Pseudomonas putida Biofilm Depends on the vWFa-Domain of LapA in Peptides-Containing Growth Medium
Abstract
:1. Introduction
2. Results
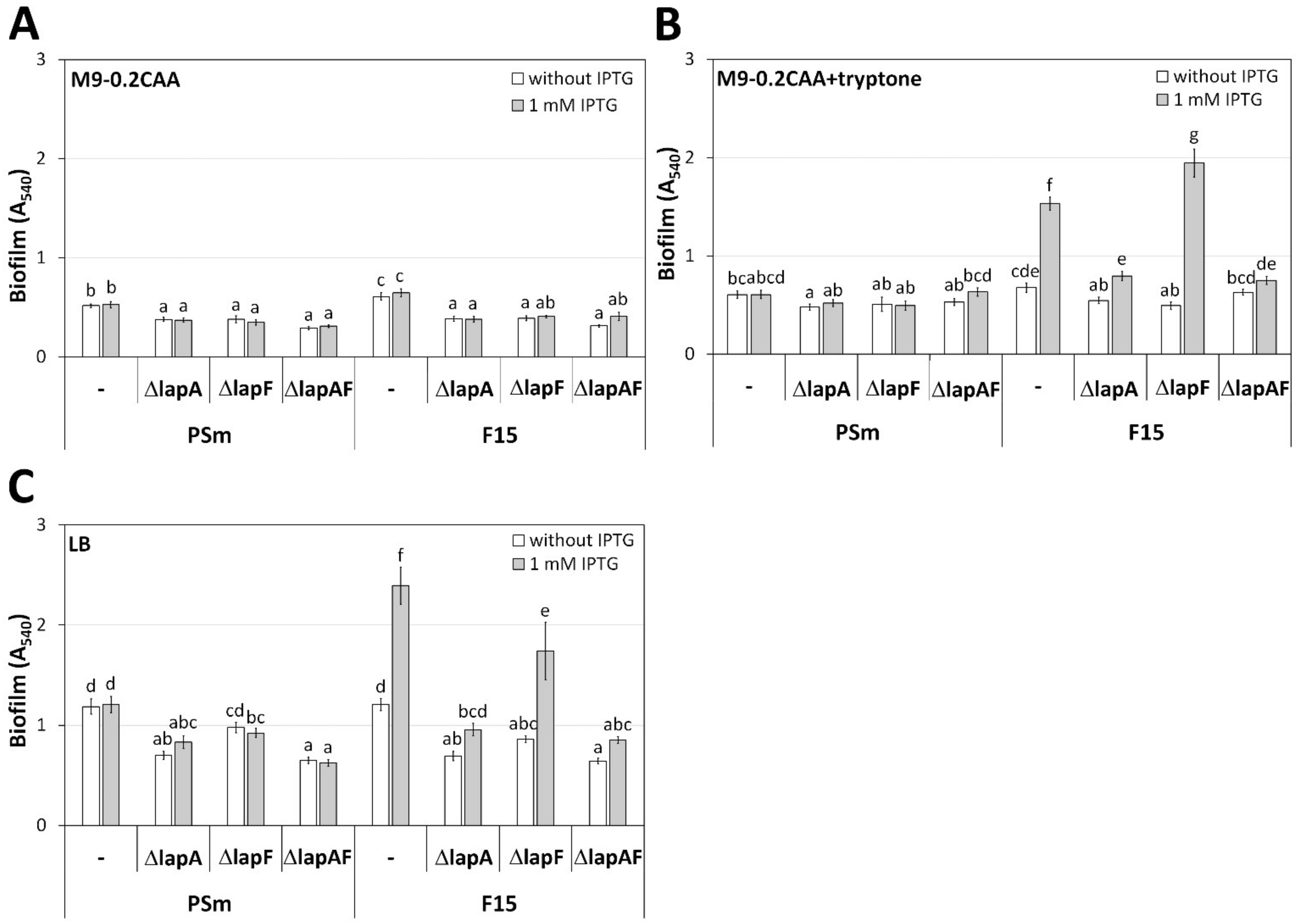
2.1. LapA Is Essential for Peptide-Dependent Biofilm
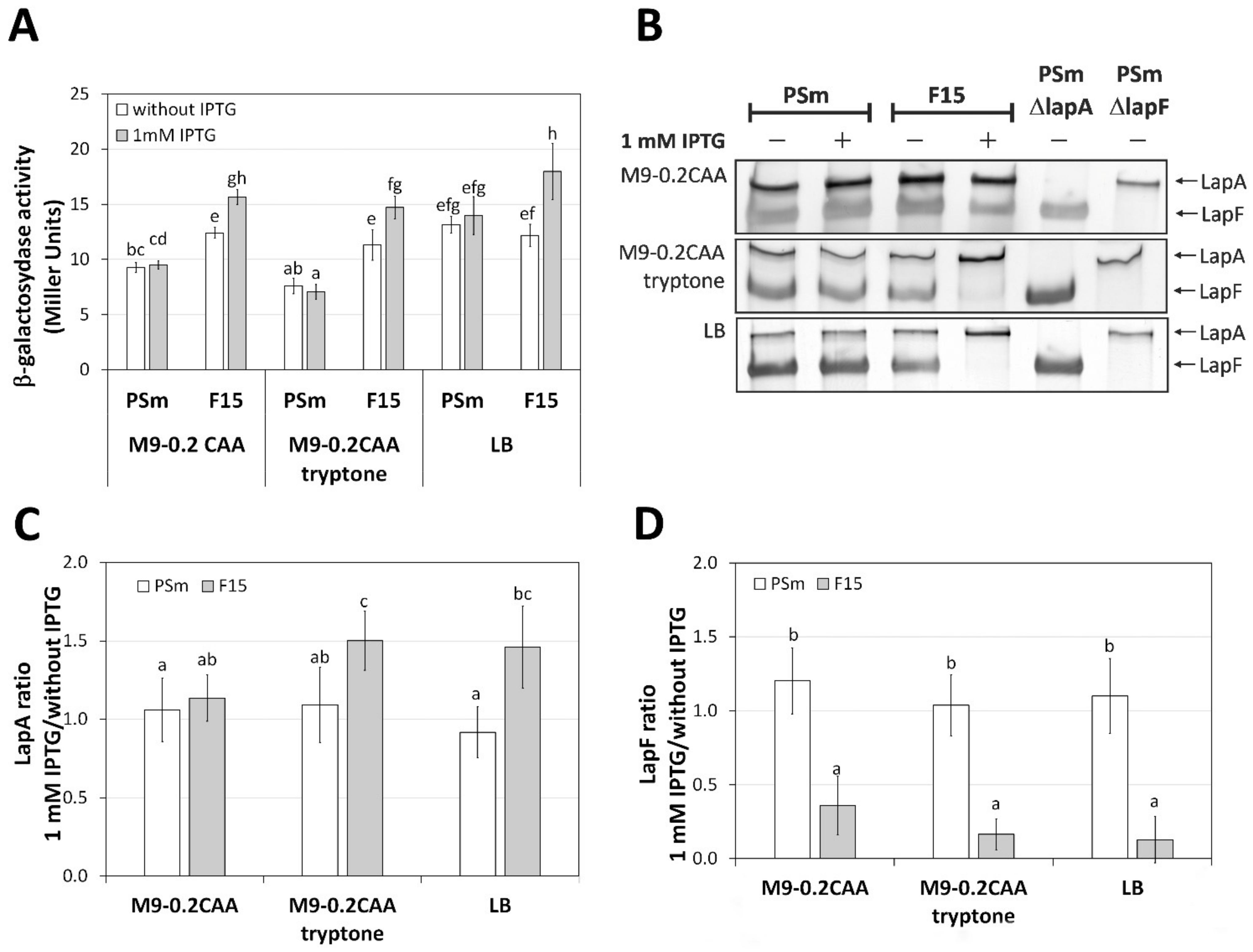
2.2. The Transcription of lapA Is Growth Medium Independent, the Amount of LapA Is Not
2.3. Construction of a Strain with a Modified lapA Transcription and a LapA Strain without a vWFa Domain
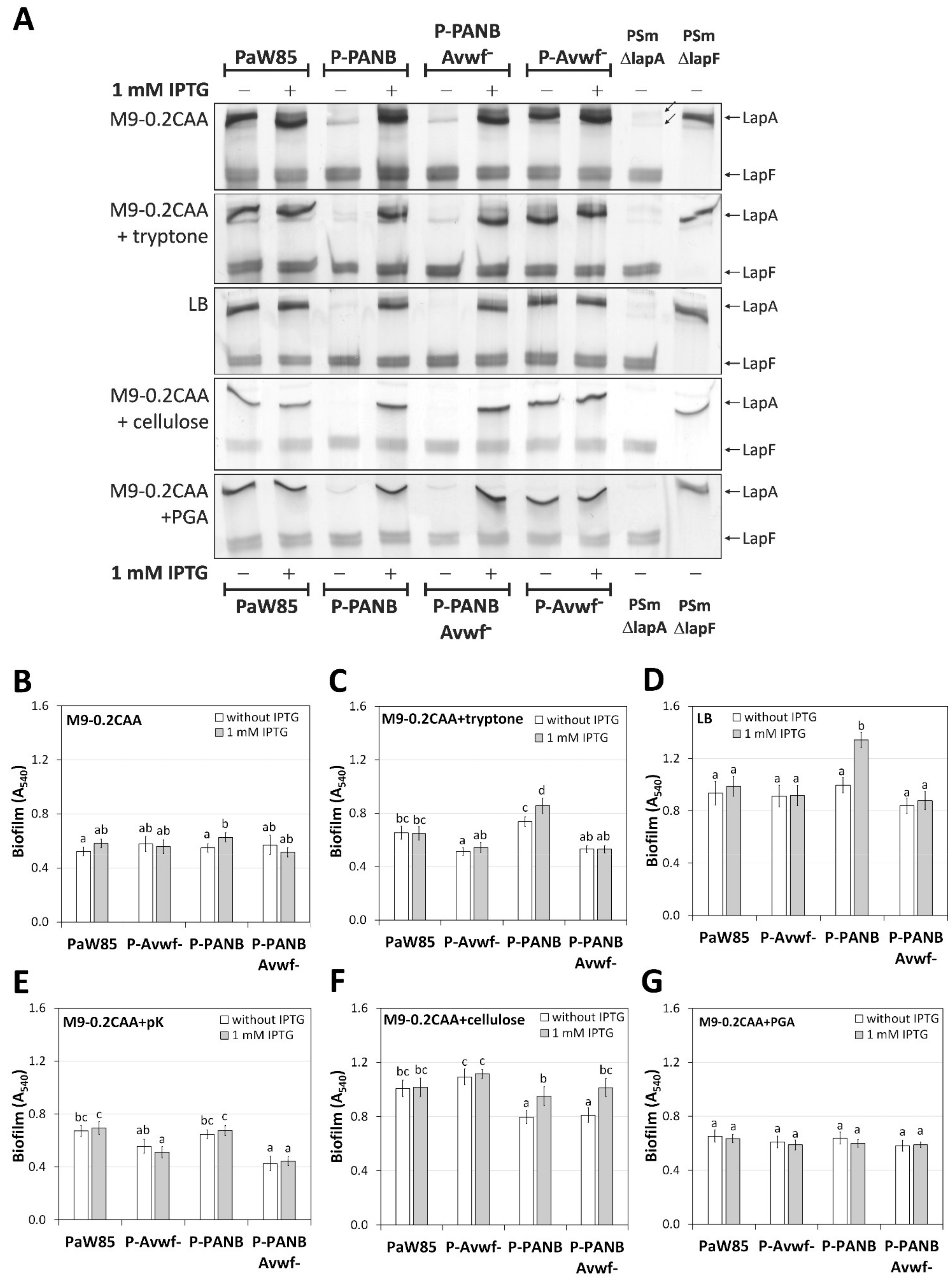
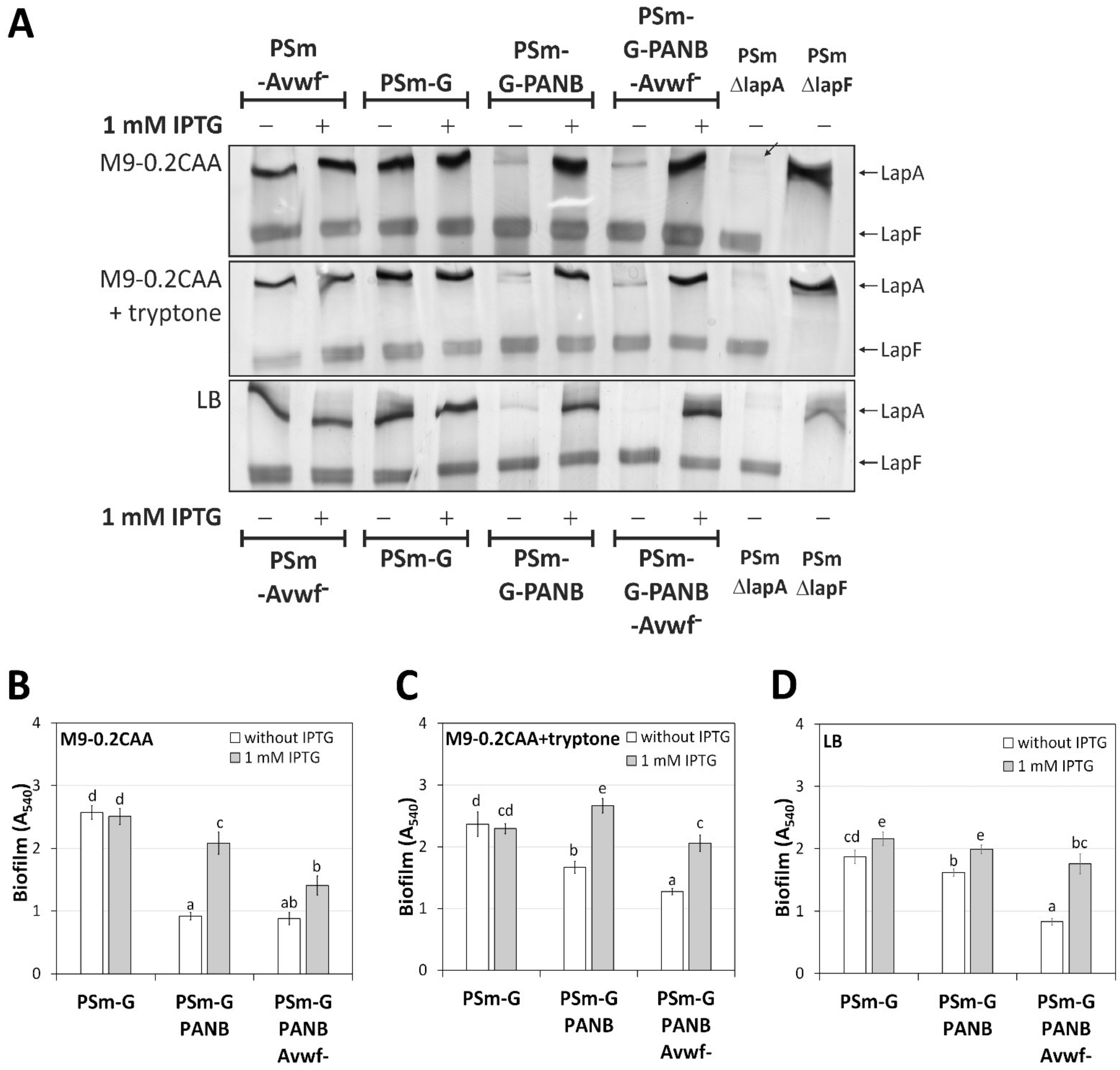
2.4. LapA Domain vWFa Is Involved in the Peptide-Dependent Biofilm of P. putida PaW85
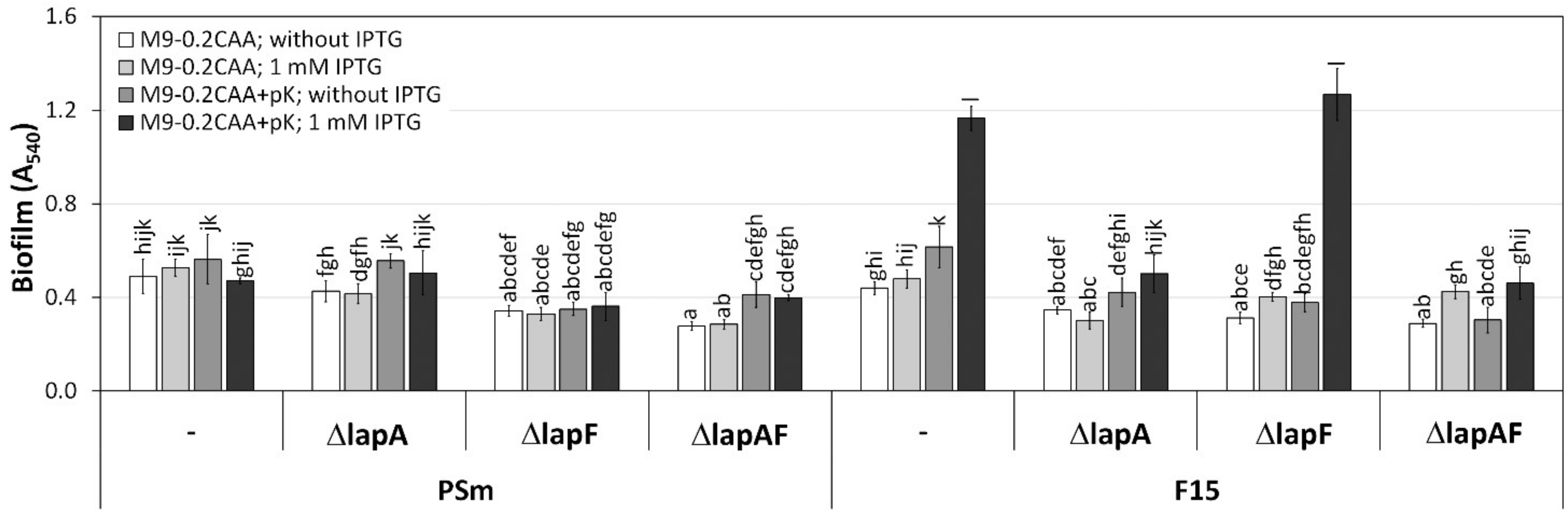
2.5. Biofilm of P. putida PSmΔlapE- and PSmΔlapG-Derived Strains
2.6. The vWFa Domain Is Essential for F15 Biofilm
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Media
4.2. DNA Manipulation
4.3. Assessment of Biofilm
4.4. Assessment of β-Galactosidase Activity
4.5. Preparation of Cell Lysates, Protein Quantification, SDS-Polyacrylamide Gel Electrophoresis, and Western Immunoblot Assay
4.6. Statistical Analysis of the Results
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moor, H.; Teppo, A.; Lahesaare, A.; Kivisaar, M.; Teras, R. Fis Overexpression Enhances Pseudomonas Putida Biofilm Formation by Regulating the Ratio of LapA and LapF. Microbiology 2014, 160, 2681–2693. [Google Scholar] [CrossRef] [PubMed]
- Lahesaare, A.; Ainelo, H.; Teppo, A.; Kivisaar, M.; Heipieper, H.J.; Teras, R. LapF and Its Regulation by FIS Affect the Cell Surface Hydrophobicity of Pseudomonas putida. PLoS ONE 2016, 11, e0166078. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gil, M.; Yousef-Coronado, F.; Espinosa-Urgel, M. LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol. Microbiol. 2010, 77, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Gjermansen, M.; Nilsson, M.; Yang, L.; Tolker-Nielsen, T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: Genetic elements and molecular mechanisms. Mol. Microbiol. 2010, 75, 815–826. [Google Scholar] [CrossRef]
- Hinsa, S.M.; Espinosa-Urgel, M.; Ramos, J.L.; O’Toole, G.A. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003, 49, 905–918. [Google Scholar] [CrossRef]
- Holland, I.B.; Schmitt, L.; Young, J. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway (Review). Mol. Membr. Biol. 2005, 22, 29–39. [Google Scholar] [CrossRef]
- Ivanov, I.E.; Boyd, C.D.; Newell, P.D.; Schwartz, M.E.; Turnbull, L.; Johnson, M.S.; Whitchurch, C.B.; O’Toole, G.A.; Camesano, T.A. Atomic force and super-resolution microscopy support a role for LapA as a cell-surface biofilm adhesin of Pseudomonas fluorescens. Res. Microbiol. 2012, 163, 685–691. [Google Scholar] [CrossRef] [Green Version]
- El-Kirat-Chatel, S.; Beaussart, A.; Boyd, C.D.; O’Toole, G.A.; Dufrêne, Y.F. Single-Cell and Single-Molecule Analysis Deciphers the Localization, Adhesion, and Mechanics of the Biofilm Adhesin LapA. ACS Chem. Biol. 2013, 9, 485–494. [Google Scholar] [CrossRef] [Green Version]
- El-Kirat-Chatel, S.; Boyd, C.D.; O’Toole, G.A.; Dufrêne, Y.F. Single-Molecule Analysis of Pseudomonas fluorescens Footprints. ACS Nano 2014, 8, 1690–1698. [Google Scholar] [CrossRef] [Green Version]
- Boyd, C.D.; Smith, T.J.; El-Kirat-Chatel, S.; Newell, P.D.; Dufrêne, Y.F.; O’Toole, G.A. Structural Features of the Pseudomonas fluorescens Biofilm Adhesin LapA Required for LapG-Dependent Cleavage, Biofilm Formation, and Cell Surface Localization. J. Bacteriol. 2014, 196, 2775–2788. [Google Scholar] [CrossRef] [Green Version]
- Collins, A.J.; Smith, T.J.; Sondermann, H.; O’Toole, G.A. From Input to Output: The Lap/c-di-GMP Biofilm Regulatory Circuit. Annu. Rev. Microbiol. 2020, 74, 607–631. [Google Scholar] [CrossRef] [PubMed]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S.L. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2015, 44, D646–D653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, A.J.; Pastora, A.B.; Smith, T.J.; O’Toole, G.A. MapA, a Second Large RTX Adhesin Conserved across the Pseudomonads, Contributes to Biofilm Formation by Pseudomonas fluorescens. J. Bacteriol. 2020, 202, e00277-20. [Google Scholar] [CrossRef] [PubMed]
- Economou, A.; Hamilton, W.; Johnston, A.; Downie, J. The Rhizobium nodulation gene nodO encodes a Ca2(+)-binding protein that is exported without N-terminal cleavage and is homologous to haemolysin and related proteins. EMBO J. 1990, 9, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, Z.M.; von Ware, J. Willebrand factor. FASEB J. 1993, 7, 308–316. [Google Scholar] [CrossRef]
- Colombatti, A.; Bonaldo, P.; Doliana, R. Type A Modules: Interacting Domains Found in Several Non-Fibrillar Collagens and in Other Extracellular Matrix Proteins. Matrix 1993, 13, 297–306. [Google Scholar] [CrossRef]
- Hartleib, J.; Köhler, N.; Dickinson, R.B.; Chhatwal, G.S.; Sixma, J.J.; Hartford, O.M.; Foster, T.J.; Peters, G.; Kehrel, E.B.; Herrmann, M. Protein A is the von Willebrand factor binding protein on Staphylococcus aureus. Blood J. Am. Soc. Hematol. 2000, 96, 2149–2156. [Google Scholar]
- Siryaporn, A.; Kuchma, S.L.; O’Toole, G.A.; Gitai, Z. Surface attachment induces Pseudomonas aeruginosa virulence. Proc. Natl. Acad. Sci. USA 2014, 111, 16860–16865. [Google Scholar] [CrossRef] [Green Version]
- Puhm, M.; Ainelo, H.; Kivisaar, M.; Teras, R. Tryptone in Growth Media Enhances Pseudomonas putida Biofilm. Microorganisms 2022, 10, 618. [Google Scholar] [CrossRef]
- Ainelo, H.; Lahesaare, A.; Teppo, A.; Kivisaar, M.; Teras, R. The promoter region of lapA and its transcriptional regulation by Fis in Pseudomonas putida. PLoS ONE 2017, 12, e0185482. [Google Scholar] [CrossRef] [Green Version]
- Lahesaare, A.; Moor, H.; Kivisaar, M.; Teras, R. Pseudomonas putida FIS Binds to the lapF Promoter In Vitro and Represses the Expression of LapF. PLoS ONE 2014, 9, e115901. [Google Scholar] [CrossRef] [PubMed]
- Herrero, M.; de Lorenzo, V.; Timmis, K.N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1990, 172, 6557–6567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosendahl, S.; Tamman, H.; Brauer, A.; Remm, M.; Hõrak, R. Chromosomal toxin-antitoxin systems in Pseudomonas putida are rather selfish than beneficial. Sci. Rep. 2020, 10, 9230. [Google Scholar] [CrossRef] [PubMed]
- Regenhardt, D.; Heuer, H.; Heim, S.; Fernandez, D.U.; Strompl, C.; Moore, E.R.B.; Timmis, K.N. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ. Microbiol. 2002, 4, 912–915. [Google Scholar] [CrossRef]
- Jakovleva, J.; Teppo, A.; Velts, A.; Saumaa, S.; Moor, H.; Kivisaar, M.; Teras, R. FIS regulates the competitiveness of Pseudomonas putida on barley roots by inducing biofilm formation. Microbiology 2012, 158, 708–720. [Google Scholar] [CrossRef] [Green Version]
- Kivistik, P.A.; Putrinš, M.; Püvi, K.; Ilves, H.; Kivisaar, M.; Hõrak, R. The ColRS Two-Component System Regulates Membrane Functions and Protects Pseudomonas putida against Phenol. J. Bacteriol. 2006, 188, 8109–8117. [Google Scholar] [CrossRef] [Green Version]
- Wong, S.M.; Mekalanos, J.J. Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2000, 97, 10191–10196. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Garcia, E.; de Lorenzo, V. Engineering multiple genomic deletions in Gram-negative bacteria: Analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 2011, 13, 2702–2716. [Google Scholar] [CrossRef]
- Martinez-Garcia, E.; Aparicio, T.; Goni-Moreno, A.; Fraile, S.; De Lorenzo, V. SEVA 2.0: An update of the Standard European Vector Architecture for de-/re-construction of bacterial functionalities. Nucleic Acids Res. 2014, 43, D1183–D1189. [Google Scholar] [CrossRef]
- Wirth, N.T.; Kozaeva, E.; Nikel, P.I. Accelerated genome engineering of Pseudomonas putida by I-Sce I―mediated recombination and CRISPR-Cas9 counterselection. Microb. Biotechnol. 2019, 13, 233–249. [Google Scholar] [CrossRef] [Green Version]
- De Lorenzo, V.; Cases, I.; Herrero, M.; Timmis, K.N. Early and late responses of TOL promoters to pathway inducers: Identification of postexponential promoters in Pseudomonas putida with lacZ-tet bicistronic reporters. J. Bacteriol. 1993, 175, 6902–6907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, T.J.; Sondermann, H.; O’Toole, G.A. Type 1 Does the Two-Step: Type 1 Secretion Substrates with a Functional Periplasmic Intermediate. J. Bacteriol. 2018, 200, e00168-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romeo, T.; Vakulskas, C.; Babitzke, P. Post-transcriptional regulation on a global scale: Form and function of CSR/RSM systems. Environ. Microbiol. 2012, 15, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gil, M.; Ramos-González, M.I.; Espinosa-Urgel, M. Roles of Cyclic Di-GMP and the GAC System in Transcriptional Control of the Genes Coding for the Pseudomonas putida Adhesins LapA and LapF. J. Bacteriol. 2014, 196, 1484–1495. [Google Scholar] [CrossRef] [Green Version]
- Huertas-Rosales, Ó.; Romero, M.; Chan, K.-G.; Hong, K.-W.; Cámara, M.; Heeb, S.; Barrientos-Moreno, L.; Molina-Henares, M.A.; Travieso, M.L.; Ramos-González, M.I.; et al. Genome-Wide Analysis of Targets for Post-Transcriptional Regulation by Rsm Proteins in Pseudomonas putida. Front. Mol. Biosci. 2021, 8, 624061. [Google Scholar] [CrossRef]
- Díaz-Salazar, C.; Calero, P.; Portero, R.E.; Jiménez-Fernández, A.; Wirebrand, L.; Velasco-Domínguez, M.G.; López-Sánchez, A.; Shingler, V.; Govantes, F. The stringent response promotes biofilm dispersal in Pseudomonas putida. Sci. Rep. 2017, 7, 18055. [Google Scholar] [CrossRef] [Green Version]
- Boyd, C.D.; Chatterjee, D.; Sondermann, H.; O’Toole, G.A. LapG, Required for Modulating Biofilm Formation by Pseudomonasfluorescens Pf0-1, Is a Calcium-Dependent Protease. J. Bacteriol. 2012, 194, 4406–4414. [Google Scholar] [CrossRef] [Green Version]
- Adams, M.H. Bacteriophages; Interscience Publishers, Inc.: New York, NY, USA, 1959. [Google Scholar]
- Bauchop, T.; Elsden, S.R. The Growth of Micro-organisms in Relation to their Energy Supply. Microbiology 1960, 23, 457–469. [Google Scholar] [CrossRef] [Green Version]
- Quan, J.; Tian, J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 2011, 6, 242–251. [Google Scholar] [CrossRef]
- Horton, R.M.; Hunt, H.D.; Ho, S.N.; Pullen, J.K.; Pease, L.R. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene 1989, 77, 61–68. [Google Scholar] [CrossRef]
- Choi, K.-H.; Kumar, A.; Schweizer, H.P. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: Application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 2006, 64, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, A. The effects of culture concentration and age, time and temperature on bacterial attachments to polystyrene. Can. J. Mircobiol. 1977, 23, 1–6. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Mikelsaar, A.-V.; Sünter, A.; Toomik, P.; Karpson, K.; Jurone, E. Development of New Monoclonal Antibodies for Immunocytochemical Characterization of Neural Stem and Differentiated Cells. In Neural Stem Cells and Therapy; Sun, T., Ed.; InTech: Rijeka, Croatia, 2012. [Google Scholar] [CrossRef] [Green Version]
- BioTek. Quantitation of Peptides and Amino Acids with a Synergy™HT Using UV Fluorescence. Available online: https://www.biotek.com/resources/docs/Synergy_HT_Quantitation_of_Peptides_and_Amino_Acids.pdf (accessed on 20 April 2022).
- Chevallet, M.; Luche, S.; Rabilloud, T. Silver staining of proteins in polyacrylamide gels. Nat. Protoc. 2006, 1, 1852–1858. [Google Scholar] [CrossRef] [Green Version]
- Teras, R.; Jakovleva, J.; Kivisaar, M. FIS negatively affects binding of Tn4652 transposase by out-competing IHF from the left end of Tn4652. Microbiology 2009, 155, 1203–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schagger, H. Tricine-SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef] [PubMed]


| Strain or Plasmid | Genotype or Description | Reference |
|---|---|---|
| E. coli | ||
| CC118 λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE (Am) recA1 λpir phage lysogen | [22] |
| P. putida | ||
| PaW85 | isogenic to KT2440 | [23,24] |
| P-PANB | PaW85; term-lacIq-Ptac-lapA | This study |
| P-PANB-Avwf− | PaW85; term-lacIq-Ptac-lapAΔS8167-8281P | This study |
| P-Avwf− | PaW85; lapAΔS8167-8281P | This study |
| PSm | PaW85, isogenic to KT2440; chromosomal mini-Tn7-ΩSm1 (Smr) | [25] |
| PSmΔlapA | PSm; ΔPP_0168 (Smr) | [1] |
| PSmΔlapF | PSm; ΔPP_0806 (Smr) | [1] |
| PSmΔlapAF | PSm; ΔPP_0168 ΔPP_0806 (Smr) | [1] |
| PSmΔlapE | PSm; ΔPP_4519 | This study |
| PSmΔlapG | PSm; ΔPP_0164 (Smr) | This study |
| PSm-Avwf− | PSm; lapAΔS8167-8281P | Thus study |
| PSm-E-PANB | PSm; ΔPP_4519 (Smr), term-lacIq-Ptac-lapA | This study |
| PSm-E-PANB-Avwf- | PSm; ΔPP_4519 (Smr), term-lacIq-Ptac-lapAΔS8167-8281P | This study |
| PSm-G-PANB | PSm; ΔPP_0164 (Smr), term-lacIq-Ptac-lapA | This study |
| PSm-G-PANB-Avwf− | PSm; ΔPP_0164 (Smr), term-lacIq Ptac-lapAΔS8167-8281P | This study |
| F15 | PaW85, isogenic to KT2440; chromosomal mini-Tn7-ΩGm-term-lacIq-Ptac-fis-T1T2 (Gmr) | [25] |
| F15ΔlapA | F15; ΔPP_0168 (Gmr) | [1] |
| F15ΔlapF | F15; ΔPP_0806 (Gmr) | [1] |
| F15ΔlapAF | F15; ΔPP_0168, ΔPP_0806 (Gmr) | [1] |
| F15-Avwf− | F15; lapAΔS8167-8281P | This study |
| Plasmids | ||
| p9_PlapA1-8 | 951 bp long promoter-area of lapA cloned in front of lacZ (Ampr) | [20] |
| p9TTBlacZ | A promoter probe vector containing lacZ reporter gene (Ampr); RK2-based vector | [26] |
| pSW | I-SceI-expressing plasmid (Ampr) | [27] |
| pGP-FpFy-Km-lacItac-lapFSD | R6K suicide vector pGP704-L containing Ptac promoter with lacI repressor gene (Ampr, Kmr) | [21] |
| pEMG | R6K suicide vector (Kmr) | [28] |
| pEMG-lapB-lapA | Derivate of pEMG containing in the opposite direction of 5′ ends of lapA and lapB genes (Kmr) | This study |
| pEMG-lapB-Pm-xylS-lacI-Ptac-lapA | Derivate of pEMG-lapB-lapA containing Pm promoter in front of lapB 5′ end and Ptac promoter in front of lapA 5′ end, xylS and lacI genes (Kmr) | This study |
| pEMG-PtacA-natB | Derivate of pEMG-lapB-lapA containing Ptac promoter in front of lapA gene and native promoter region in front of lapB gene, lacI gene (Kmr) | This study |
| pSEVA228S | oriV(RK2), xylS, Pm→I-SceI; Kmr | [29] |
| pGNW2 | Derivative of pEMG carrying P14g→msfGFP (Kmr) | [30] |
| pGNW-lapA-Avwf− | Derivate of pGNW2 containing lapA DNA for deletion of vWFa domain in lapA gene | This study |
| pUTmini-Tn5 Km2 | Suicide vector, source of Km resistance gene (Ampr, Kmr) | [31] |
| pEMG-ΔlapE | Derivate of pEMG containing lapE (PP_4519) flanking DNAs (Kmr) | This study |
| pEMG-ΔlapG | Derivate of pEMG containing lapG (PP_0164) flanking DNAs (Kmr) | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puhm, M.; Hendrikson, J.; Kivisaar, M.; Teras, R. Pseudomonas putida Biofilm Depends on the vWFa-Domain of LapA in Peptides-Containing Growth Medium. Int. J. Mol. Sci. 2022, 23, 5898. https://doi.org/10.3390/ijms23115898
Puhm M, Hendrikson J, Kivisaar M, Teras R. Pseudomonas putida Biofilm Depends on the vWFa-Domain of LapA in Peptides-Containing Growth Medium. International Journal of Molecular Sciences. 2022; 23(11):5898. https://doi.org/10.3390/ijms23115898
Chicago/Turabian StylePuhm, Marge, Johanna Hendrikson, Maia Kivisaar, and Riho Teras. 2022. "Pseudomonas putida Biofilm Depends on the vWFa-Domain of LapA in Peptides-Containing Growth Medium" International Journal of Molecular Sciences 23, no. 11: 5898. https://doi.org/10.3390/ijms23115898
APA StylePuhm, M., Hendrikson, J., Kivisaar, M., & Teras, R. (2022). Pseudomonas putida Biofilm Depends on the vWFa-Domain of LapA in Peptides-Containing Growth Medium. International Journal of Molecular Sciences, 23(11), 5898. https://doi.org/10.3390/ijms23115898





