A Genomic Information Management System for Maintaining Healthy Genomic States and Application of Genomic Big Data in Clinical Research
Abstract
:1. Introduction
2. Part 1: Genomic Information Management for Individuals
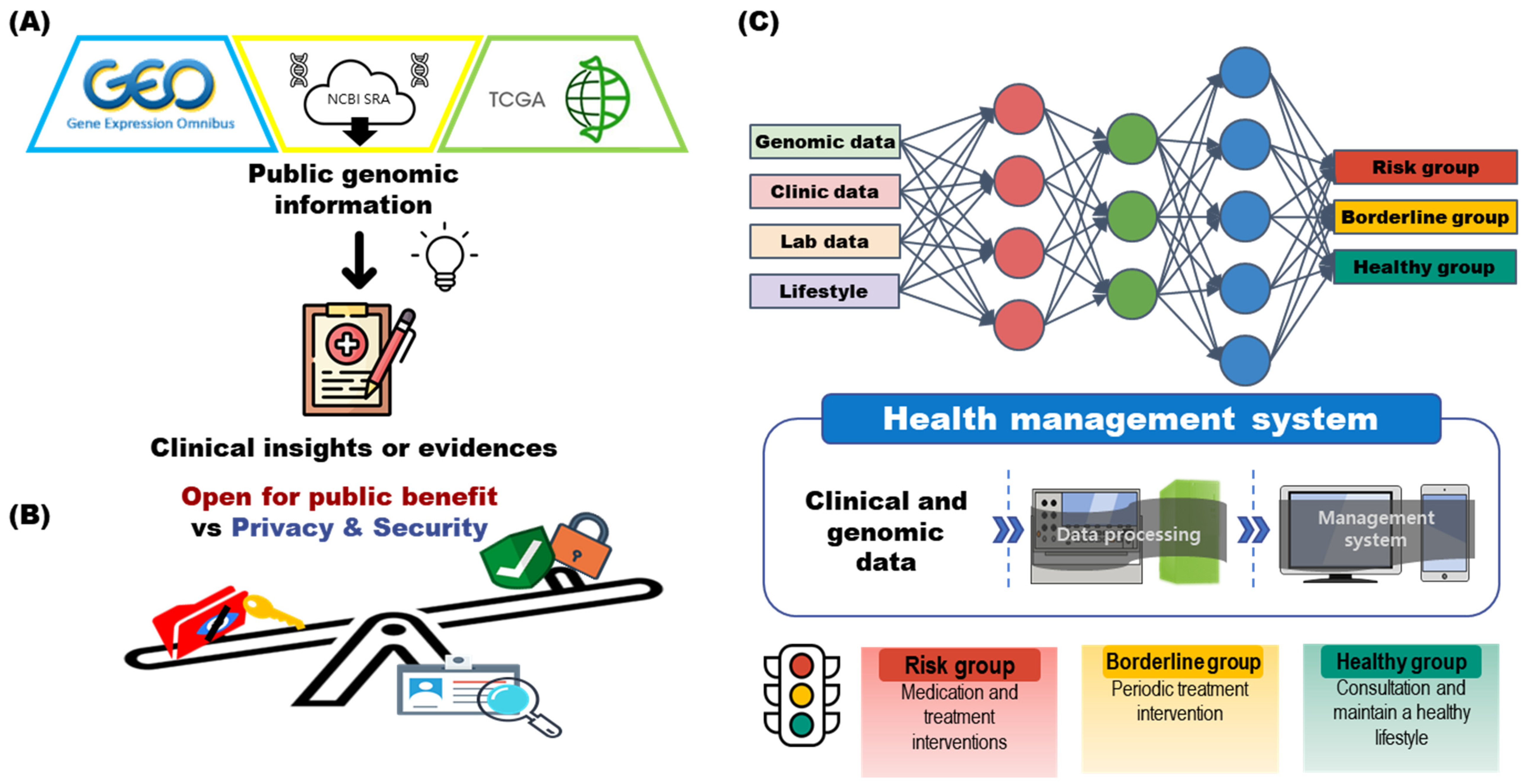
2.1. Is It Currently Possible to Develop a Health Management System Based on Genomic Information?
2.2. Analyzing Health Status through Genomic Information
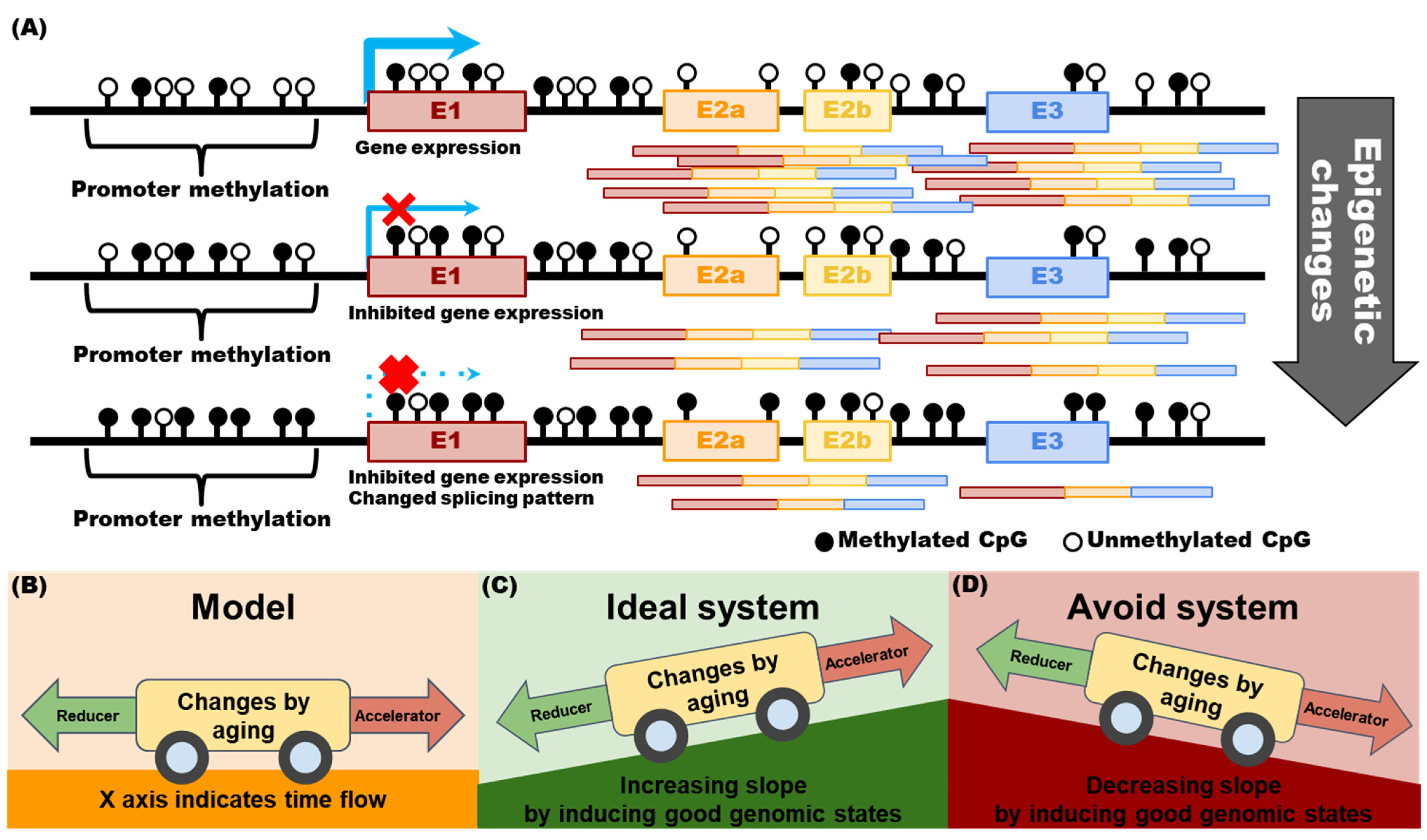
2.3. Why Should Genomic Information Be Obtained over a Lifetime?
2.4. Part 1 Subconclusions
3. Part 2: Big Data-Based Genomic Information Management System for Clinical Research
3.1. Clinical Decisions and Research Using Genomic Big Data
3.2. Storing and Indexing Genomic Information
3.3. Regulation of Genomic Information: In Terms of Personal Information and Privacy Protection
3.4. Part 2 Subconclusions
4. Future Perspective
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kulynych, J.; Greely, H.T. Clinical genomics, big data, and electronic medical records: Reconciling patient rights with research when privacy and science collide. J. Law Biosci. 2017, 4, 94–132. [Google Scholar] [CrossRef] [Green Version]
- Auffray, C.; Balling, R.; Barroso, I.; Bencze, L.; Benson, M.; Bergeron, J.; Bernal-Delgado, E.; Blomberg, N.; Bock, C.; Conesa, A. Making sense of big data in health research: Towards an EU action plan. Genome Med. 2016, 8, 71. [Google Scholar] [CrossRef]
- Pramanik, P.K.D.; Pal, S.; Mukhopadhyay, M. Healthcare big data: A comprehensive overview. Res. Anthol. Big Data Anal. Archit. Appl. 2022, 119–147. [Google Scholar]
- Phillips, K.A.; Trosman, J.R.; Kelley, R.K.; Pletcher, M.J.; Douglas, M.P.; Weldon, C.B. Genomic sequencing: Assessing the health care system, policy, and big-data implications. Health Aff. 2014, 33, 1246–1253. [Google Scholar] [CrossRef] [Green Version]
- He, K.Y.; Ge, D.; He, M.M. Big data analytics for genomic medicine. Int. J. Mol. Sci. 2017, 18, 412. [Google Scholar] [CrossRef] [Green Version]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Koh, E.J.; Hwang, S.Y. Multi-omics approaches for understanding environmental exposure and human health. Mol. Cell. Toxicol. 2019, 15, 1–7. [Google Scholar] [CrossRef]
- Castaneda, C.; Nalley, K.; Mannion, C.; Bhattacharyya, P.; Blake, P.; Pecora, A.; Goy, A.; Suh, K.S. Clinical decision support systems for improving diagnostic accuracy and achieving precision medicine. J. Clin. Bioinform. 2015, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Bhuiyan, M.; Rahman, A.; Ullah, M.; Das, A.K. iHealthcare: Predictive model analysis concerning big data applications for interactive healthcare systems. Appl. Sci. 2019, 9, 3365. [Google Scholar] [CrossRef] [Green Version]
- Sharafoddini, A.; Dubin, J.A.; Lee, J. Patient similarity in prediction models based on health data: A scoping review. JMIR Med. Inform. 2017, 5, e6730. [Google Scholar] [CrossRef] [Green Version]
- Shukla, D.; Patel, S.B.; Sen, A.K. A literature review in health informatics using data mining techniques. Int. J. Softw. Hardw. Res. Eng. 2014, 2, 123–129. [Google Scholar]
- Shah, P.; Kendall, F.; Khozin, S.; Goosen, R.; Hu, J.; Laramie, J.; Ringel, M.; Schork, N. Artificial intelligence and machine learning in clinical development: A translational perspective. NPJ Digit. Med. 2019, 2, 69. [Google Scholar] [CrossRef] [Green Version]
- Dwyer, D.B.; Falkai, P.; Koutsouleris, N. Machine learning approaches for clinical psychology and psychiatry. Annu. Rev. Clin. Psychol. 2018, 14, 91–118. [Google Scholar] [CrossRef]
- Kim, M.; Lee, D.-W.; Kim, K.; Kim, J.-H. Hierarchical structured data logging system for effective lifelog management in ubiquitous environment. Multimed. Tools Appl. 2015, 74, 3561–3577. [Google Scholar] [CrossRef]
- Barbarino, J.M.; Whirl-Carrillo, M.; Altman, R.B.; Klein, T.E. PharmGKB: A worldwide resource for pharmacogenomic information. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018, 10, e1417. [Google Scholar] [CrossRef] [Green Version]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM. org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef] [Green Version]
- Ashbury, F.D.; Thompson, K.; Williams, C.; Williams, K. Challenges adopting next-generation sequencing in community oncology practice. Curr. Opin. Oncol. 2021, 33, 507–512. [Google Scholar] [CrossRef]
- Vestergaard, L.K.; Oliveira, D.N.; Høgdall, C.K.; Høgdall, E.V. Next generation sequencing technology in the clinic and its challenges. Cancers 2021, 13, 1751. [Google Scholar] [CrossRef]
- Mardis, E.R. The impact of next-generation sequencing on cancer genomics: From discovery to clinic. Cold Spring Harb. Perspect. Med. 2019, 9, a036269. [Google Scholar] [CrossRef]
- Döring, M.; Büch, J.; Friedrich, G.; Pironti, A.; Kalaghatgi, P.; Knops, E.; Heger, E.; Obermeier, M.; Däumer, M.; Thielen, A. geno2pheno [ngs-freq]: A genotypic interpretation system for identifying viral drug resistance using next-generation sequencing data. Nucleic Acids Res. 2018, 46, W271–W277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatakeyama, M.; Opitz, L.; Russo, G.; Qi, W.; Schlapbach, R.; Rehrauer, H. SUSHI: An exquisite recipe for fully documented, reproducible and reusable NGS data analysis. BMC Bioinform. 2016, 17, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, Y.; Jeon, S.; Blazyte, A.; Kim, Y.J.; Lee, J.J.; Bhak, Y.; Cho, Y.S.; Park, Y.; Noh, E.-K.; Manica, A.; et al. Welfare Genome Project: A Participatory Korean Personal Genome Project With Free Health Check-Up and Genetic Report Followed by Counseling. Front. Genet. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Inouye, M.; Abraham, G.; Nelson, C.P.; Wood, A.M.; Sweeting, M.J.; Dudbridge, F.; Lai, F.Y.; Kaptoge, S.; Brozynska, M.; Wang, T. Genomic risk prediction of coronary artery disease in 480,000 adults: Implications for primary prevention. J. Am. Coll. Cardiol. 2018, 72, 1883–1893. [Google Scholar] [CrossRef]
- Luttropp, K.; Nordfors, L.; Ekström, T.J.; Lind, L. Physical activity is associated with decreased global DNA methylation in Swedish older individuals. Scand. J. Clin. Lab. Investig. 2013, 73, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Madrigano, J.; Baccarelli, A.A.; Mittleman, M.A.; Sparrow, D.; Vokonas, P.S.; Tarantini, L.; Schwartz, J. Aging and epigenetics: Longitudinal changes in gene-specific DNA methylation. Epigenetics 2012, 7, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.F.; Cardarelli, R.; Carroll, J.; Zhang, S.; Fulda, K.G.; Gonzalez, K.; Vishwanatha, J.K.; Morabia, A.; Santella, R.M. Physical activity and global genomic DNA methylation in a cancer-free population. Epigenetics 2011, 6, 293–299. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Han, B.-G.; Group, K. Cohort profile: The Korean genome and epidemiology study (KoGES) consortium. Int. J. Epidemiol. 2017, 46, e20. [Google Scholar] [CrossRef]
- Pimpão, R.C.; Dew, T.; Figueira, M.E.; McDougall, G.J.; Stewart, D.; Ferreira, R.B.; Santos, C.N.; Williamson, G. Urinary metabolite profiling identifies novel colonic metabolites and conjugates of phenolics in healthy volunteers. Mol. Nutr. Food Res. 2014, 58, 1414–1425. [Google Scholar] [CrossRef]
- Zhou, Y.; Wylie, K.M.; El Feghaly, R.E.; Mihindukulasuriya, K.A.; Elward, A.; Haslam, D.B.; Storch, G.A.; Weinstock, G.M. Metagenomic approach for identification of the pathogens associated with diarrhea in stool specimens. J. Clin. Microbiol. 2016, 54, 368–375. [Google Scholar] [CrossRef] [Green Version]
- Kadayifci, F.Z.; Zheng, S.; Pan, Y.-X. Molecular mechanisms underlying the link between diet and DNA methylation. Int. J. Mol. Sci. 2018, 19, 4055. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, A.M.; Ali, M.M. Methyl donor micronutrients that modify DNA methylation and cancer outcome. Nutrients 2019, 11, 608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voisin, S.; Eynon, N.; Yan, X.; Bishop, D. Exercise training and DNA methylation in humans. Acta Physiol. 2015, 213, 39–59. [Google Scholar] [CrossRef] [PubMed]
- Hibler, E.; Huang, L.; Andrade, J.; Spring, B. Impact of a diet and activity health promotion intervention on regional patterns of DNA methylation. Clin. Epigenetics 2019, 11, 133. [Google Scholar] [CrossRef] [Green Version]
- Learmonth, Y.; Paul, L.; Miller, L.; Mattison, P.; McFadyen, A. The effects of a 12-week leisure centre-based, group exercise intervention for people moderately affected with multiple sclerosis: A randomized controlled pilot study. Clin. Rehabil. 2012, 26, 579–593. [Google Scholar] [CrossRef] [Green Version]
- Morabia, A.; Zhang, F.F.; Kappil, M.A.; Flory, J.; Mirer, F.E.; Santella, R.M.; Wolff, M.; Markowitz, S.B. Biologic and epigenetic impact of commuting to work by car or using public transportation: A case-control study. Prev. Med. 2012, 54, 229–233. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.F.; Santella, R.M.; Wolff, M.; Kappil, M.A.; Markowitz, S.B.; Morabia, A. White blood cell global methylation and IL-6 promoter methylation in association with diet and lifestyle risk factors in a cancer-free population. Epigenetics 2012, 7, 606–614. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, K.; Takeoka, M.; Mori, M.; Hashimoto, S.; Sakurai, A.; Nose, H.; Higuchi, K.; Itano, N.; Shiohara, M.; Oh, T. Exercise effects on methylation of ASC gene. Int. J. Sports Med. 2010, 31, 671–675. [Google Scholar] [CrossRef] [Green Version]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.-B.; Gao, Y. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [Green Version]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, 3156. [Google Scholar] [CrossRef] [Green Version]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 2019, 11, 303. [Google Scholar] [CrossRef]
- Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y. An epigenetic biomarker of aging for lifespan and healthspan. Aging 2018, 10, 573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.-E.; Geem, Z.W.; Na, K.-S. Prediction of suicide among 372,813 individuals under medical check-up. J. Psychiatr. Res. 2020, 131, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Artac, M.; Dalton, A.R.; Majeed, A.; Car, J.; Millett, C. Effectiveness of a national cardiovascular disease risk assessment program (NHS Health Check): Results after one year. Prev. Med. 2013, 57, 129–134. [Google Scholar] [CrossRef]
- Gim, J.-A. Integrative approaches of DNA methylation patterns according to age, sex, and longitudinal changes. 2022; Preprint version. [Google Scholar]
- Ko, Y.K.; Kim, H.; Lee, Y.; Lee, Y.-S.; Gim, J.-A. DNA Methylation Patterns According to Fatty Liver Index and Longitudinal Changes from the Korean Genome and Epidemiology Study (KoGES). Curr. Issues Mol. Biol. 2022, 44, 1149–1168. [Google Scholar] [CrossRef]
- Marioni, R.E.; Suderman, M.; Chen, B.H.; Horvath, S.; Bandinelli, S.; Morris, T.; Beck, S.; Ferrucci, L.; Pedersen, N.L.; Relton, C.L. Tracking the epigenetic clock across the human life course: A meta-analysis of longitudinal cohort data. J. Gerontol. Ser. A 2019, 74, 57–61. [Google Scholar] [CrossRef] [Green Version]
- SI, J.; MU, D.; Sun, L.; Qiao, Z.; Yang, K. Analysis and forecast of clinical decision support system for diabetes mellitus based on big data technique. Int. J. Biomed. Eng. 2017, 6, 216–220. [Google Scholar]
- Casal-Guisande, M.; Comesaña-Campos, A.; Dutra, I.; Cerqueiro-Pequeño, J.; Bouza-Rodríguez, J.-B. Design and Development of an Intelligent Clinical Decision Support System Applied to the Evaluation of Breast Cancer Risk. J. Pers. Med. 2022, 12, 169. [Google Scholar] [CrossRef]
- Roncato, R.; Dal Cin, L.; Mezzalira, S.; Comello, F.; De Mattia, E.; Bignucolo, A.; Giollo, L.; D’Errico, S.; Gulotta, A.; Emili, L. FARMAPRICE: A pharmacogenetic clinical decision support system for precise and cost-effective therapy. Genes 2019, 10, 276. [Google Scholar] [CrossRef] [Green Version]
- Roosan, D.; Hwang, A.; Law, A.V.; Chok, J.; Roosan, M.R. The inclusion of health data standards in the implementation of pharmacogenomics systems: A scoping review. Pharmacogenomics 2020, 21, 1191–1202. [Google Scholar] [CrossRef]
- Eckelt, F.; Remmler, J.; Kister, T.; Wernsdorfer, M.; Richter, H.; Federbusch, M.; Adler, M.; Kehrer, A.; Voigt, M.; Cundius, C. Improved patient safety through a clinical decision support system in laboratory medicine. Der Internist 2020, 61, 452–459. [Google Scholar] [CrossRef] [Green Version]
- Rubinstein, M.; Hirsch, R.; Bandyopadhyay, K.; Madison, B.; Taylor, T.; Ranne, A.; Linville, M.; Donaldson, K.; Lacbawan, F.; Cornish, N. Effectiveness of practices to support appropriate laboratory test utilization: A laboratory medicine best practices systematic review and meta-analysis. Am. J. Clin. Pathol. 2018, 149, 197–221. [Google Scholar] [CrossRef] [PubMed]
- Souza-Pereira, L.; Pombo, N.; Ouhbi, S.; Felizardo, V.; Garcia, N. Clinical decision support systems for chronic diseases: A systematic literature review. Comput. Methods Programs Biomed. 2020, 195, 105565. [Google Scholar] [CrossRef] [PubMed]
- Altay, E.V.; Alatas, B. A novel clinical decision support system for liver fibrosis using evolutionary multi-objective method based numerical association analysis. Med. Hypotheses 2020, 144, 110028. [Google Scholar] [CrossRef]
- Hamedan, F.; Orooji, A.; Sanadgol, H.; Sheikhtaheri, A. Clinical decision support system to predict chronic kidney disease: A fuzzy expert system approach. Int. J. Med. Inform. 2020, 138, 104134. [Google Scholar] [CrossRef] [PubMed]
- Helmons, P.J.; Suijkerbuijk, B.O.; Nannan Panday, P.V.; Kosterink, J.G. Drug-drug interaction checking assisted by clinical decision support: A return on investment analysis. J. Am. Med. Inform. Assoc. 2015, 22, 764–772. [Google Scholar] [CrossRef] [Green Version]
- Nair, P.C.; Gupta, D.; Indira Devi, B. Automatic Symptom Extraction from Unstructured Web Data for Designing Healthcare Systems. In Emerging Research in Computing, Information, Communication and Applications; Springer: Singapore, 2022; pp. 599–608. [Google Scholar]
- Shah, A.M.; Muhammad, W.; Lee, K.; Naqvi, R.A. Examining Different Factors in Web-Based Patients’ Decision-Making Process: Systematic Review on Digital Platforms for Clinical Decision Support System. Int. J. Environ. Res. Public Health 2021, 18, 11226. [Google Scholar] [CrossRef]
- Sung, H.-K.; Jung, B.; Kim, K.H.; Sung, S.-H.; Sung, A.-D.-M.; Park, J.-K. Trends and future direction of the clinical decision support system in traditional Korean Medicine. J. Pharmacopunct. 2019, 22, 260. [Google Scholar] [CrossRef]
- Dash, S.; Shakyawar, S.K.; Sharma, M.; Kaushik, S. Big data in healthcare: Management, analysis and future prospects. J. Big Data 2019, 6, 54. [Google Scholar] [CrossRef] [Green Version]
- Tai, A.M.; Albuquerque, A.; Carmona, N.E.; Subramanieapillai, M.; Cha, D.S.; Sheko, M.; Lee, Y.; Mansur, R.; McIntyre, R.S. Machine learning and big data: Implications for disease modeling and therapeutic discovery in psychiatry. Artif. Intell. Med. 2019, 99, 101704. [Google Scholar] [CrossRef]
- Mirza, B.; Wang, W.; Wang, J.; Choi, H.; Chung, N.C.; Ping, P. Machine learning and integrative analysis of biomedical big data. Genes 2019, 10, 87. [Google Scholar] [CrossRef] [Green Version]
- Mayo, C.S.; Matuszak, M.M.; Schipper, M.J.; Jolly, S.; Hayman, J.A.; Ten Haken, R.K. Big data in designing clinical trials: Opportunities and challenges. Front. Oncol. 2017, 7, 187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aiello, M.; Cavaliere, C.; D’Albore, A.; Salvatore, M. The challenges of diagnostic imaging in the era of big data. J. Clin. Med. 2019, 8, 316. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Gim, J.-A.; Oh, C.; Ha, S.; Lee, H.; Choi, H.; Im, H.-J. Association of metabolic and genetic heterogeneity in head and neck squamous cell carcinoma with prognostic implications: Integration of FDG PET and genomic analysis. EJNMMI Res. 2019, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, M.F.; Esmaeli, B. Molecular characteristics of uveal melanoma: Insights from the cancer genome atlas (TCGA) project. Cancers 2019, 11, 1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, R.N.; Moon, H.-G.; Han, W.; Noh, D.-Y. Perspective insight into future potential fusion gene transcript biomarker candidates in breast cancer. Int. J. Mol. Sci. 2018, 19, 502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef] [Green Version]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [Green Version]
- Brazma, A.; Parkinson, H.; Sarkans, U.; Shojatalab, M.; Vilo, J.; Abeygunawardena, N.; Holloway, E.; Kapushesky, M.; Kemmeren, P.; Lara, G.G.; et al. ArrayExpress—A public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003, 31, 68–71. [Google Scholar] [CrossRef] [Green Version]
- Leinonen, R.; Sugawara, H.; Shumway, M.; International Nucleotide Sequence Database Collaboration. The Sequence Read Archive. Nucleic Acids Res. 2010, 39, D19–D21. [Google Scholar] [CrossRef] [Green Version]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Seol, Y.-J.; Lee, T.-H.; Park, D.-S.; Kim, C.-K. NABIC: A New Access Portal to Search, Visualize, and Share Agricultural Genomics Data. Evol. Bioinform. 2016, 12, EBO.S34493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [Green Version]
- Baiden-Amissah, R.E.M.; Annibali, D.; Tuyaerts, S.; Amant, F. Endometrial Cancer Molecular Characterization: The Key to Identifying High-Risk Patients and Defining Guidelines for Clinical Decision-Making? Cancers 2021, 13, 3988. [Google Scholar] [CrossRef] [PubMed]
- Clayton, E.W.; Evans, B.J.; Hazel, J.W.; Rothstein, M.A. The law of genetic privacy: Applications, implications, and limitations. J. Law Biosci. 2019, 6, 1–36. [Google Scholar] [CrossRef] [Green Version]
- Wei, X. Hospital Information System Management and Security Maintenance; Springer: Berlin/Heidelberg, Germany, 2011; pp. 418–421. [Google Scholar]
- Thapa, C.; Camtepe, S. Precision health data: Requirements, challenges and existing techniques for data security and privacy. Comput. Biol. Med. 2021, 129, 104130. [Google Scholar] [CrossRef]
- Puppala, M.; He, T.; Yu, X.; Chen, S.; Ogunti, R.; Wong, S.T. Data security and privacy management in healthcare applications and clinical data warehouse environment. In Proceedings of the 2016 IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI), Las Vegas, NV, USA, 24–27 February 2016; pp. 5–8. [Google Scholar]
- De Maria Marchiano, R.; Di Sante, G.; Piro, G.; Carbone, C.; Tortora, G.; Boldrini, L.; Pietragalla, A.; Daniele, G.; Tredicine, M.; Cesario, A. Translational research in the era of precision medicine: Where we are and where we will go. J. Pers. Med. 2021, 11, 216. [Google Scholar] [CrossRef]
- Wang, Y.; Li, G.; Ma, M.; He, F.; Song, Z.; Zhang, W.; Wu, C. GT-WGS: An efficient and economic tool for large-scale WGS analyses based on the AWS cloud service. BMC Genom. 2018, 19, 89–98. [Google Scholar] [CrossRef] [Green Version]
- Kang, W.; Kadri, S.; Puranik, R.; Wurst, M.N.; Patil, S.A.; Mujacic, I.; Benhamed, S.; Niu, N.; Zhen, C.J.; Ameti, B. System for informatics in the molecular pathology laboratory: An open-source end-to-end solution for next-generation sequencing clinical data management. J. Mol. Diagn. 2018, 20, 522–532. [Google Scholar] [CrossRef]
- McGraw, D.; Mandl, K.D. Privacy protections to encourage use of health-relevant digital data in a learning health system. Npj Digit. Med. 2021, 4, 2. [Google Scholar] [CrossRef]
- Price, W.N.; Cohen, I.G. Privacy in the age of medical big data. Nat. Med. 2019, 25, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, H.R.; Kim, S.; Kim, E.; Kim, S.Y.; Park, H.-Y. Public Attitudes Toward Precision Medicine: A Nationwide Survey on Developing a National Cohort Program for Citizen Participation in the Republic of Korea. Front. Genet. 2020, 11, 283. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gim, J.-A. A Genomic Information Management System for Maintaining Healthy Genomic States and Application of Genomic Big Data in Clinical Research. Int. J. Mol. Sci. 2022, 23, 5963. https://doi.org/10.3390/ijms23115963
Gim J-A. A Genomic Information Management System for Maintaining Healthy Genomic States and Application of Genomic Big Data in Clinical Research. International Journal of Molecular Sciences. 2022; 23(11):5963. https://doi.org/10.3390/ijms23115963
Chicago/Turabian StyleGim, Jeong-An. 2022. "A Genomic Information Management System for Maintaining Healthy Genomic States and Application of Genomic Big Data in Clinical Research" International Journal of Molecular Sciences 23, no. 11: 5963. https://doi.org/10.3390/ijms23115963
APA StyleGim, J.-A. (2022). A Genomic Information Management System for Maintaining Healthy Genomic States and Application of Genomic Big Data in Clinical Research. International Journal of Molecular Sciences, 23(11), 5963. https://doi.org/10.3390/ijms23115963




