of Intracellular Microviscosity in Human Breast Cancer Cells. Int. J. Mol. Sci. 2022, 23, 5687. https://doi.org/10.3390/ijms23105687
Jurgutis D, Jarockyte G, Poderys V, Dodonova-Vaitkuniene J, Tumkevicius S, Vysniauskas A, Rotomskis R, Karabanovas V. Exploring BODIPY-Based Sensor for Imaging of Intracellular Microviscosity in Human Breast Cancer Cells. International Journal of Molecular Sciences. 2022; 23(10):5687. https://doi.org/10.3390/ijms23105687
Chicago/Turabian StyleJurgutis, Dziugas, Greta Jarockyte, Vilius Poderys, Jelena Dodonova-Vaitkuniene, Sigitas Tumkevicius, Aurimas Vysniauskas, Ricardas Rotomskis, and Vitalijus Karabanovas. 2022. "Exploring BODIPY-Based Sensor for Imaging of Intracellular Microviscosity in Human Breast Cancer Cells" International Journal of Molecular Sciences 23, no. 10: 5687. https://doi.org/10.3390/ijms23105687






