Short-Chain Fatty Acids Augment Differentiation and Function of Human Induced Regulatory T Cells
Abstract
1. Introduction
2. Results
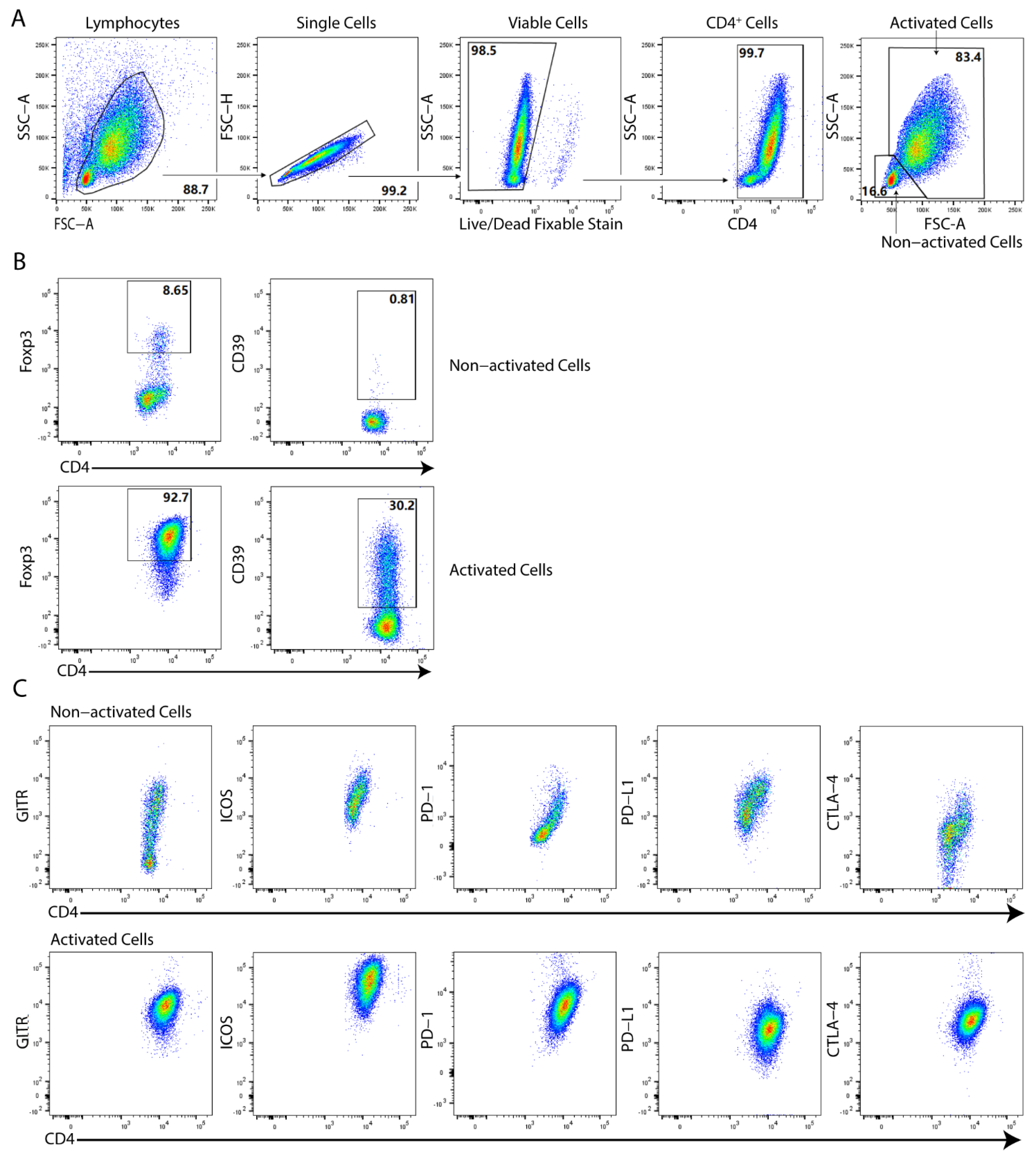
2.1. Differential Activation Patterns in the Generation of Human Tregs In Vitro via SCFAs
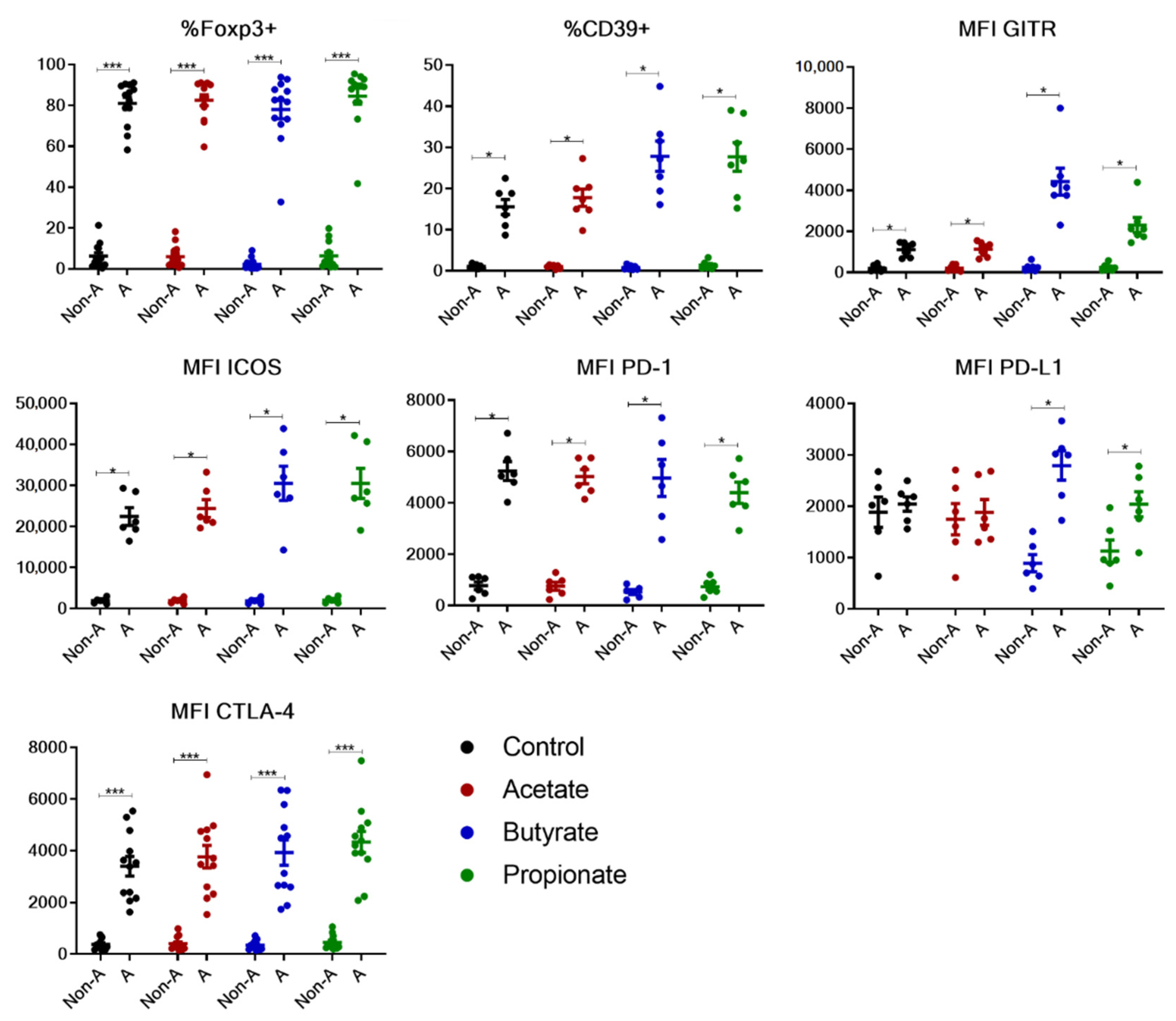
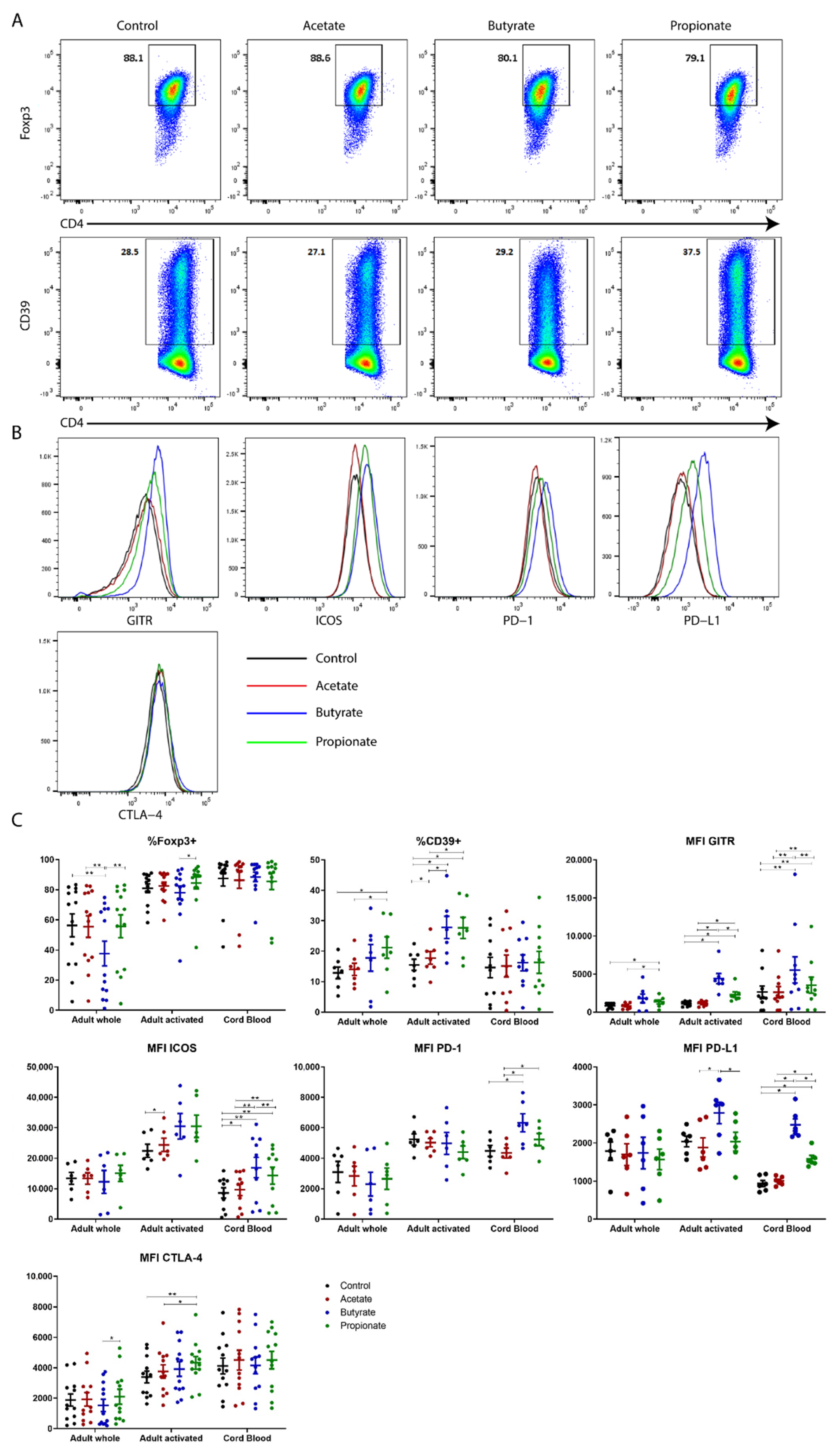
2.2. Butyrate and Propionate, but Not Acetate, Potentiate the Expression of Phenotypic Markers of Human TGF-β-Induced Tregs In Vitro
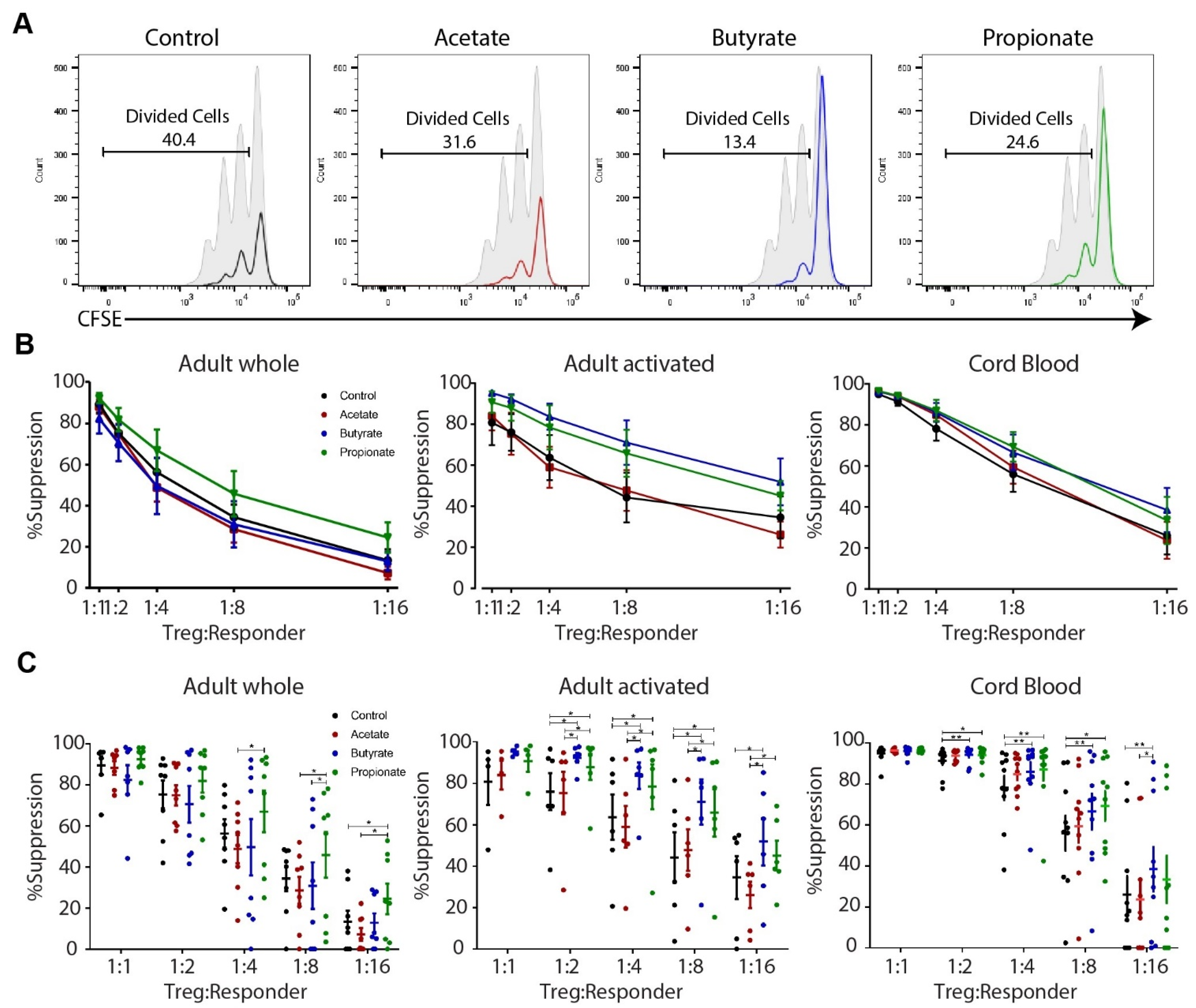
2.3. Butyrate and Propionate, but Not Acetate, Enhance the Suppressive Capacity of Human TGF-β-Induced Tregs In Vitro
2.4. Butyrate and Propionate Affect Histone Acetylation Levels at Important Regulatory Regions of Genes Encoding Molecules Crucial for Treg Function
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Mononuclear Cell Isolation
4.3. Fluorescence-Activated Cell Sorting (FACS)
4.4. In Vitro Cultures
4.5. Suppression Assay
4.6. Flow Cytometry
4.7. Chromatin Immunoprecipitation–Quantitative Polymerase Chain Reaction (ChIP-qPCR)
4.8. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharabi, A.; Tsokos, M.G.; Ding, Y.; Malek, T.R.; Klatzmann, D.; Tsokos, G.C. Regulatory T cells in the treatment of disease. Nat. Rev. Drug Discov. 2018, 17, 823–844. [Google Scholar] [CrossRef] [PubMed]
- Kasper, I.R.; Apostolidis, S.A.; Sharabi, A.; Tsokos, G.C. Empowering Regulatory T Cells in Autoimmunity. Trends Mol. Med. 2016, 22, 784–797. [Google Scholar] [CrossRef] [PubMed]
- Foussat, A.; Gregoire, S.; Clerget-Chossat, N.; Terrada, C.; Asnagli, H.; Lemoine, F.M.; Klatzmann, D.; LeHoang, P.; Forte, M.; Bodaghi, B. Regulatory T Cell Therapy for Uveitis: A New Promising Challenge. J. Ocul. Pharmacol. Ther. 2017, 33, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Tordesillas, L.; Berin, M.C.; Sampson, H.A. Immunology of Food Allergy. Immunity 2017, 47, 32–50. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Khare, A.; Krishnamoorthy, N.; Qi, Z.; Ray, P. Regulatory T cells in many flavors control asthma. Mucosal Immunol. 2010, 3, 216–229. [Google Scholar] [CrossRef] [PubMed]
- MacBeth, M.; Joetham, A.; Gelfand, E.; Schedel, M. Plasticity of Naturally Occurring Regulatory T Cells in Allergic Airway Disease Is Modulated by the Transcriptional Activity of Il-6. Int. J. Mol. Sci. 2021, 22, 4582. [Google Scholar] [CrossRef]
- Joetham, A.; Schedel, M.; O’Connor, B.P.; Kim, S.; Takeda, K.; Abbott, J.; Gelfand, E.W. Inducible and naturally occurring regulatory T cells enhance lung allergic responses through divergent transcriptional pathways. J. Allergy Clin. Immunol. 2016, 139, 1331–1342. [Google Scholar] [CrossRef][Green Version]
- Zhang, H.; Kong, H.; Zeng, X.; Guo, L.; Sun, X.; He, S. Subsets of regulatory T cells and their roles in allergy. J. Transl. Med. 2014, 12, 125. [Google Scholar] [CrossRef]
- Morales, M.A.G.; Montero-Vargas, J.M.; Vizuet-De-Rueda, J.C.; Teran, L.M. New Insights into the Role of PD-1 and Its Ligands in Allergic Disease. Int. J. Mol. Sci. 2021, 22, 11898. [Google Scholar] [CrossRef]
- García, E.-M.; Galicia-Carreón, J.; Novak, N. In vitro Conversion into CD4+CD25+Foxp3+ Induced Regulatory T Cells Is Reduced in Atopic Dermatitis Patients. Int. Arch. Allergy Immunol. 2020, 181, 353–356. [Google Scholar] [CrossRef]
- Fujimura, T.; Yonekura, S.; Taniguchi, Y.; Horiguchi, S.; Saito, A.; Yasueda, H.; Nakayama, T.; Takemori, T.; Taniguchi, M.; Sakaguchi, M.; et al. The Induced Regulatory T Cell Level, Defined as the Proportion of IL-10+Foxp3+ Cells among CD25+CD4+ Leukocytes, Is a Potential Therapeutic Biomarker for Sublingual Immunotherapy: A Preliminary Report. Int. Arch. Allergy Immunol. 2010, 153, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Paris, J.L.; de la Torre, P.; Flores, A.I. New Therapeutic Approaches for Allergy: A Review of Cell Therapy and Bio- or Nano-Material-Based Strategies. Pharmaceutics 2021, 13, 2149. [Google Scholar] [CrossRef] [PubMed]
- Verstegen, R.E.M.; Kostadinova, A.I.; Merenciana, Z.; Garssen, J.; Folkerts, G.; Hendriks, R.W.; Willemsen, L.E.M. Dietary Fibers: Effects, Underlying Mechanisms and Possible Role in Allergic Asthma Management. Nutrients 2021, 13, 4153. [Google Scholar] [CrossRef] [PubMed]
- Calzada, D.; Cremades-Jimeno, L.; López-Ramos, M.; Cárdaba, B. Peptide Allergen Immunotherapy: A New Perspective in Olive-Pollen Allergy. Pharmaceutics 2021, 13, 1007. [Google Scholar] [CrossRef] [PubMed]
- Dembele, M.; Tao, S.; Massoud, A.H.; Miah, S.M.S.; Lelias, S.; De Groot, A.S.; Mazer, B.D. Tregitopes Improve Asthma by Promoting Highly Suppressive and Antigen-Specific Tregs. Front. Immunol. 2021, 12, 634509. [Google Scholar] [CrossRef]
- Satitsuksanoa, P.; Jansen, K.; Głobińska, A.; Van De Veen, W.; Akdis, M. Regulatory Immune Mechanisms in Tolerance to Food Allergy. Front. Immunol. 2018, 9, 2939. [Google Scholar] [CrossRef]
- Liu, G.; Liu, M.; Wang, J.; Mou, Y.; Che, H. The Role of Regulatory T Cells in Epicutaneous Immunotherapy for Food Allergy. Front. Immunol. 2021, 12, 660974. [Google Scholar] [CrossRef]
- Pandiyan, P.; Bhaskaran, N.; Zou, M.; Schneider, E.; Jayaraman, S.; Huehn, J. Microbiome Dependent Regulation of Tregs and Th17 Cells in Mucosa. Front. Immunol. 2019, 10, 426. [Google Scholar] [CrossRef]
- Yap, Y.-A.; Mariño, E. An Insight into the Intestinal Web of Mucosal Immunity, Microbiota, and Diet in Inflammation. Front. Immunol. 2018, 9, 2617. [Google Scholar] [CrossRef]
- Statovci, D.; Aguilera, M.; Mac Sharry, J.; Melgar, S. The Impact of Western Diet and Nutrients on the Microbiota and Immune Response at Mucosal Interfaces. Front. Immunol. 2017, 8, 838. [Google Scholar] [CrossRef]
- Tanaka, M.; Nakayama, J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 2017, 66, 515–522. [Google Scholar] [CrossRef]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; De Los Reyes-Gavilán, C.G.; Salazar, N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef] [PubMed]
- Richards, L.B.; Li, M.; Folkerts, G.; Henricks, P.A.; Garssen, J.; Van Esch, B.C. Butyrate and Propionate Restore the Cytokine and House Dust Mite Compromised Barrier Function of Human Bronchial Airway Epithelial Cells. Int. J. Mol. Sci. 2020, 22, 65. [Google Scholar] [CrossRef] [PubMed]
- Macia, L.; Mackay, C.R. Dysfunctional microbiota with reduced capacity to produce butyrate as a basis for allergic diseases. J. Allergy Clin. Immunol. 2019, 144, 1513–1515. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; McKenzie, C.; Vuillermin, P.J.; Goverse, G.; Vinuesa, C.G.; Mebius, R.E.; Macia, L.; Mackay, C.R. Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell Rep. 2016, 15, 2809–2824. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; DeRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Hsu, P.; Nanan, R. Foetal immune programming: Hormones, cytokines, microbes and regulatory T cells. J. Reprod. Immunol. 2014, 104–105, 2–7. [Google Scholar] [CrossRef]
- Miyara, M.; Yoshioka, Y.; Kitoh, A.; Shima, T.; Wing, K.; Niwa, A.; Parizot, C.; Taflin, C.; Heike, T.; Valeyre, D.; et al. Functional Delineation and Differentiation Dynamics of Human CD4+ T Cells Expressing the FoxP3 Transcription Factor. Immunity 2009, 30, 899–911. [Google Scholar] [CrossRef]
- Allan, S.E.; Crome, S.; Crellin, N.K.; Passerini, L.; Steiner, T.; Bacchetta, R.; Roncarolo, M.G.; Levings, M. Activation-induced FOXP3 in human T effector cells does not suppress proliferation or cytokine production. Int. Immunol. 2007, 19, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Gavin, M.A.; Torgerson, T.R.; Houston, E.; Deroos, P.; Ho, W.Y.; Stray-Pedersen, A.; Ocheltree, E.L.; Greenberg, P.D.; Ochs, H.D.; Rudensky, A.Y. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc. Natl. Acad. Sci. USA 2006, 103, 6659–6664. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, M.G.; Ronchetti, S.; Ricci, E.; Alunno, A.; Gerli, R.; Nocentini, G.; Riccardi, C. GITR+ regulatory T cells in the treatment of autoimmune diseases. Autoimmun. Rev. 2014, 14, 117–126. [Google Scholar] [CrossRef] [PubMed]
- McHugh, R.S.; Whitters, M.J.; A Piccirillo, C.; A Young, D.; Shevach, E.M.; Collins, M.; Byrne, M.C. CD4+CD25+ Immunoregulatory T Cells: Gene Expression Analysis Reveals a Functional Role for the Glucocorticoid-Induced TNF Receptor. Immunity 2002, 16, 311–323. [Google Scholar] [CrossRef]
- Hamaoka, T.; Shimizu, J.; Suda, T.; Fujiwara, H. Induction of tumor-specific in vivo protective immunity by immunization with tumor antigen-pulsed antigen-presenting cells. Princess Takamatsu Symp. 1988, 19, 265–275. [Google Scholar] [PubMed]
- Borsellino, G.; Kleinewietfeld, M.; DI Mitri, D.; Sternjak, A.; Diamantini, A.; Giometto, R.; Höpner, S.; Centonze, D.; Bernardi, G.; Dell’Acqua, M.L.; et al. Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: Hydrolysis of extracellular ATP and immune suppression. Blood 2007, 110, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Herrath, J.; Chemin, K.; Albrecht, I.; Catrina, A.I.; Malmström, V. Surface expression of CD39 identifies an enriched Treg-cell subset in the rheumatic joint, which does not suppress IL-17A secretion. Eur. J. Immunol. 2014, 44, 2979–2989. [Google Scholar] [CrossRef]
- Deaglio, S.; Dwyer, K.M.; Gao, W.; Friedman, D.; Usheva, A.; Erat, A.; Chen, J.-F.; Enjyoji, K.; Linden, J.; Oukka, M.; et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J. Exp. Med. 2007, 204, 1257–1265. [Google Scholar] [CrossRef]
- Redpath, S.A.; van der Werf, N.; Cervera, A.M.; MacDonald, A.S.; Gray, D.; Maizels, R.M.; Taylor, M.D. ICOS controls Foxp3 + regulatory T-cell expansion, maintenance and IL-10 production during helminth infection. Eur. J. Immunol. 2013, 43, 705–715. [Google Scholar] [CrossRef]
- Zheng, J.; Chan, P.L.; Liu, Y.; Qin, G.; Xiang, Z.; Lam, K.-T.; Lewis, D.B.; Lau, Y.-L.; Tu, W. ICOS Regulates the Generation and Function of Human CD4+ Treg in a CTLA-4 Dependent Manner. PLoS ONE 2013, 8, e82203. [Google Scholar] [CrossRef]
- Krupnick, A.S.; Gelman, A.E.; Barchet, W.; Richardson, S.; Kreisel, F.H.; Turka, L.A.; Colonna, M.; Patterson, G.A.; Kreisel, D. Cutting Edge: Murine Vascular Endothelium Activates and Induces the Generation of Allogeneic CD4+25+Foxp3+ Regulatory T Cells. J. Immunol. 2005, 175, 6265–6270. [Google Scholar] [CrossRef] [PubMed]
- Francisco, L.M.; Salinas, V.H.; Brown, K.E.; Vanguri, V.K.; Freeman, G.J.; Kuchroo, V.K.; Sharpe, A.H. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J. Exp. Med. 2009, 206, 3015–3029. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Putnam, A.L.; Xu-Yu, Z.; Szot, G.L.; Lee, M.R.; Zhu, S.; Gottlieb, P.A.; Kapranov, P.; Gingeras, T.R.; de St Groth, B.F.; et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J. Exp. Med. 2006, 203, 1701–1711. [Google Scholar] [CrossRef] [PubMed]
- Baecher-Allan, C.; Wolf, E.; Hafler, D.A. MHC Class II Expression Identifies Functionally Distinct Human Regulatory T Cells. J. Immunol. 2006, 176, 4622–4631. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Eriksson, M.; Shang, M.-M.; Weyd, H.; Tegnér, J. Comparative Analysis of Protocols to Induce Human CD4+Foxp3+ Regulatory T Cells by Combinations of IL-2, TGF-beta, Retinoic Acid, Rapamycin and Butyrate. PLoS ONE 2016, 11, e0148474. [Google Scholar] [CrossRef]
- Wang, J.; Huizinga, T.W.J.; Toes, R.E.M. De Novo Generation and Enhanced Suppression of Human CD4+CD25+ Regulatory T Cells by Retinoic Acid. J. Immunol. 2009, 183, 4119–4126. [Google Scholar] [CrossRef]
- Candía, E.; Reyes, P.; Covian, C.; Rodríguez, F.; Wainstein, N.; Morales, J.; Mosso, C.; Rosemblatt, M.; Fierro, J.A. Single and combined effect of retinoic acid and rapamycin modulate the generation, activity and homing potential of induced human regulatory T cells. PLoS ONE 2017, 12, e0182009. [Google Scholar] [CrossRef]
- Long, S.A.; Buckner, J.H. Combination of rapamycin and IL-2 increases de novo induction of human CD4+CD25+FOXP3+ T cells. J. Autoimmun. 2008, 30, 293–302. [Google Scholar] [CrossRef]
- Jeffery, L.E.; Burke, F.; Mura, M.; Zheng, Y.; Qureshi, O.S.; Hewison, M.; Walker, L.S.K.; Lammas, D.A.; Raza, K.; Sansom, D.M. 1,25-Dihydroxyvitamin D3 and IL-2 Combine to Inhibit T Cell Production of Inflammatory Cytokines and Promote Development of Regulatory T Cells Expressing CTLA-4 and FoxP3. J. Immunol. 2009, 183, 5458–5467. [Google Scholar] [CrossRef]
- Hsu, P.; Santner-Nanan, B.; Hu, M.; Skarratt, K.; Lee, C.H.; O Stormon, M.; Wong, M.; Fuller, S.J.; Nanan, R. IL-10 Potentiates Differentiation of Human Induced Regulatory T Cells via STAT3 and Foxo1. J. Immunol. 2015, 195, 3665–3674. [Google Scholar] [CrossRef]
- Tran, D.Q.; Ramsey, H.; Shevach, E.M. Induction of FOXP3 expression in naive human CD4+FOXP3− T cells by T-cell receptor stimulation is transforming growth factor-β–dependent but does not confer a regulatory phenotype. Blood 2007, 110, 2983–2990. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, M.; Spreafico, R.; Saidin, S.; Chua, C.; Moshref, M.; Leong, J.Y.; Tan, Y.K.; Thumboo, J.; Van Loosdregt, J.; Albani, S. Ex Vivo–Expanded but Not In Vitro–Induced Human Regulatory T Cells Are Candidates for Cell Therapy in Autoimmune Diseases Thanks to Stable Demethylation of the FOXP3 Regulatory T Cell–Specific Demethylated Region. J. Immunol. 2014, 194, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Shevach, E.M.; Thornton, A.M. tTregs, pTregs, and iTregs: Similarities and differences. Immunol. Rev. 2014, 259, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Caccamo, N.; Joosten, S.A.; Ottenhoff, T.H.M.; Dieli, F. Atypical Human Effector/Memory CD4+ T Cells with a Naive-Like Phenotype. Front. Immunol. 2018, 9, 2832. [Google Scholar] [CrossRef]
- Larbi, A.; Fulop, T. From “truly naïve” to “exhausted senescent” T cells: When markers predict functionality. Cytom. Part A 2013, 85, 25–35. [Google Scholar] [CrossRef]
- Braber, I.D.; Mugwagwa, T.; Vrisekoop, N.; Westera, L.; Mögling, R.; de Boer, A.B.; Willems, N.; Schrijver, E.H.; Spierenburg, G.; Gaiser, K.; et al. Maintenance of Peripheral Naive T Cells Is Sustained by Thymus Output in Mice but Not Humans. Immunity 2012, 36, 288–297. [Google Scholar] [CrossRef]
- Silva, S.L.; Sousa, A.E. Establishment and Maintenance of the Human Naïve CD4+ T-Cell Compartment. Front. Pediatr. 2016, 4, 119. [Google Scholar] [CrossRef]
- van den Broek, T.; Borghans, J.A.M.; Van Wijk, F. The full spectrum of human naive T cells. Nat. Rev. Immunol. 2018, 18, 363–373. [Google Scholar] [CrossRef]
- Pulko, V.; Davies, J.S.; Martinez, C.; Lanteri, M.C.; Busch, M.C.L.M.P.; Diamond, M.S.; Knox, K.; Bush, E.C.; Sims, P.; Sinari, S.; et al. Human memory T cells with a naive phenotype accumulate with aging and respond to persistent viruses. Nat. Immunol. 2016, 17, 966–975. [Google Scholar] [CrossRef]
- Qi, Q.; Liu, Y.; Cheng, Y.; Glanville, J.; Zhang, D.; Lee, J.-Y.; Olshen, R.A.; Weyand, C.M.; Boyd, S.D.; Goronzy, J.J. Diversity and clonal selection in the human T-cell repertoire. Proc. Natl. Acad. Sci. USA 2014, 111, 13139–13144. [Google Scholar] [CrossRef]
- Goronzy, J.J.; Fang, F.; Cavanagh, M.M.; Fengqin, F.; Weyand, C.M. Naive T Cell Maintenance and Function in Human Aging. J. Immunol. 2015, 194, 4073–4080. [Google Scholar] [CrossRef] [PubMed]
- Teteloshvili, N.; Kluiver, J.; Van Der Geest, K.S.M.; Van Der Lei, R.J.; Jellema, P.; Pawelec, G.; Brouwer, E.; Kroesen, B.-J.; Boots, A.M.H.; van den Berg, A. Age-Associated Differences in MiRNA Signatures Are Restricted to CD45RO Negative T Cells and Are Associated with Changes in the Cellular Composition, Activation and Cellular Ageing. PLoS ONE 2015, 10, e0137556. [Google Scholar] [CrossRef]
- Potaczek, D.P.; Harb, H.; Michel, S.; Alhamwe, B.A.; Renz, H.; Tost, J. Epigenetics and allergy: From basic mechanisms to clinical applications. Epigenomics 2017, 9, 539–571. [Google Scholar] [CrossRef] [PubMed]
- Alaskhar Alhamwe, B.; Khalaila, R.; Wolf, J.; Von Bülow, V.; Harb, H.; Alhamdan, F.; Hii, C.S.; Prescott, S.L.; Ferrante, A.; Renz, H.; et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin. Immunol. 2018, 14, 39. [Google Scholar] [CrossRef] [PubMed]
- Alhamwe, B.A.; Alhamdan, F.; Ruhl, A.; Potaczek, D.P.; Renz, H. The role of epigenetics in allergy and asthma development. Curr. Opin. Allergy Clin. Immunol. 2020, 20, 48–55. [Google Scholar] [CrossRef]
- Delgoffe, G.M.; Kole, T.P.; Zheng, Y.; Zarek, P.E.; Matthews, K.L.; Xiao, B.; Worley, P.F.; Kozma, S.C.; Powell, J.D. The mTOR Kinase Differentially Regulates Effector and Regulatory T Cell Lineage Commitment. Immunity 2009, 30, 832–844. [Google Scholar] [CrossRef]
- Dennis, P.B.; Jaeschke, A.; Saitoh, M.; Fowler, B.; Kozma, S.C.; Thomas, G. Mammalian TOR: A Homeostatic ATP Sensor. Science 2001, 294, 1102–1105. [Google Scholar] [CrossRef]
- Timperi, E.; Barnaba, V. CD39 Regulation and Functions in T Cells. Int. J. Mol. Sci. 2021, 22, 8068. [Google Scholar] [CrossRef]
- Rissiek, A.; Baumann, I.; Cuapio, A.; Mautner, A.; Kolster, M.; Arck, P.C.; Dodge-Khatami, A.; Mittrücker, H.-W.; Koch-Nolte, F.; Haag, F.; et al. The expression of CD39 on regulatory T cells is genetically driven and further upregulated at sites of inflammation. J. Autoimmun. 2015, 58, 12–20. [Google Scholar] [CrossRef]
- Timperi, E.; Folgori, L.; Amodio, D.; De Luca, M.; Chiurchiù, S.; Piconese, S.; Di Cesare, S.; Pacella, I.; Martire, C.; Bonatti, G.; et al. Expansion of activated regulatory T cells inversely correlates with clinical severity in septic neonates. J. Allergy Clin. Immunol. 2016, 137, 1617–1620. [Google Scholar] [CrossRef]
- Harb, H.; Amarasekera, M.; Ashley, S.; Tulic, M.K.; Pfefferle, P.I.; Potaczek, D.P.; Martino, D.; Kesper, D.A.; Prescott, S.L.; Renz, H. Epigenetic Regulation in Early Childhood: A Miniaturized and Validated Method to Assess Histone Acetylation. Int. Arch. Allergy Immunol. 2015, 168, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Abbring, S.; Wolf, J.; Ayechu-Muruzabal, V.; Diks, M.A.; Alhamdan, F.; Harb, H.; Renz, H.; Garn, H.; Potaczek, D.P.; Van Esch, B.C.; et al. Raw Cow’s Milk Reduces Allergic Symptoms in a Murine Model for Food Allergy—A Potential Role for Epigenetic Modifications. Nutrients 2019, 11, 1721. [Google Scholar] [CrossRef] [PubMed]
- Alhamwe, B.A.; Meulenbroek, L.A.P.M.; Veening-Griffioen, D.H.; Wehkamp, T.M.D.; Alhamdan, F.; Miethe, S.; Harb, H.; Hogenkamp, A.; Knippels, L.M.J.; Von Strandmann, E.P.; et al. Decreased Histone Acetylation Levels at Th1 and Regulatory Loci after Induction of Food Allergy. Nutrients 2020, 12, 3193. [Google Scholar] [CrossRef] [PubMed]
- Haring, M.; Offermann, S.; Danker, T.; Horst, I.; Peterhansel, C.; Stam, M. Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods 2007, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Hu, M. Human Regulatory T Cell Physiology—Lessons Learnt from Newborns and Adults. Ph.D. Thesis, The University of Sydney, Sydney, Australia, 2018. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, M.; Alashkar Alhamwe, B.; Santner-Nanan, B.; Miethe, S.; Harb, H.; Renz, H.; Potaczek, D.P.; Nanan, R.K. Short-Chain Fatty Acids Augment Differentiation and Function of Human Induced Regulatory T Cells. Int. J. Mol. Sci. 2022, 23, 5740. https://doi.org/10.3390/ijms23105740
Hu M, Alashkar Alhamwe B, Santner-Nanan B, Miethe S, Harb H, Renz H, Potaczek DP, Nanan RK. Short-Chain Fatty Acids Augment Differentiation and Function of Human Induced Regulatory T Cells. International Journal of Molecular Sciences. 2022; 23(10):5740. https://doi.org/10.3390/ijms23105740
Chicago/Turabian StyleHu, Mingjing, Bilal Alashkar Alhamwe, Brigitte Santner-Nanan, Sarah Miethe, Hani Harb, Harald Renz, Daniel P. Potaczek, and Ralph K. Nanan. 2022. "Short-Chain Fatty Acids Augment Differentiation and Function of Human Induced Regulatory T Cells" International Journal of Molecular Sciences 23, no. 10: 5740. https://doi.org/10.3390/ijms23105740
APA StyleHu, M., Alashkar Alhamwe, B., Santner-Nanan, B., Miethe, S., Harb, H., Renz, H., Potaczek, D. P., & Nanan, R. K. (2022). Short-Chain Fatty Acids Augment Differentiation and Function of Human Induced Regulatory T Cells. International Journal of Molecular Sciences, 23(10), 5740. https://doi.org/10.3390/ijms23105740








