Unraveling the Binding Mechanism of Alzheimer’s Drugs with Irisin: Spectroscopic, Calorimetric, and Computational Approaches
Abstract
:1. Introduction
2. Results and Discussion
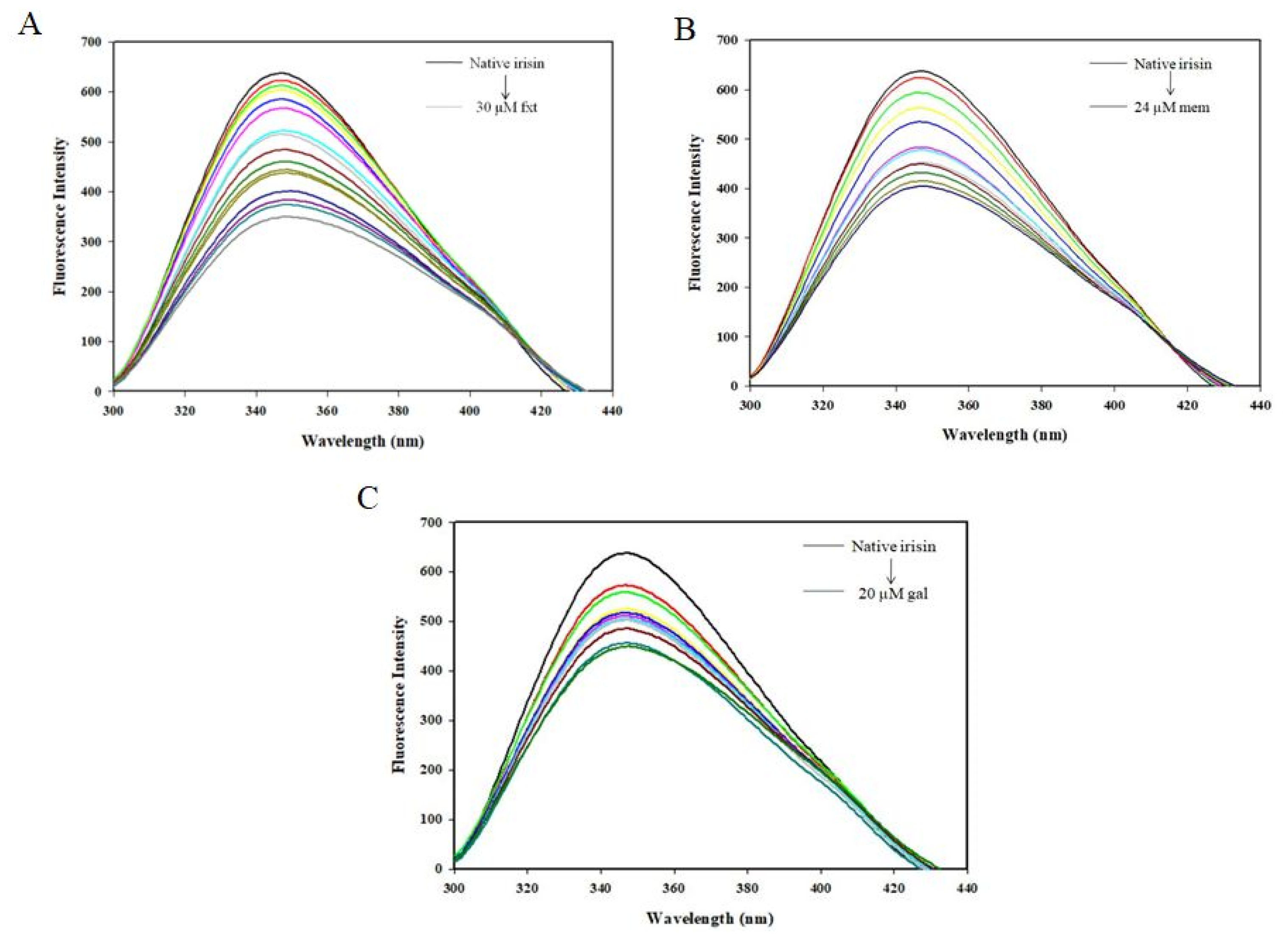
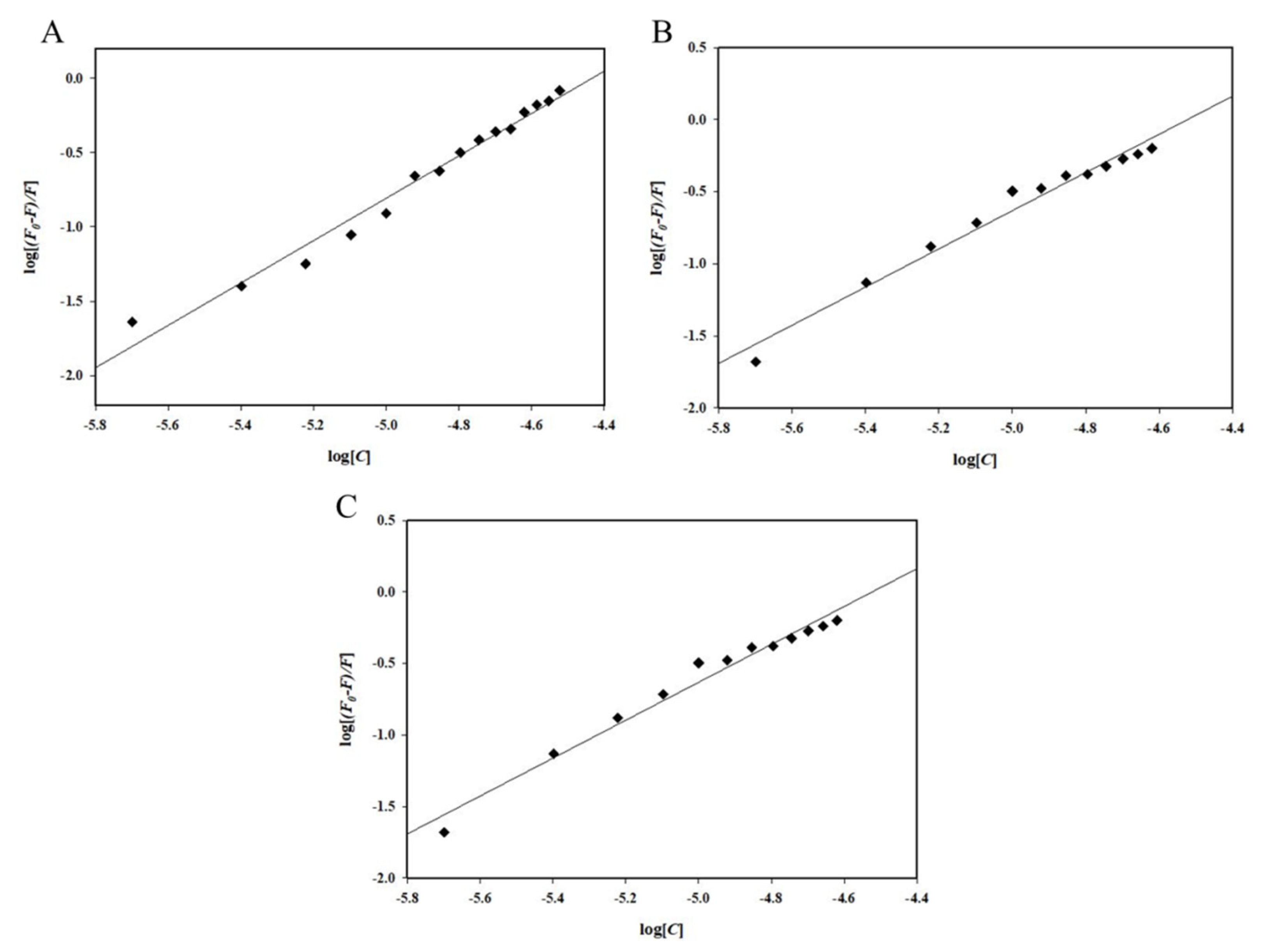
2.1. Fluorescence Spectroscopic Measurements
2.2. Isothermal Titration Calorimetry (ITC)
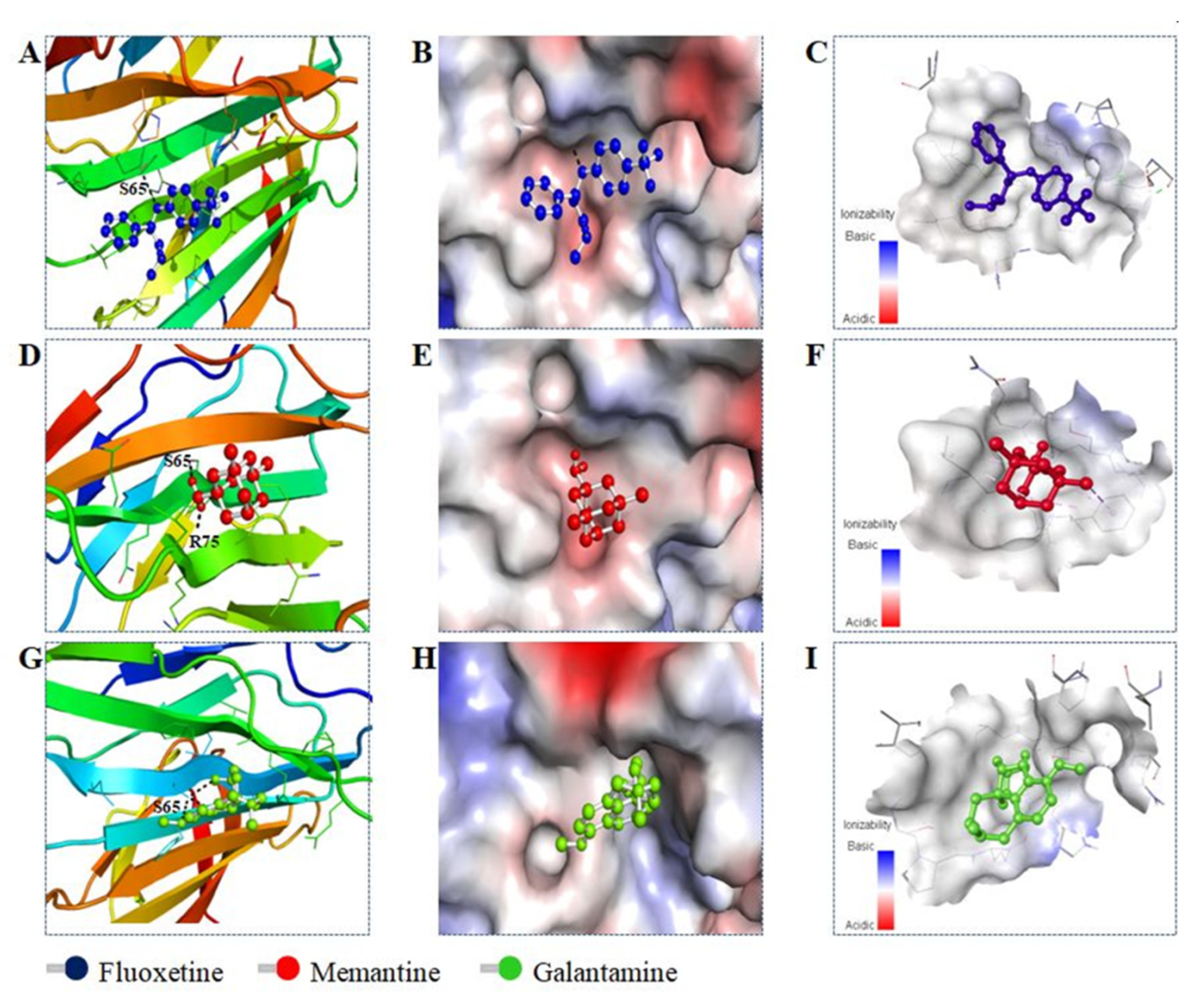
2.3. Molecular Docking
3. Materials and Methods
3.1. Expression and Purification of Irisin
3.2. Fluorescence Spectroscopic Measurements
3.3. ITC Measurements
3.4. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waseem, R.; Shamsi, A.; Kazim, S.N.; Islam, A. An insight into mitochondrial dysfunction and its implications in neurological diseases. Curr. Drug Targets 2021, 22, 1585–1595. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.L.; Tong, G.; Ballard, C. Treatment combinations for Alzheimer’s disease: Current and future pharmacotherapy options. J. Alzheimer’s Dis. 2019, 67, 779–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goedert, M.; Spillantini, M.G. A century of Alzheimer’s disease. Science 2006, 314, 777–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, S.-Y.; Fu, W.-M. Drug candidates in clinical trials for Alzheimer’s disease. J. Biomed. Sci. 2017, 24, 47. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.-X.; Liu, F.; Iqbal, K. Multifactorial hypothesis and multi-targets for Alzheimer’s disease. J. Alzheimer’s Dis. 2018, 64, S107–S117. [Google Scholar] [CrossRef]
- Fish, P.V.; Steadman, D.; Bayle, E.D.; Whiting, P. New approaches for the treatment of Alzheimer’s disease. Bioorganic Med. Chem. Lett. 2019, 29, 125–133. [Google Scholar] [CrossRef]
- Marucci, G.; Buccioni, M.; Dal Ben, D.; Lambertucci, C.; Volpini, R.; Amenta, F. Efficacy of acetylcholinesterase inhibitors in Alzheimer’s disease. Neuropharmacology 2021, 190, 108352. [Google Scholar] [CrossRef]
- Yoo, J.-W.; Irvine, D.J.; Discher, D.E.; Mitragotri, S. Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat. Rev. Drug Discov. 2011, 10, 521–535. [Google Scholar] [CrossRef]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef] [Green Version]
- Kratz, F.; Ehling, G.; Kauffmann, H.; Unger, C. Acute and repeat-dose toxicity studies of the (6-maleimidocaproyl) hydrazone derivative of doxorubicin (DOXO-EMCH), an albumin-binding prodrug of the anticancer agent doxorubicin. Hum. Exp. Toxicol. 2007, 26, 19–35. [Google Scholar] [CrossRef]
- Wenk, E.; Merkle, H.P.; Meinel, L. Silk fibroin as a vehicle for drug delivery applications. J. Control. Release 2011, 150, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Szwed, M.; Matusiak, A.; Laroche-Clary, A.; Robert, J.; Marszalek, I.; Jozwiak, Z. Transferrin as a drug carrier: Cytotoxicity, cellular uptake and transport kinetics of doxorubicin transferrin conjugate in the human leukemia cells. Toxicol. Vitr. 2014, 28, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, M.V.; Frozza, R.L.; de Freitas, G.B.; Zhang, H.; Kincheski, G.C.; Ribeiro, F.C.; Gonçalves, R.A.; Clarke, J.R.; Beckman, D.; Staniszewski, A. Exercise-linked FNDC5/irisin rescues synaptic plasticity and memory defects in Alzheimer’s models. Nat. Med. 2019, 25, 165–175. [Google Scholar] [CrossRef]
- Waseem, R.; Shamsi, A.; Mohammad, T.; Hassan, M.; Kazim, S.N.; Chaudhary, A.A.; Rudayni, H.A.; Al-Zharani, M.; Ahmad, F.; Islam, A. FNDC5/Irisin: Physiology and Pathophysiology. Molecules 2022, 27, 1118. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Deng, X.; Huang, W.; Yu, J.-H.; Wang, J.-X.; Wang, J.-P.; Yang, S.-B.; Liu, X.; Wang, L.; Zhang, Y. Irisin protects against neuronal injury induced by oxygen-glucose deprivation in part depends on the inhibition of ROS-NLRP3 inflammatory signaling pathway. Mol. Immunol. 2017, 91, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Wrann, C.D.; White, J.P.; Salogiannnis, J.; Laznik-Bogoslavski, D.; Wu, J.; Ma, D.; Lin, J.D.; Greenberg, M.E.; Spiegelman, B.M. Exercise induces hippocampal BDNF through a PGC-1α/FNDC5 pathway. Cell Metab. 2013, 18, 649–659. [Google Scholar] [CrossRef] [Green Version]
- Noda, Y.; Kuzuya, A.; Tanigawa, K.; Araki, M.; Kawai, R.; Ma, B.; Sasakura, Y.; Maesako, M.; Tashiro, Y.; Miyamoto, M. Fibronectin type III domain-containing protein 5 interacts with APP and decreases amyloid β production in Alzheimer’s disease. Mol. Brain 2018, 11, 61. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Jiang, H. Efficacy and adverse effects of memantine treatment for Alzheimer’s disease from randomized controlled trials. Neurol. Sci. 2015, 36, 1633–1641. [Google Scholar] [CrossRef]
- Alhumaydhi, F.A.; Aljasir, M.A.; Aljohani, A.S.; Alsagaby, S.A.; Alwashmi, A.S.; Shahwan, M.; Hassan, M.I.; Islam, A.; Shamsi, A. Probing the interaction of memantine, an important Alzheimer’s drug, with human serum albumin: In silico and in vitro approach. J. Mol. Liq. 2021, 340, 116888. [Google Scholar] [CrossRef]
- Tariot, P. Current status and new developments with galantamine in the treatment of Alzheimer’s disease. Expert Opin. Pharmacother. 2001, 2, 2027–2049. [Google Scholar] [CrossRef]
- Khan, M.S.; Husain, F.M.; Alhumaydhi, F.A.; Alwashmi, A.S.; Rehman, M.T.; Alruwetei, A.M.; Hassan, M.I.; Islam, A.; Shamsi, A. Exploring the molecular interactions of Galantamine with human Transferrin: In-silico and in vitro insight. J. Mol. Liq. 2021, 335, 116227. [Google Scholar] [CrossRef]
- Benfield, P.; Heel, R.C.; Lewis, S.P. Fluoxetine. Drugs 1986, 32, 481–508. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Shahwan, M.; Shamsi, A.; Alhumaydhi, F.A.; Alsagaby, S.A.; Al Abdulmonem, W.; Abdullaev, B.; Yadav, D.K. Elucidating the Interactions of Fluoxetine with Human Transferrin Employing Spectroscopic, Calorimetric, and In Silico Approaches: Implications of a Potent Alzheimer’s Drug. ACS Omega 2022, 7, 9015. [Google Scholar] [CrossRef] [PubMed]
- Waseem, R.; Anwar, S.; Khan, S.; Shamsi, A.; Hassan, M.; Anjum, F.; Shafie, A.; Islam, A.; Yadav, D.K. MAP/Microtubule Affinity Regulating Kinase 4 Inhibitory Potential of Irisin: A New Therapeutic Strategy to Combat Cancer and Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 10986. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, A.; Mohammad, T.; Khan, M.S.; Shahwan, M.; Husain, F.M.; Rehman, M.; Hassan, M.; Ahmad, F.; Islam, A. Unraveling binding mechanism of Alzheimer’s drug rivastigmine tartrate with human transferrin: Molecular docking and multi-spectroscopic approach towards neurodegenerative diseases. Biomolecules 2019, 9, 495. [Google Scholar] [CrossRef] [Green Version]
- Waseem, R.; Shamsi, A.; Mohammad, T.; Alhumaydhi, F.A.; Kazim, S.N.; Hassan, M.I.; Ahmad, F.; Islam, A. Multispectroscopic and Molecular Docking Insight into Elucidating the Interaction of Irisin with Rivastigmine Tartrate: A Combinational Therapy Approach to Fight Alzheimer’s Disease. ACS Omega 2021, 6, 7910–7921. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Quenching of fluorescence. In Principles of Fluorescence Spectroscopy; Springer: Boston, MA, USA, 1983; pp. 257–301. [Google Scholar]
- Waseem, R.; Shamsi, A.; Shahbaz, M.; Khan, T.; Kazim, S.N.; Ahmad, F.; Hassan, M.I.; Islam, A. Effect of pH on the structure and stability of irisin, a multifunctional protein: Multispectroscopic and molecular dynamics simulation approach. J. Mol. Struct. 2022, 1252, 132141. [Google Scholar] [CrossRef]
- Shamsi, A.; Al Shahwan, M.; Ahamad, S.; Hassan, M.I.; Ahmad, F.; Islam, A. Spectroscopic, calorimetric and molecular docking insight into the interaction of Alzheimer’s drug donepezil with human transferrin: Implications of Alzheimer’s drug. J. Biomol. Struct. Dyn. 2020, 38, 1094–1102. [Google Scholar] [CrossRef]
- Anwar, S.; Shamsi, A.; Shahbaaz, M.; Queen, A.; Khan, P.; Hasan, G.M.; Islam, A.; Alajmi, M.F.; Hussain, A.; Ahmad, F. Rosmarinic acid exhibits anticancer effects via MARK4 inhibition. Sci. Rep. 2020, 10, 10300. [Google Scholar] [CrossRef]
- Bae, K.H.; Lee, F.; Xu, K.; Keng, C.T.; Tan, S.Y.; Tan, Y.J.; Chen, Q.; Kurisawa, M. Microstructured dextran hydrogels for burst-free sustained release of PEGylated protein drugs. Biomaterials 2015, 63, 146–157. [Google Scholar] [CrossRef]
- Garrait, G.; Beyssac, E.; Subirade, M. Development of a novel drug delivery system: Chitosan nanoparticles entrapped in alginate microparticles. J. Microencapsul. 2014, 31, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Hubbell, J.A.; Chilkoti, A. Nanomaterials for drug delivery. Science 2012, 337, 303–305. [Google Scholar] [CrossRef]
- Jao, D.; Xue, Y.; Medina, J.; Hu, X. Protein-based drug-delivery materials. Materials 2017, 10, 517. [Google Scholar] [CrossRef]
- Kratz, F.; Elsadek, B. Clinical impact of serum proteins on drug delivery. J. Control. Release 2012, 161, 429–445. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, A.; Ahmed, A.; Bano, B. Probing the interaction of anticancer drug temsirolimus with human serum albumin: Molecular docking and spectroscopic insight. J. Biomol. Struct. Dyn. 2018, 36, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Liu, R. Phenotypic characterization of the binding of tetracycline to human serum albumin. Biomacromolecules 2011, 12, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Valeur, B. Molecular Fluorescence: Principles and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2002. [Google Scholar]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer US: Boston, MA, USA, 2006. [Google Scholar]
- Feng, X.-Z.; Lin, Z.; Yang, L.-J.; Wang, C.; Bai, C.-L. Investigation of the interaction between acridine orange and bovine serum albumin. Talanta 1998, 47, 1223–1229. [Google Scholar] [CrossRef]
- Anwar, S.; DasGupta, D.; Shafie, A.; Alhumaydhi, F.A.; Alsagaby, S.A.; Shahwan, M.; Anjum, F.; Al Abdulmonem, W.; Sharaf, S.E.; Hassan, M.I. Implications of Tempol in Pyruvate Dehydrogenase Kinase 3 Targeted Anticancer therapeutics: Computational, Spectroscopic and Calorimetric Studies. J. Mol. Liq. 2022, 350, 118581. [Google Scholar] [CrossRef]
- Shamsi, A.; Mohammad, T.; Anwar, S.; Alajmi, M.F.; Hussain, A.; Hassan, M.I.; Ahmad, F.; Islam, A. Probing the interaction of Rivastigmine Tartrate, an important Alzheimer’s drug, with serum albumin: Attempting treatment of Alzheimer’s disease. Int. J. Biol. Macromol. 2020, 148, 533–542. [Google Scholar] [CrossRef]
- Yousuf, M.; Shamsi, A.; Queen, A.; Shahbaaz, M.; Khan, P.; Hussain, A.; Alajmi, M.F.; Rizwanul Haque, Q.M.; Imtaiyaz Hassan, M. Targeting cyclin-dependent kinase 6 by vanillin inhibits proliferation of breast and lung cancer cells: Combined computational and biochemical studies. J. Cell. Biochem. 2021, 122, 897–910. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System. 2002. Available online: http://www.pymol.org (accessed on 12 May 2022).
- Jacob, R.B.; Andersen, T.; McDougal, O.M. Accessible high-throughput virtual screening molecular docking software for students and educators. PLoS Comput. Biol. 2012, 8, e1002499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biovia, D.S. Discovery Studio Modeling Environment, Release 4.5; Dassault Systèmes Biovia: San Diego, CA, USA, 2015. [Google Scholar]

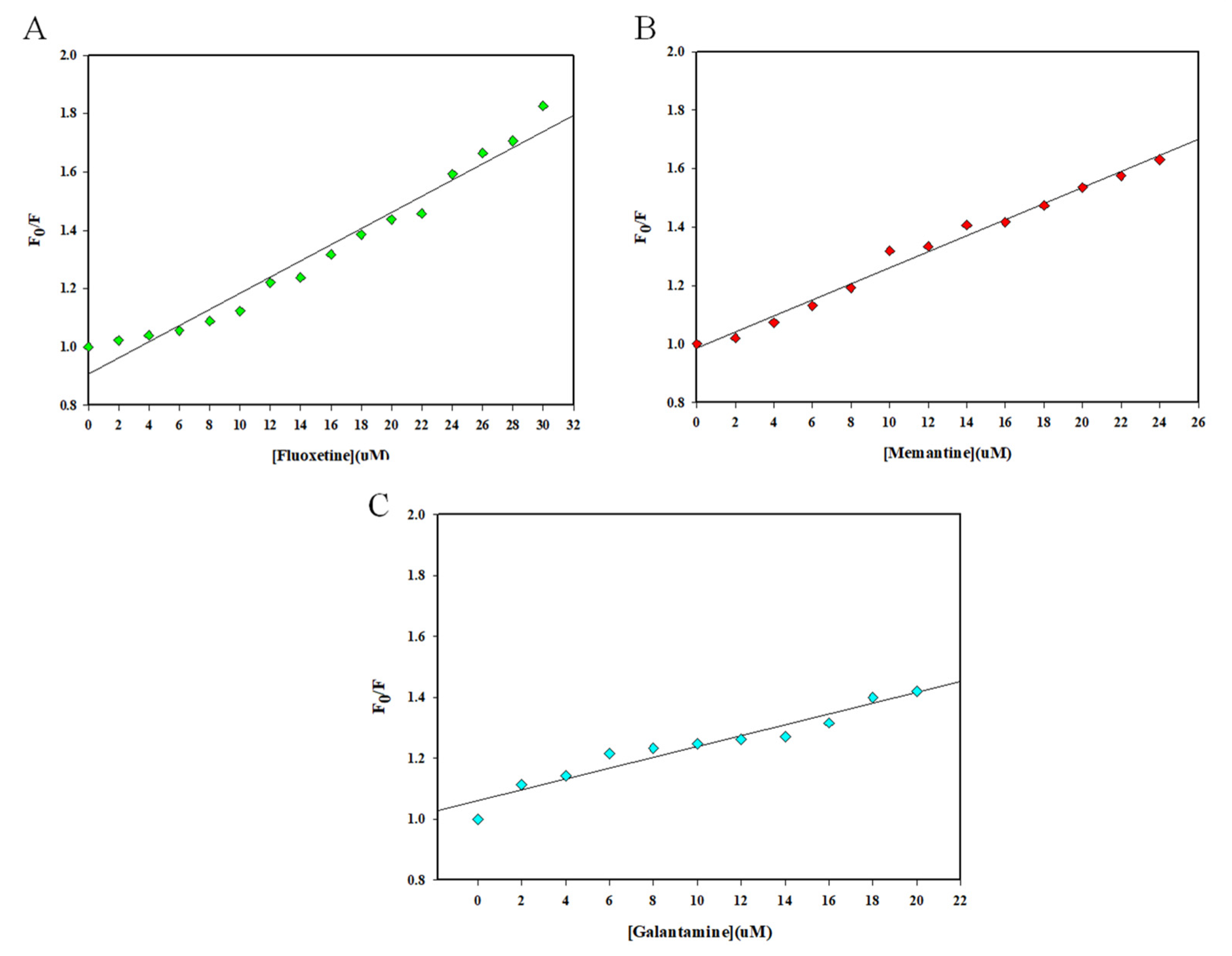


| Irisin–Drug | Ksv (104M−1) | R2 |
|---|---|---|
| Irisin–Fluoxetine | 2.77 | 0.97 |
| Irisin–Memantine | 2.73 | 0.98 |
| Irisin–Galantamine | 1.77 | 0.94 |
| Irisin–Drug | K | n |
|---|---|---|
| Irisin–Fluoxetine | 0.21 × 107M−1 | 1.43 |
| Irisin–Memantine | 9.78 × 105M−1 | 1.32 |
| Irisin–Galantamine | 0.14 × 103M−1 | 0.56 |
| Irisin–Fluoxetine System | ||
|---|---|---|
| Ka (Association Constant), M−1 | ∆H (Enthalpy Change), cal/mol | ∆S (cal/mol/deg) |
| Ka1 = 6.41 × 106 ± 9.66 × 104 | ∆H1 = −1.23 × 104 ± 979.4 | ∆S1 = −10.3 |
| Irisin–Memantine system | ||
| Ka1 = 8.97 × 104 ± 2.9 × 103 | ∆H1 = 5744 ± 2.52 × 103 | ∆S1 = 41.9 |
| Ka2 = 1.07 × 105 ± 4.2 × 103 | ∆H2 = −1.357 × 105 ± 7.41 × 103 | ∆S2 = −432 |
| Ka3 = 9.91 × 104 ± 5.5 × 103 | ∆H3 = 1.92 × 105 ± 1.74 × 104 | ∆S3 = 669 |
| Ka4 = 1.06 × 105 ± 6.1 × 103 | ∆H4 = −1.99 × 105 ± 1.65 × 104 | ∆S4 = −645 |
| Irisin–Galantamine system | ||
| Ka1 = 1.05 × 105 ± 1.7 × 104 | ∆H1 = 6763 ± 6.18 × 103 | ∆S1 = 45.7 |
| Ka2 = 1.24 × 105 ± 2.3 × 104 | ∆H2 = −2.121 × 105 ± 3.19 × 104 | ∆S2 = −688 |
| Ka3 = 6.16 × 104 ± 9.1 × 103 | ∆H3 = 1.88 × 105 ± 6.12 × 104 | ∆S3 = 655 |
| Ka4 = 5.54 × 104 ± 1.0 × 104 | ∆H4 = −3.076 × 105 ± 4.49 × 104 | ∆S4 = −1.01 × 103 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waseem, R.; Shamsi, A.; Khan, T.; Hassan, M.I.; Kazim, S.N.; Shahid, M.; Islam, A. Unraveling the Binding Mechanism of Alzheimer’s Drugs with Irisin: Spectroscopic, Calorimetric, and Computational Approaches. Int. J. Mol. Sci. 2022, 23, 5965. https://doi.org/10.3390/ijms23115965
Waseem R, Shamsi A, Khan T, Hassan MI, Kazim SN, Shahid M, Islam A. Unraveling the Binding Mechanism of Alzheimer’s Drugs with Irisin: Spectroscopic, Calorimetric, and Computational Approaches. International Journal of Molecular Sciences. 2022; 23(11):5965. https://doi.org/10.3390/ijms23115965
Chicago/Turabian StyleWaseem, Rashid, Anas Shamsi, Tanzeel Khan, Md. Imtaiyaz Hassan, Syed Naqui Kazim, Mohammad Shahid, and Asimul Islam. 2022. "Unraveling the Binding Mechanism of Alzheimer’s Drugs with Irisin: Spectroscopic, Calorimetric, and Computational Approaches" International Journal of Molecular Sciences 23, no. 11: 5965. https://doi.org/10.3390/ijms23115965
APA StyleWaseem, R., Shamsi, A., Khan, T., Hassan, M. I., Kazim, S. N., Shahid, M., & Islam, A. (2022). Unraveling the Binding Mechanism of Alzheimer’s Drugs with Irisin: Spectroscopic, Calorimetric, and Computational Approaches. International Journal of Molecular Sciences, 23(11), 5965. https://doi.org/10.3390/ijms23115965









