Genome-Wide Identification, Classification, Expression and Duplication Analysis of bZIP Family Genes in Juglans regia L.
Abstract
:1. Introduction
2. Results
2.1. Identification, Chromosomal Localization and Physicochemical Property Analysis of bZIP Gene Family Members in Walnut
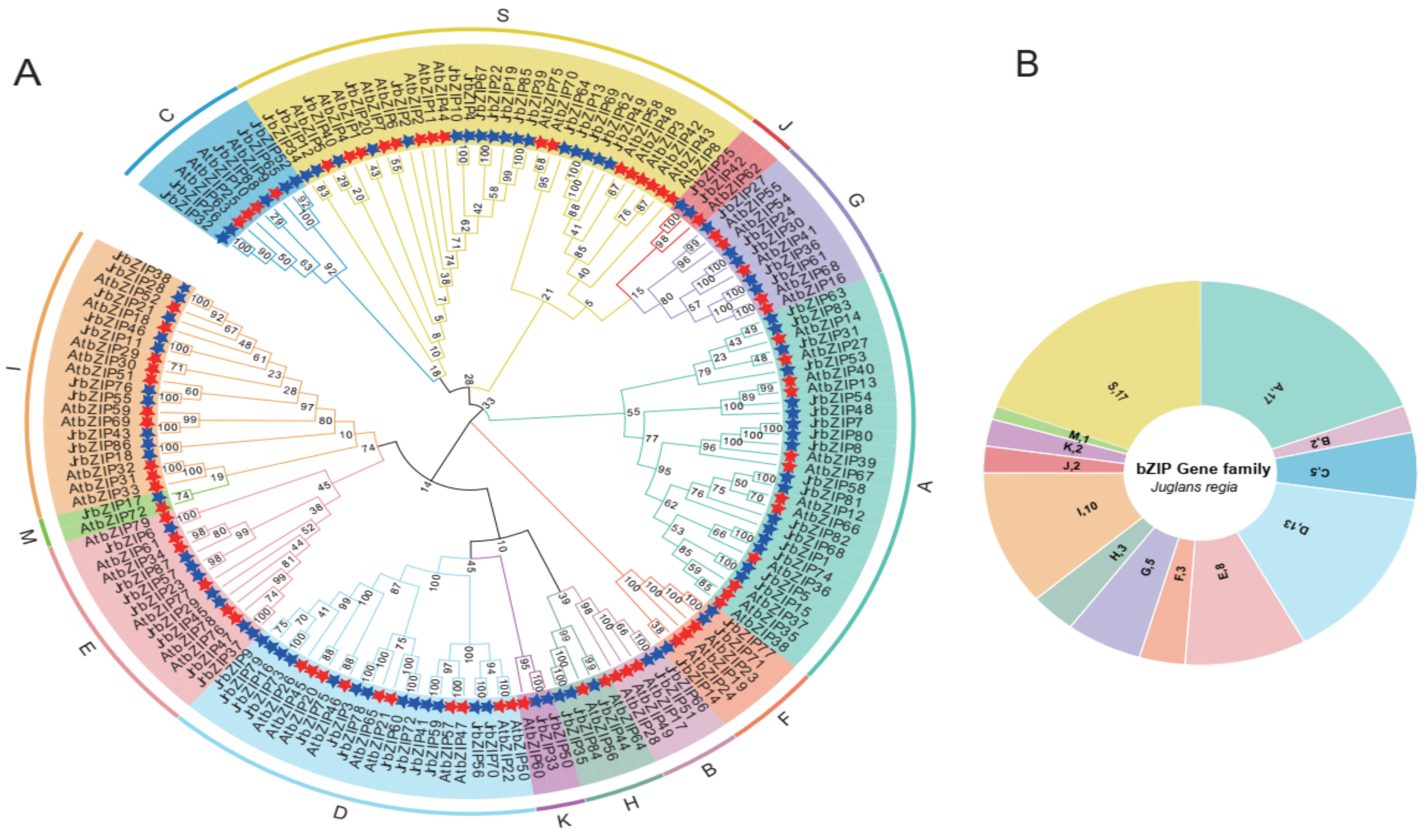
2.2. Phylogenetic Analysis of Walnut bZIP Family Members
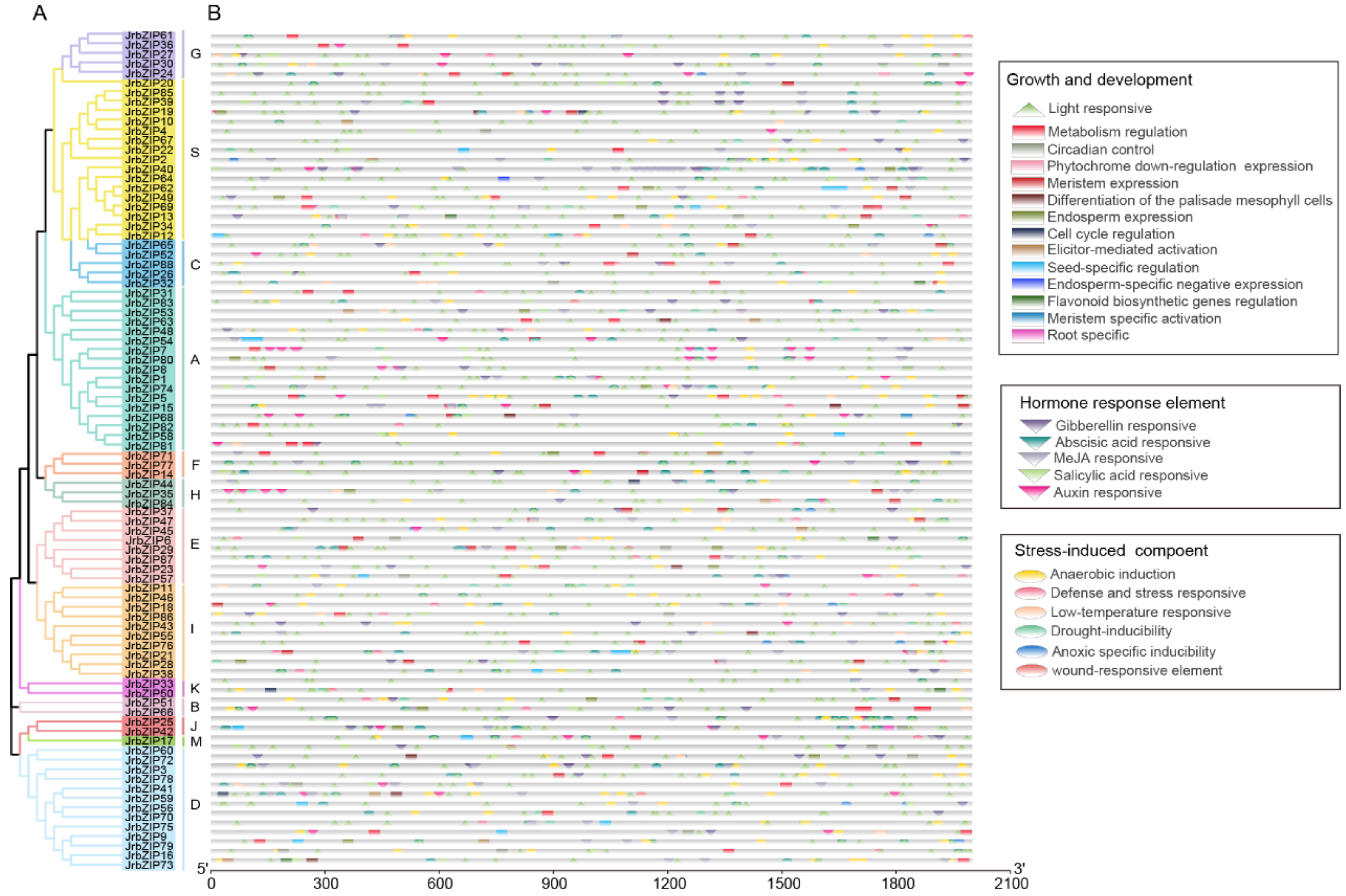
2.3. Gene Structure and Motifs Analysis of JrbZIP Members
2.4. Cis-Elements Analysis in JrBZIPs Promoter Regions
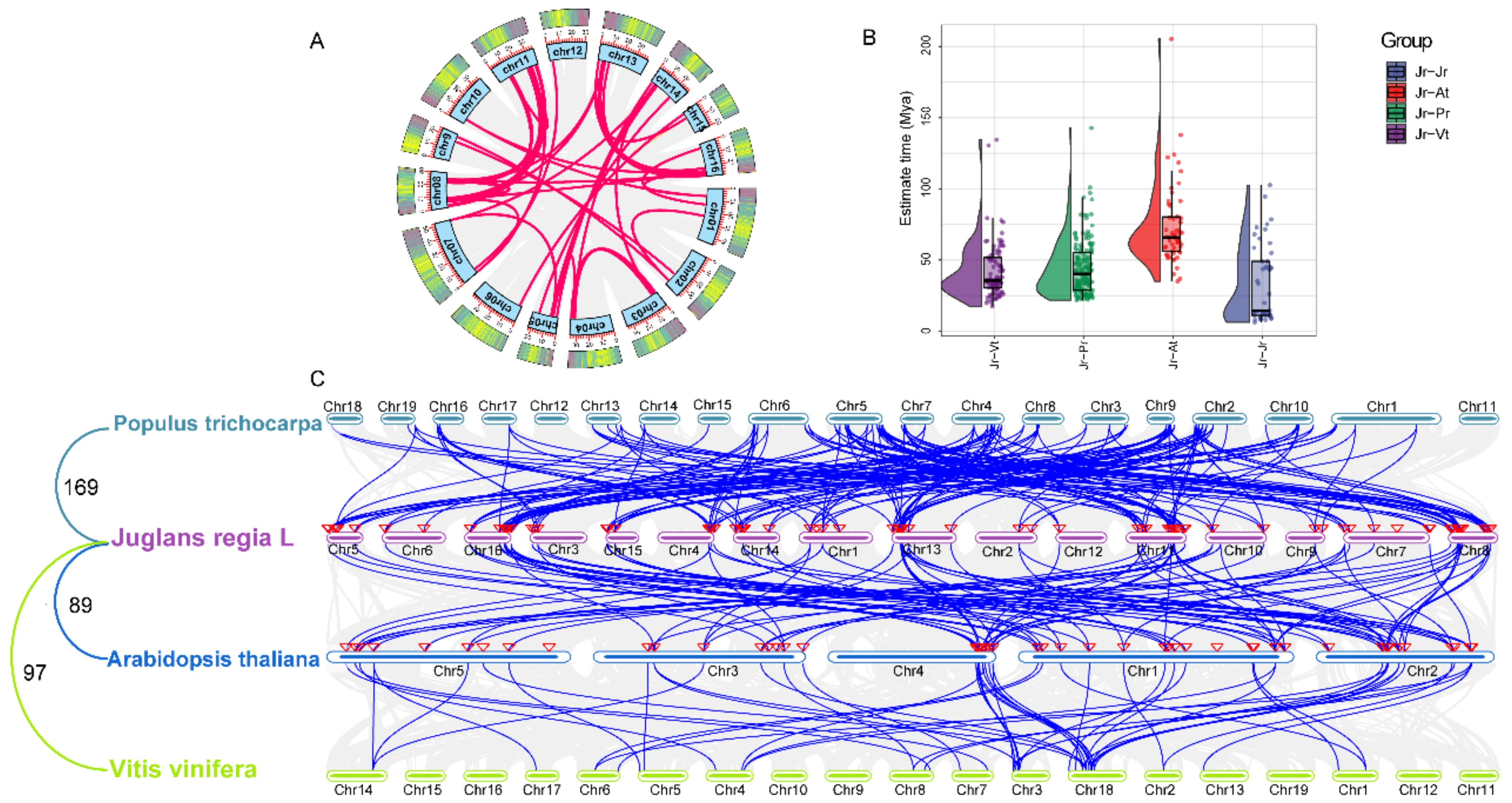
2.5. Chromosomal Distribution and Synteny Analysis of JrbZIPs
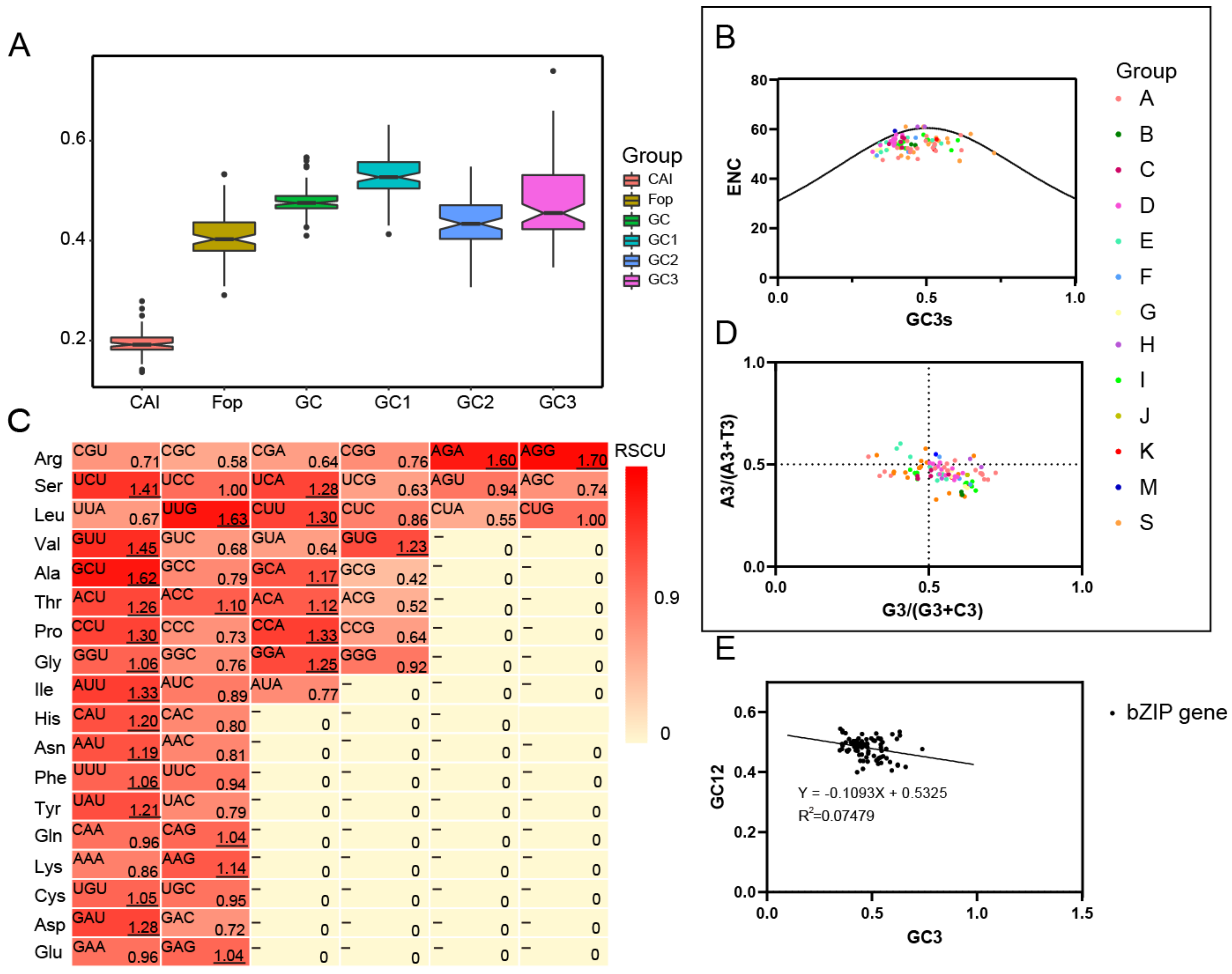
2.6. Analysis of Base Number and Codon Usage Bias of JrBZIPs
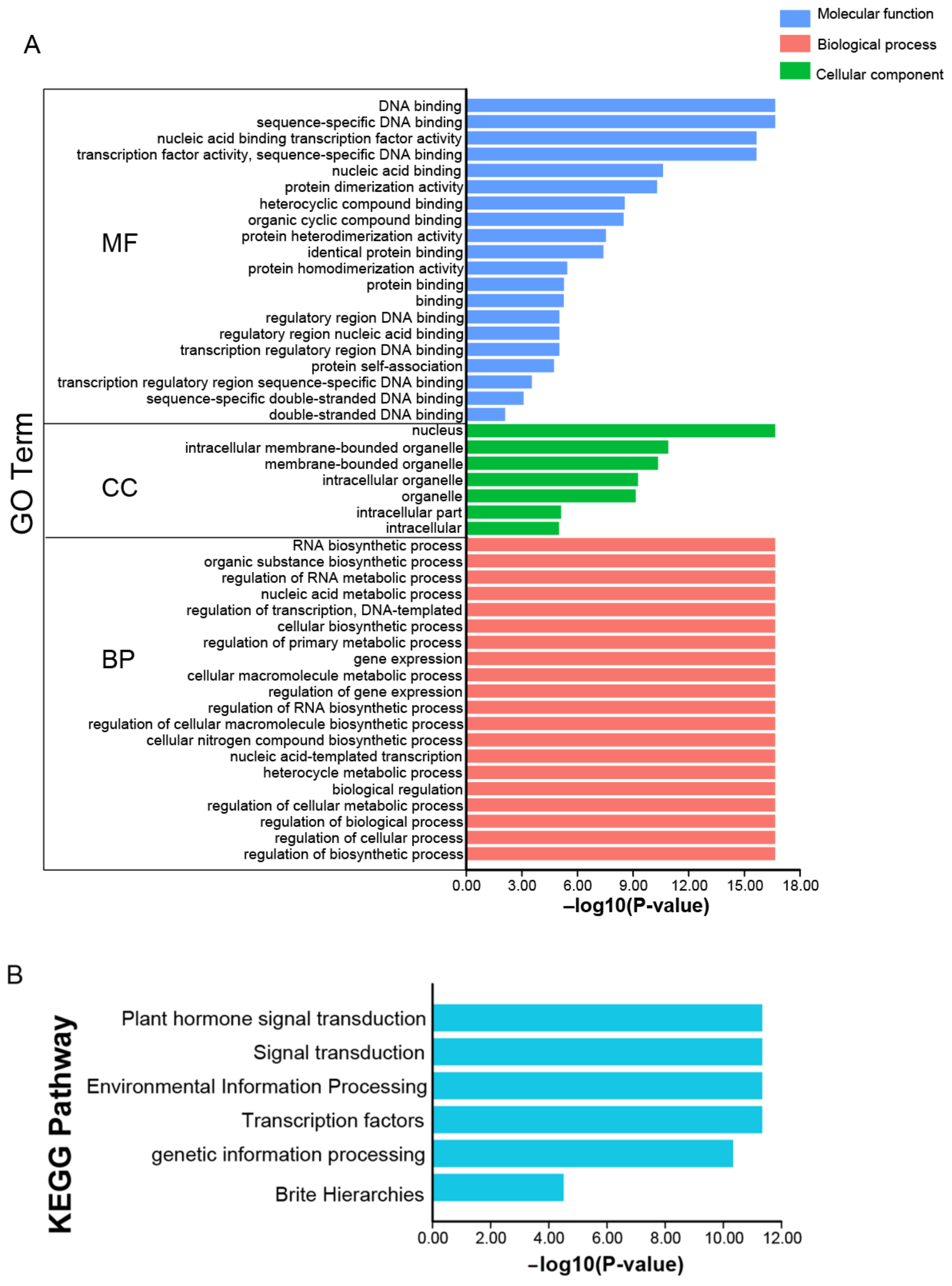
2.7. Functional Annotations of JrbZIP Genes
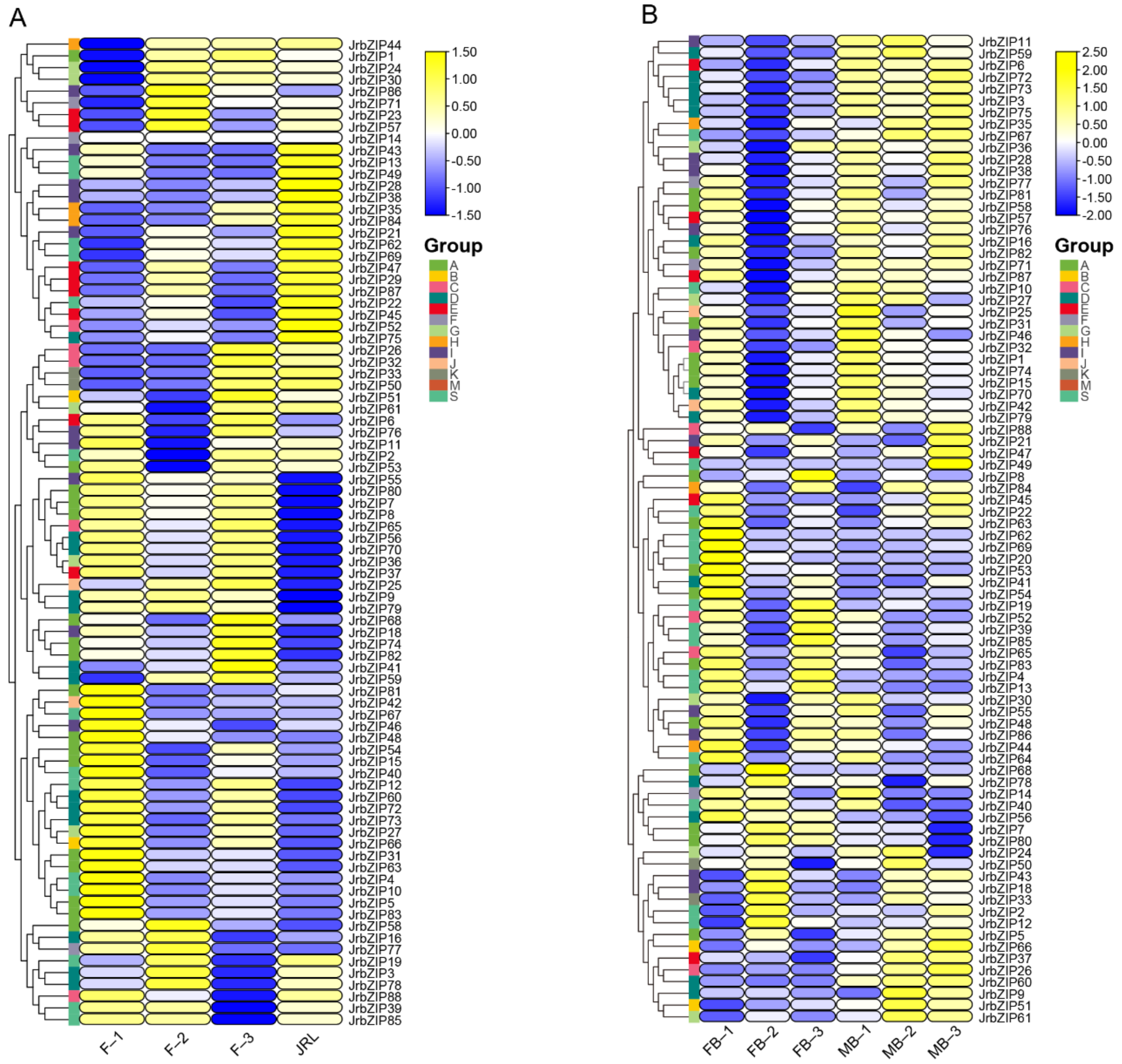
2.8. Expression Profile Analysis of Walnut bZIP Genes
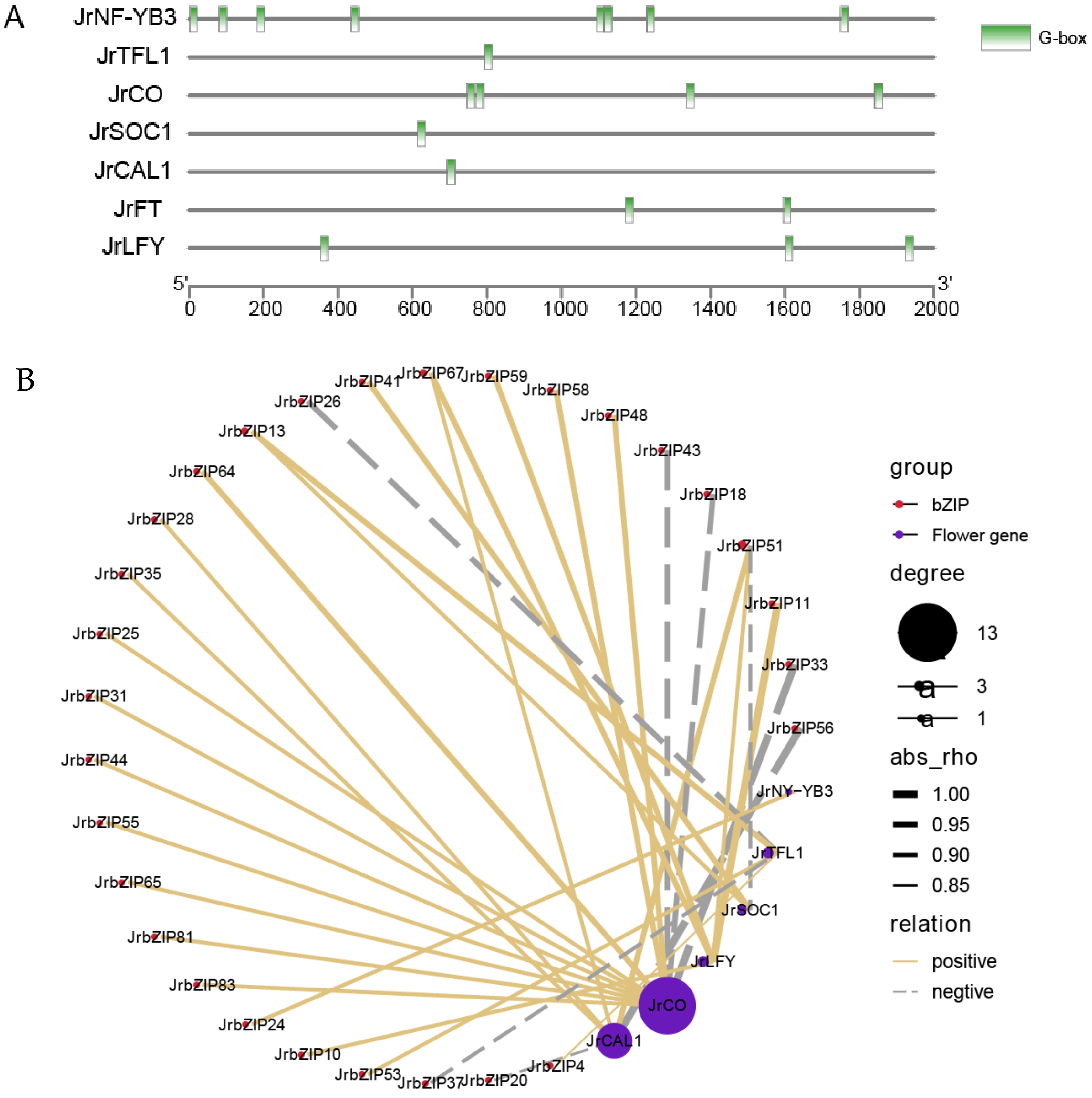
2.9. Analysis of JrbZIP Protein–Protein Interaction
2.10. Analysis of the Role of JrbZIPs in Flowering
2.11. Verification of JrbZIP Expression Pattern by qRT-PCR
3. Discussion
3.1. Identification of bZIP Genes
3.2. bZIP Genes Clustering and Evolutionary Analysis
3.3. Structural Diversity of bZIP Genes
3.4. Specific Expression Patterns of bZIP Genes
4. Materials and Methods
4.1. Plant Materials
4.2. Database Search for bZIP Members in Juglans regia L.
4.3. Phylogenetic Analysis and Classification
4.4. Sequence Analysis
4.5. Chromosomal Distribution and Synteny Analysis of JrbZIPs
4.6. Analysis of Codon Usage Pattern of JrbZIP Genes
4.7. GO and KEGG Enrichment Analyses of JrbZIP Genes
4.8. Expression Pattern Analysis of JrbZIP Genes
4.9. Protein Interaction Analysis Network of JrbZIP Protein
4.10. Quantitative Real-Time PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kong, S.; Park, S.Y.; Lee, Y.H. Systematic characterization of the bZIP transcription factor gene family in the rice blast fungus, M agnaporthe oryzae. Environ. Microbiol. 2015, 17, 1425–1443. [Google Scholar] [CrossRef] [PubMed]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Izawa, T.; Foster, R.; Chua, N.H. Plant bZIP protein DNA binding specificity. J. Mol. Biol. 1993, 230, 1131–1144. [Google Scholar] [CrossRef]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef] [Green Version]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef] [Green Version]
- Gibalová, A.; Reňák, D.; Matczuk, K.; Dupl’áková, N.; Cháb, D.; Twell, D.; Honys, D. AtbZIP34 is required for Arabidopsis pollen wall patterning and the control of several metabolic pathways in developing pollen. Plant Mol. Biol. 2009, 70, 581–601. [Google Scholar]
- Tzfira, T.; Vaidya, M.; Citovsky, V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001, 20, 3596–3607. [Google Scholar] [CrossRef] [Green Version]
- Gangappa, S.N.; Srivastava, A.K.; Maurya, J.P.; Ram, H.; Chattopadhyay, S. Z-box binding transcription factors (ZBFs): A new class of transcription factors in Arabidopsis seedling development. Mol. Plant 2013, 6, 1758–1768. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Ram, H.; Abbas, N.; Chattopadhyay, S. Molecular interactions of GBF1 with HY5 and HYH proteins during lightmediated seedling development in Arabidopsis thaliana. J. Biol. Chem. 2012, 287, 25995–26009. [Google Scholar] [CrossRef] [Green Version]
- Alonso, R.; Oñate-Sánchez, L.; Weltmeier, F.; Ehlert, A.; Diaz, I.; Dietrich, K.; Dröge-Laser, W. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 2009, 21, 1747–1761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartmann, L.; Pedrotti, L.; Weiste, C.; Fekete, A.; Schierstaedt, J.; Göttler, J.; Dröge-Laser, W. Crosstalk between two bZIP signaling pathways orchestrates salt-induced metabolic reprogramming in Arabidopsis roots. Plant Cell 2015, 27, 2244–2260. [Google Scholar] [CrossRef] [PubMed]
- Dröge-Laser, W.; Weiste, C. The C/S1 bZIP network: A regulatory hub orchestrating plant energy homeostasis. Trends Plant Sci. 2018, 23, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 2017, 254, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Luo, X.; Meng, Y.; Yang, W. Toward a molecular understanding of abscisic acid actions in floral transition. Plant Cell Physiol. 2018, 59, 215–221. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2018, 309, 1052–1056. [Google Scholar] [CrossRef]
- Liu, H.; Huang, X.; Ma, B.; Zhang, T.; Sang, N.; Zhuo, L.; Zhu, J. Components and functional diversification of florigen activation complexes in cotton. Plant Cell Physiol. 2021, 62, 1542–1555. [Google Scholar] [CrossRef]
- Jang, S.; Li, H.Y.; Kuo, M.L. Ectopic expression of Arabidopsis FD and FD PARALOGUE in rice results in dwarfism with size reduction of spikelets. Sci. Rep. 2017, 7, 44477. [Google Scholar] [CrossRef]
- Ma, H.; Liu, C.; Li, Z.; Ran, Q.; Xie, G.; Wang, B.; Fang, S.; Chu, J.; Zhang, J. ZmbZIP4 Contributes to Stress Resistance in Maize by Regulating ABA Synthesis and Root Development. Plant Physiol. 2018, 178, 753–770. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.T.; Jia, L.J.; Wang, H.Y.; Zhao, P.; Wang, W.Y.; Liu, N.; Song, S.W.; Wu, Y.; Su, L.; Zhang, J.; et al. The potato transcription factor StbZIP 61 regulates dynamic biosynthesis of salicylic acid in defense against Phytophthora infestans infection. Plant J. 2018, 95, 1055–1068. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Jia, R.; Dou, W.; Qi, J.; Qin, X.; Fu, Y.; He, Y.; Chen, S. CsBZIP40, a BZIP transcription factor in sweet orange, plays a positive regulatory role in citrus bacterial canker response and tolerance. PLoS ONE 2019, 14, e0223498. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Labuckas, D.; Lamarque, A.; Maestri, D. Walnut (Juglans regia L.): Genetic resources, chemistry, by-products. J. Sci. Food Agric. 2010, 90, 1959–1967. [Google Scholar] [CrossRef] [PubMed]
- Jahanban-Esfahlan, A.; Ostadrahimi, A.; Tabibiazar, M.; Amarowicz, R. A comparative review on the extraction, antioxidant content and antioxidant potential of different parts of walnut (Juglans regia L.) fruit and tree. Molecules 2019, 24, 2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, N.; An, Y.; Li, J.; Wang, L. Cloning and characterization of a homologue of the FLORICAULA/LEAFY gene in Ficus carica L., FcLFY, and its role in flower bud differentiation. Sci. Hortic. 2020, 261, 109014. [Google Scholar] [CrossRef]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Fu, F.; Zhang, H.; Song, F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genom. 2015, 16, 771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baloglu, M.C.; Eldem, V.; Hajyzadeh, M.; Unver, T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 2014, 9, e96014. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Gao, W.; Li, H.; Wang, Y.; Li, D.; Xue, C.; Liu, Z.; Liu, M.; Zhao, J. Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genom. 2020, 21, 483. [Google Scholar] [CrossRef]
- Liao, Y.; Zou, H.F.; Wei, W.; Hao, Y.J.; Tian, A.G.; Huang, J.; Liu, Y.F.; Zhang, J.S.; Chen, S.Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, D.; Jia, L.; Huang, X.; Ma, G.; Wang, S.; Zhu, M.; Zhang, A.; Guan, M.; Lu, K.; et al. Genome-Wide Identification and Structural Analysis of bZIP Transcription Factor Genes in Brassica napus. Genes 2017, 8, 288. [Google Scholar] [CrossRef] [Green Version]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021, 21, 122. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Sun, J.; Chen, Y.; Zhu, P.; Zhang, L.; Wu, S.; Ma, D.; Cao, Q.; Li, Z.; Xu, T. Genome-wide identification, structural and gene expression analysis of the bZIP transcription factor family in sweet potato wild relative Ipomoea trifida. BMC Genet. 2019, 20, 41. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Quan, S.; Zhang, Z.; Kang, C.; Liu, J.; Niu, J. Genome-wide Identification, Characterization and Expression profile of TALE gene family in (Juglans regia L.). Sci. Hortic. 2022, 297, 110945. [Google Scholar] [CrossRef]
- Chuang, C.F.; Running, M.P.; Williams, R.W.; Meyerowitz, E.M. The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 1999, 13, 334–344. [Google Scholar] [CrossRef] [Green Version]
- Iven, T.; Strathmann, A.; Böttner, S.; Zwafink, T.; Heinekamp, T.; Guivarc, H.A.; Roitsch, T.; Droge-Laser, W. Homo-and heterodimers of tobacco bZIP proteins counteract as positive or negative regulators of transcription during pollen development. Plant J. 2010, 63, 155–166. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.Y.; Meng, D.; Li, M.; Cheng, L. Genome-wide identification and expression analysis of the bZIP gene family in apple (Malus domestica). Tree Genet. Genomes 2016, 12, 1–17. [Google Scholar] [CrossRef]
- Yong, X.; Zheng, T.; Zhuo, X.; Ahmad, S.; Li, L.; Li, P.; Zhang, Q. Genome-wide identification, characterisation, and evolution of ABF/AREB subfamily in nine Rosaceae species and expression analysis in mei (Prunus mume). PeerJ 2021, 9, e10785. [Google Scholar] [CrossRef]
- Shu, K.; Chen, Q.; Wu, Y.; Liu, R.; Zhang, H.; Wang, S.; Tang, S.; Yang, W.; Xie, Q. ABSCISIC ACID-INSENSITIVE 4 negatively regulates flowering through directly promoting Arabidopsis FLOWERING LOCUS C transcription. J. Exp. Bot. 2016, 67, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Kaneko-Suzuki, M.; Kurihara-Ishikawa, R.; Okushita-Terakawa, C.; Kojima, C.; Nagano-Fujiwara, M.; Ohki, I.; Tsuji, H.; Shimamoto, K.; Taoka, K.I. TFL1-like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol. 2018, 59, 458–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parmentier-Line, C.M.; Coleman, G.D. Constitutive expression of the Poplar FD-like basic leucine zipper transcription factor alters growth and bud development. Plant Biotechnol. J. 2016, 14, 260–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.; Xia, J.; Jiang, Y.; Bao, Y.; Chen, H.; Wang, D.; Zhang, D.; Yu, J.; Cang, J. Genome-Wide Identification and Analysis of bZIP Gene Family and Resistance of TaABI5 (TabZIP96) under Freezing Stress in Wheat (Triticum aestivum). Int. J. Mol. Sci. 2022, 23, 2351. [Google Scholar] [CrossRef] [PubMed]
- Soding, J. Protein homology detection by HMM-HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press Inc.: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 3, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Feng, R.; Ma, R.; Shen, Z.; Cai, Z.; Song, Z.; Peng, B.; Yu, M. Genome-wide analysis of basic helix-loop-helix superfamily members in peach. PLoS ONE 2018, 13, e0195974. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, M.A.; Haubold, B.; Mitchell-Olds, T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 2000, 17, 1483–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharp, P.M.; Li, W.H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986, 24, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Quan, S.; Niu, J.; Zhou, L.; Xu, H.; Ma, L.; Qin, Y. Stages identifying and transcriptome profiling of the floral transition in Juglans regia. Sci. Rep. 2019, 9, 7092. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. Correction to ‘The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets’. Nucleic Acids Res. 2021, 49, 10800. [Google Scholar] [CrossRef]
- Zhou, L.; Niu, J.; Quan, S. Identification of appropriate reference genes for RT-qPCR analysis in Juglans regia L. PLoS ONE 2018, 13, e0209424. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Quan, S.; Niu, J.; Guo, C.; Kang, C.; Liu, J.; Yuan, X. Genome-Wide Identification, Classification, Expression and Duplication Analysis of bZIP Family Genes in Juglans regia L. Int. J. Mol. Sci. 2022, 23, 5961. https://doi.org/10.3390/ijms23115961
Zhang Z, Quan S, Niu J, Guo C, Kang C, Liu J, Yuan X. Genome-Wide Identification, Classification, Expression and Duplication Analysis of bZIP Family Genes in Juglans regia L. International Journal of Molecular Sciences. 2022; 23(11):5961. https://doi.org/10.3390/ijms23115961
Chicago/Turabian StyleZhang, Zhongrong, Shaowen Quan, Jianxin Niu, Caihua Guo, Chao Kang, Jinming Liu, and Xing Yuan. 2022. "Genome-Wide Identification, Classification, Expression and Duplication Analysis of bZIP Family Genes in Juglans regia L." International Journal of Molecular Sciences 23, no. 11: 5961. https://doi.org/10.3390/ijms23115961
APA StyleZhang, Z., Quan, S., Niu, J., Guo, C., Kang, C., Liu, J., & Yuan, X. (2022). Genome-Wide Identification, Classification, Expression and Duplication Analysis of bZIP Family Genes in Juglans regia L. International Journal of Molecular Sciences, 23(11), 5961. https://doi.org/10.3390/ijms23115961





