Abstract
We studied the effects of stimulation of the medial septal area on the gene expression in the dorsal and ventral hippocampus. Rats under urethane anesthesia were implanted with a recording electrode in the right hippocampus and stimulating electrode in the dorsal medial septum (dMS) or medial septal nucleus (MSN). After one-hour-long deep brain stimulation, we collected ipsi- and contralateral dorsal and ventral hippocampi. Quantitative PCR showed that deep brain stimulation did not cause any changes in the intact contralateral dorsal and ventral hippocampi. A comparison of ipsi- and contralateral hippocampi in the control unstimulated animals showed that electrode implantation in the ipsilateral dorsal hippocampus led to a dramatic increase in the expression of immediate early genes (c-fos, arc, egr1, npas4), neurotrophins (ngf, bdnf) and inflammatory cytokines (il1b and tnf, but not il6) not only in the area close to implantation site but also in the ventral hippocampus. Moreover, the stimulation of MSN but not dMS further increased the expression of c-fos, egr1, npas4, bdnf, and tnf in the ipsilateral ventral but not dorsal hippocampus. Our data suggest that the activation of medial septal nucleus can change the gene expression in ventral hippocampal cells after their priming by other stimuli.
Keywords:
deep brain stimulation; septum; dorsal hippocampus; ventral hippocampus; early genes; bdnf; ngf; inflammation 1. Introduction
The source of cholinergic innervation of the hippocampus, a brain area critically involved in memory formation, is the medial septal area (MSA), which consists of the medial septal nucleus and the diagonal band of Broca. The cholinergic neurons in this area are involved in the regulation of synaptic plasticity and rhythmogenesis in the neocortex and hippocampus [1,2]. Moreover, the activity of MSA neurons is important for the memory formation, learning, and spatial navigation [3,4,5,6,7,8,9,10]. Current views on the septohippocampal interaction, which are predominantly based on the electrophysiological analysis of neuronal activity in the MSA and hippocampus, suggest that acetylcholine released by MSA neurons induces short-term changes in the electrophysiological characteristics of hippocampal neurons and hippocampal synapses [11,12,13,14] and long-term changes in the efficacy of synaptic transmission in the hippocampus [1]. The ability of acetylcholine to induce long-term changes suggests that it can activate some signaling cascades that may finally result in changes in the expression of genes that are responsible for the development of long-term synaptic effects.
It was shown that the activation of muscarinic acetylcholine receptors induced transcription of the early gene cyr61/ccn1 in HEK 293 cells [15,16]. The administration of M1 agonist pilocarpine, which is known to induce seizure activity, increased c-fos mRNA induction in many forebrain structures including piriform cortex, nucleus accumbens, amygdala, hippocampus, and neocortex [17]. It was shown that nicotinic acetylcholine receptors can also modulate gene expression in the hippocampal neurons via activation of the transcription factor CREB [18,19]. However, currently, it is largely unclear whether the activation of cholinergic neurons in MSA may lead to changes in the gene expression in the hippocampus. It was shown that the injection of potent glutamate agonist quisqualate into MSA leads to postponed elevation of mRNA expression of bdnf and ngf in the hippocampus [20]. Recently, it was shown that repeated electrical stimulation of MSA may induce the expression of c-fos in the dentate gyrus [21]. However, MSA is a heterogeneous structure, and it is known that the activation of different parts may result in different effects in the hippocampus [13,22], and the aforementioned studies did not analyze the consequences of activation of different parts of the MSA.
Deep brain stimulation (DBS) of the medial septal nucleus is considered as an approach that can have beneficial effects under pathological conditions [23]. In animal studies, deep brain stimulation of the medial septum improved spatial memory during cholinergic dysfunction [24]. Furthermore, it was shown that selective cholinergic activation triggers a robust network effect in the septo-hippocampal system during an inactive behavioral state [25]. One of possible long-term mechanisms that are triggered by deep brain stimulation of MSA may be related to the expression of immediate early genes, such as c-fos [21].
The aim of the present study was to examine the effect of deep brain stimulation at the different depths of the MSA, which correspond to the dorsal medial septum (dMS) and cholinergic medial septal nucleus (MSN), on the expression of genes in different parts of hippocampal formation.
2. Results
2.1. Electrical Stimulation of MSA at Different Depths Induces Different Field Response in the Hippocampus
Stimulation of the dorsal medial septum (dMS) at the depth of 3.5–4.5 mm evoked field responses in the CA1 region of the hippocampus. As depicted in Figure 1A, a negative waveform, having a latency of 3 msec, was recorded by a bipolar electrode positioned in the molecular layer of the hippocampus. After relocation of the stimulation electrode to the depth of 6.5 mm ventral to dura, this response disappears (Figure 1B). A position at this depth failed to elicit stable field responses. Nevertheless, in some animals, we observed positive/negative waves of excitation (Figure 1C). The amplitude of the responses evoked from this depth was lower compared with the amplitude at the higher coordinate.
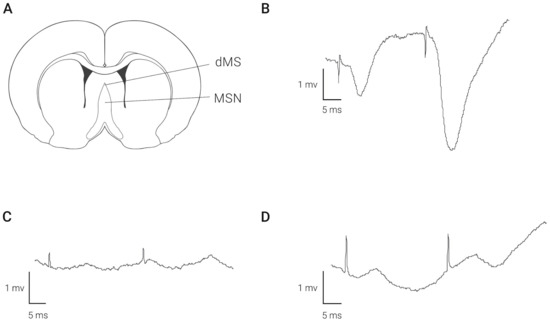
Figure 1.
Field responses recorded in the hippocampal CA1 area following stimulation of medial septal area at different depths. Panel (A) shows localization of stimulating electrodes. Examples of fEPSP after stimulation of dMS (B) and MSN (C,D) are shown.
2.2. Electrical Stimulation of MSA Does Not Affect EEG in the Hippocampus
To evaluate the possible general effect that may induce deep brain stimulation on the functioning of the hippocampus, we compared basic EEG characteristics in the hippocampus in the control and after dMS and MSN stimulation. We found that stimulation at both depths of the medial septum did not cause any significant effect on the rhythmic activity in the hippocampus (Figure 2).
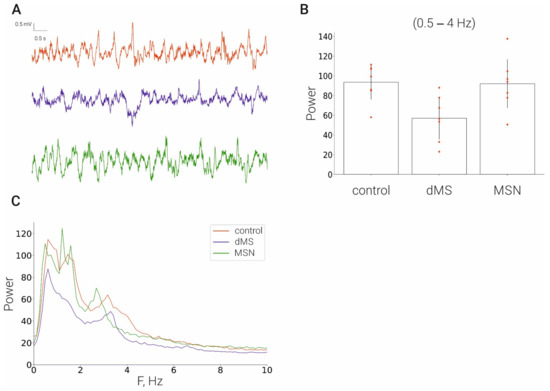
Figure 2.
Effect of stimulation of the medial septum on the hippocampal EEG. (A), LFP time series of: control rats (red line), dMS rats (blue), MSN rats (green); (B), corresponding power spectra; (C), corresponding averaged power in frequency range [0.5–4] Hz.
2.3. Analysis of Gene Expression in the Hippocampus after Ultraslow Deep Brain Stimulation of MSA
In our experiments, we analyzed mRNA expression of several groups of genes that, according to the literature, may respond to elevation of acetylcholine level and, hence, to MSA stimulation. The first group consisted of immediate early genes such as c-fos [21], arc, egr1, npas4, and cyr61 [15,16]. The second group included two major growth factors bdnf and ngf [20], and the third group included genes that encoded inflammatory cytokines il1b, il6, and tnf [26,27]. We compared the level of the mentioned mRNAs in the groups of animals, which were implanted with stimulation electrode in the dorsal medial septum (dMS) or MSN, and the control animals that were implanted with electrode in the MSN but were not stimulated. Our expression analysis included the four following structures: left dorsal and ventral hippocampi and right dorsal and ventral hippocampi. To analyze the effect of deep brain stimulation, we compared mRNA levels between experimental groups in each part of the hippocampus separately. In this comparison, we considered left hippocampi in the control unstimulated animals as the basal level of expression. We also compared mRNA expression between left and right parts of the hippocampus to elucidate additional effects that may arise from implantation of the recording electrode in the right dorsal hippocampus.
2.3.1. Immediate Early Genes
We found that deep brain stimulation (DBS) of dMS or MSN did not induce any substantial influence on the expression of the studied immediate early genes in both parts of the left (intact) hippocampus (Figure 3). Electrode implantation in the control animals induced a strong increase in the expression of c-fos and arc in both dorsal and ventral parts of the right hippocampus compared to the corresponding parts of the left hippocampus. Importantly, DBS induced a significant increase in the right ventral hippocampus in the expression of c-fos and did not affect arc expression in both parts of the right hippocampus. The DBS-induced increase in c-fos expression occurred only after DBS of MSN but not dMS.
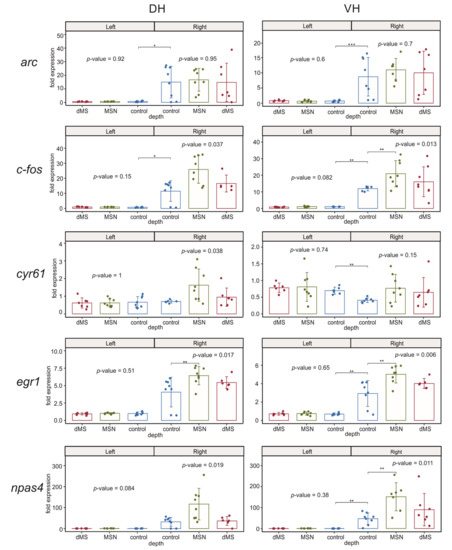
Figure 3.
Changes in the expression of early genes after stimulation of the dorsal medium septum (dMS) and medial septal nucleus (MSN) in dorsal and ventral parts (DH and VH, respectively) of the left (intact) and right (damaged by implanted electrode) hippocampi. p-values are shown for Kruskal–Wallis test for comparison of three groups in one hemisphere. Interhemispheric comparisons were performed only in the control group using the Mann–Whitney test. For the intergroup comparisons, *, **, and *** mark significant differences at 0.005 ≤ p < 0.025, p < 0.005, and p < 0.0005 (post hoc Dunn test). For interhemispheric comparisons, *, **, and *** mark significant differences at p ≤ 0.05, p < 0.01, and p < 0.001 (Mann–Whitney test).
Electrode implantation also resulted in a significant increase in the expression of egr1 and npas4 in the right ventral hippocampus compared to the left ventral hippocampus. When we compared the dorsal parts of the left and right hippocampus in the control animals, we found the elevation of expression of these genes in the right dorsal hippocampus was not significant due to very high dispersion of mRNA expression between samples of the right dorsal hippocampus. However, post hoc analysis of the egr1 expression in the right hemisphere showed that the increase after MSN stimulation but not dMS stimulation was significant for both genes (Figure 3). In the right ventral hippocampus, the effect of DBS was significant for both genes, and post hoc analysis revealed a significant increase in the expression of both npas4 and egr1 after DBS of MSN compared to control animals.
The expression of putative acetylcholine-dependent gene cyr61/ccn1 in the right hippocampus was completely independent of DBS at any level of the medial septum; however, electrode implantation led to a significant decrease in the expression of this gene in the right ventral hippocampus compared to the left ventral hippocampus.
2.3.2. Neurotrophins
Analysis of expression of ngf showed that the mRNA expression of this neurotrophin is not influenced by the DBS of any area of the medial septum. However, hippocampal damage resulting from electrode implantation led to an increase in the ngf expression in both right dorsal and ventral parts of hippocampus (Figure 4).
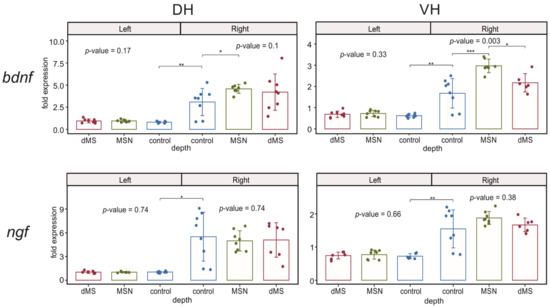
Figure 4.
Changes in the expression of neurotrophins after stimulation of the dorsal medium septum and medial septal nucleus in dorsal and ventral parts (DH and VH, respectively) of the left (intact) and right (damaged by implanted electrode) hippocampi. p-values are shown for Kruskal–Wallis test for comparison of three groups in one hemisphere. Interhemispheric comparisons were performed only in the control group using the Mann–Whitney test. For the intergroup comparisons, *, **, and *** mark significant differences at 0.005 ≤ p < 0.025, p < 0.005, and p < 0.0005 (post hoc Dunn test). For interhemispheric comparisons, *, **, and *** mark significant differences at p ≤ 0.05, p < 0.01, and p < 0.001 (Mann–Whitney test).
While bdnf expression in the undamaged left hippocampus was also not affected by DBS, an increase in the bdnf expression in the both parts of the right hippocampus caused by electrode implantation was significantly enhanced by DBS at the level of MSN but not at the level of dMS only in the ventral part, whereas the increase in the right dorsal hippocampus was insensitive to DBS (Figure 4).
2.3.3. Inflammatory Cytokines
In our experiments, we analyzed changes in the expression of three cytokines involved in the regulation of inflammatory response il1b, il6, and tnf (Figure 5). The expression of all studied cytokines was not affected by DBS in the left undamaged hemisphere in both hippocampal parts. The expression of il6 appeared to be insensitive to the damage of the right dorsal hippocampus by implanted electrode, and the subsequent stimulation of dMS and MSN did not influence its expression as well. In contrast, the expression of il1b was enhanced in the right dorsal hippocampus, which reflects the development of acute inflammation after electrode implantation into this hippocampal part. Stimulation of the medial septal area at both depths induced a trend to augmentation of expression of il1b; however, it did not reach the level of significance due to high dispersion among samples in both dMS and MSN groups. The picture of tnf expression in the right hippocampus was even more complex. It appeared that electrode implantation resulted in a significant increase in the tnf expression in the right ventral hippocampus and only a weak trend to enhancement in the right dorsal hippocampus despite the fact that implantation caused damage in the dorsal part. Stimulation of the septum at both levels induced a trend to an increase in the tnf expression in right dorsal hippocampus and did not affect the tnf level in the right ventral hippocampus.
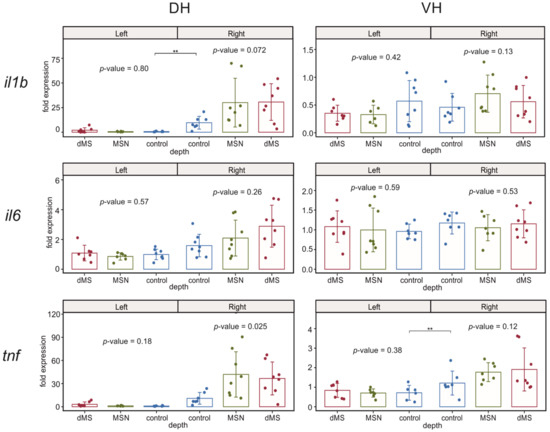
Figure 5.
Changes in the expression of inflammatory cytokines after stimulation of the dorsal medium septum and medial septal nucleus in dorsal and ventral parts (DH and VH, respectively) of the left (intact) and right (damaged by implanted electrode) hippocampi. p-values are shown for Kruskal–Wallis test for comparison of three groups in one hemisphere. Interhemispheric comparisons were performed only in the control group using the Mann–Whitney test. For interhemispheric comparisons, ** mark significant differences at p < 0.01 (Mann–Whitney test).
3. Discussion
Septohippocampal interaction is frequently discussed in terms of the generation of hippocampal rhythms and plasticity of hippocampal synapses, which are critical for learning and memory formation [28]. Previous studies showed that memory formation is associated with changes in the expression of immediate early genes, suggesting that the activation of medial septal neurons may be one of factors that induces changes in the gene expression in the hippocampal cells. In our study, we used the stimulation of two parts of the medial septum to evaluate its effect on the functioning of the hippocampus. We found, in agreement with previous reports [22], that the stimulation of dMS induces field responses in the hippocampus, and electrode implantation in the medial septal nucleus practically does not induce any field response in the hippocampus, which is probably reflecting the fact that the electrode reached the area that predominantly contains cholinergic neurons [13,29]. Stimulation of both parts of the medial septum did not induce any effect on the EEG pattern in the hippocampus, supporting previous data that medial septum stimulation has a very narrow temporal window for influencing characteristics of hippocampal neurons [13].
We found that the stimulation of dMS and MSN had no effect on the expression of all studied genes in the left dorsal and ventral hippocampi, which were not damaged by the electrode implantation. In contrast, in the right hippocampus, which was damaged by the electrode implantation, we found that the stimulation of MSN but not dMS modulated the expression of several studied genes. First, it should be mentioned that stimulation of the medial septum induces acetylcholine release in both the left and right hippocampi [30], and the observed asymmetry of response is not related to any asymmetry of acetylcholine release. Second, the electrode implantation per se induced not only local effect in the dorsal hippocampus, where it was implanted, but also a distant effect in the ventral hippocampus. At first glance, electrode implantation should have a local effect in the dorsal hippocampus where it should induce an inflammatory response. Indeed, we observed an increase in the expression of proinflammatory cytokines typical of acute inflammatory process in the dorsal hippocampus (il1b and tnf). However, the increase in the expression of il1b was localized to the dorsal hippocampus, whereas tnf expression increased in the ventral part of the right hippocampus, suggesting that an additional process occurred that spreads the effect from the dorsal to ventral part of the hippocampus without spreading to the contralateral hippocampus. It is well known that the hippocampal tissue is very sensitive to the induction of spreading depression, and its injury may induce spreading depression [31]. After induction, spreading depression will travel along the longitudinal axis of the hippocampus and will not be induced in the contralateral hemisphere. It was previously shown that in the neocortex, spreading depression increases the expression of c-fos [32], arc [33], egr1 [34], npas4 [35], bdnf [33], il1b [36], and tnf [36]. Our data are in agreement with these observations because in our case, the expression of all these genes, except il1b, increased in the right ventral hippocampus compared to the left ventral hippocampus. Additionally, we showed that the expression of early gene cyr61/ccn1 in the ventral hippocampus decreases after putative spreading depression. Presumably, spreading depression resulted from tissue damage after electrode implantation in the right dorsal hippocampus and spread to the ventral hippocampus, where it altered the expression of the majority of studied genes.
As we mentioned above, stimulation of the medial septal area at different depths induces different field responses in the hippocampus. Stimulation in the dorsal medial septum induced a clear field excitatory postsynaptic response in CA1 area; however, it had no effect on the expression of any of the studied genes in any of the studied hippocampal areas. In contrast, stimulation of the medial septal area at a depth of 6.5 mm induced a barely visible field response; however, it modulated the expression of a majority of the studied genes in the ventral hippocampus. This deep septal area is the location of the medial septal nucleus rich in the cholinergic neurons. Presumably, stimulation of this area led to activation of the septal network and release of acetylcholine in the hippocampus.
One of our unexpected findings is that deep brain stimulation by trains of paired pulses can induce changes in the expression of genes only in the hippocampus that was potentially affected by spreading depression after electrode implantation. The latter means that the sensitivity of hippocampal cells was shifted by spreading depression, and even weak stimulus coming from the MSN was able to enhance the expression of immediate early genes c-fos, egr1, and npas4 but not arc and cyr61. The expression of inflammatory cytokines was largely left unchanged, except for the expression of tnf in the right dorsal hippocampus, where we observed only a trend to an increase after MSN stimulation. The effect of septal stimulation on the expression of bdnf and ngf also was not the same. While ngf expression remained insensitive to MSN and dMS stimulation in both hippocampi, the bdnf expression increased specifically in the right ventral hippocampus after MSN but not dMS stimulation. Taken together, these data suggest that MSN stimulation has a specific modulatory effect on not all transcriptional machinery but is likely to specifically activate the transcription of some genes in cells located in specific part of the hippocampus. The mechanism of sensibilization of hippocampal cells to the putative acetylcholine release from septal fibers after the passing of spreading depression in the hippocampus may include epigenetic changes that occurred after a massive release of various transmitters (glutamate, dopamine, NO, etc. [37,38,39,40]) and an increase in the density of muscarinic acetylcholine receptors after spreading depression [41]. The selective sensitivity of the ventral hippocampus to MSN DBS, compared to the dorsal part, is unclear but may be related to a well-known difference in the expression of various proteins between these hippocampal parts [42,43].
An important outcome of our results is that a train of paired stimuli activating MSN can alter the expression of a number of genes in the hippocampal cells that were previously sensitized by another stimulus (in our case, spreading depression). These results suggest that the induction of long-term effects in the hippocampal cells after MSN activity requires the specific activation of hippocampal cells before the arrival of MSN input. If the activation of hippocampal cells is strong enough to alter the epigenetic state of cells, then several trains of paired activation of MSN, which previously had no effect, will be able to change gene expression. From a physiological viewpoint, our DBS protocol seems to be very weak because septal neurons have predominantly bursting activity at frequencies above 4 Hz [44]; however, even this activity can produce a long-term effect in the cells that were “made ready” for receiving this stimulus. It is possible to hypothesize that under physiological conditions, a situation with the induction of changes in the gene expression by the activity of septal neurons will be similar, i.e., the activity of hippocampal cells in combination with inputs to these cells from other brain parts can sensibilize these cells to MSN activity, which can lead to changes in the expression of genes.
From a functional viewpoint, the early genes c-fos and egr1 are transcriptional regulators, whose expression can trigger intracellular cascades leading to the expression of other genes, including arc and bdnf [45], which, in turn, are important regulators of synaptic plasticity [45,46]. Npas4 is also a transcription factor that is activated by neuronal depolarization and modulates the expression of genes responsible for the regulation of the excitation/inhibition balance, including bdnf [47]. The enhancement of expression of these early transcription factors after MSN stimulation suggests that the activity of septal neurons can intensify the response of hippocampal cells to other stimuli and trigger or prolong plastic processes in these cells under normal conditions by the augmentation of expression of early genes involved in nerve cell plasticity. Stress is one of the well-known inducers of plastic changes in the CNS. Previously, it was shown that acute stress may induce the expression of early genes including egr1 [46] and c-fos [48], and it was proposed that the activation of septal cholinergic neurons may be one of inductors of expression of c-fos [48]. Here, we show that the activation of the medial septal input to the hippocampus can, at least, enhance the expression of these early genes, pointing to the possibility that cholinergic activity indeed can be one of factors that can upregulate the expression of early genes in the hippocampus under stress conditions.
Among the genes we studied, cyr61 and ngf were the most probable candidates for being acetylcholine-dependent because it was previously shown that cyr61/ccn1 is an early gene whose expression may be activated in culture by acetylcholine [49], and ngf expression may be induced in the hippocampus by the activation of MSA [20]. We found that the level of mRNA of these genes is insensitive to the stimulation of either dMS or MSN. We believe that at higher stimulation frequencies, the enhancing effect of MSN activity may appear.
It is also important to note that there are some data that acetylcholine may serve as a regulator of inflammatory processes in the nervous tissue by influencing the function of glial cells [26,27]. However, our data suggest that the low-frequency activity of septal neurons can hardly affect the expression of inflammatory cytokines during acute inflammation. There may be several reasons for the absence of this effect. First, according to single-cell RNAseq data [50,51,52,53], in the mouse brain, all types of acetylcholine receptors are predominantly expressed in neurons, whereas the studied pro-inflammatory cytokines are expressed in glial and vascular cells. However, the modulatory effect of acetylcholine was mainly described in cultured glial cells, which may differ from the brain cells in vivo by the expression profile of acetylcholine receptors. Second, a more substantial increase in the level of acetylcholine may be required to produce noticeable changes in the level of inflammatory cytokines. Third, the protective effect of acetylcholine may appear under chronic inflammatory conditions when the sensitivity of glial cells to this mediator is changed. Anyway, under conditions of acute inflammation, slow septal activity can hardly be used to modulate inflammation in the brain.
4. Materials and Methods
The experiments were performed with adult male Wistar rats (250–350 g) received from the Research Center of Biomedical Technology RAMS, nursery “Pushchino.” A total of 24 rats were involved in the study (n = 8/group). Animals were housed under standard vivarium conditions at 21 ± 1 °C with a 12 h light/dark cycle; food and water were provided ad libitum. All experiments were performed in accordance with the ethical principles stated in the EU Directive 2010/63/EU for animal experiments and were approved by the Ethical Committee of the Institute of Higher Nervous Activity and Neurophysiology of the Russian Academy of Sciences.
4.1. Stereotaxic Surgery and Electrophysiology
Rats were anesthetized with urethane (1.75 g/kg, i.p.) and mounted in a Kopf stereotaxic frame for surgical preparation for the recording session. A stimulating nickel–chrome electrode (diameter 80 μm) was implanted into the medial septal area (0.5 mm posterior, 0.0 mm lateral to bregma, approximately 3.5–4.5 mm or 6.5 mm ventral to dura). A recording electrode was placed into the CA1 area (2.7 mm posterior, 1.5 lateral to bregma, 2.2 mm ventral to dura) [54]. One electrode under the skin served as a ground and as a reference electrode.
The fEPSP amplitude in the CA1 field evoked by paired MS stimulation (interstimulus interval 30 ms; intertrain time 20 s at intensity of 100–300 μA; 10 paired stimulations) was recorded every 10 min for 1 h. The intensity of the testing paired pulse stimulation was set to evoke 40–50% of the maximum fEPSP amplitude. In our experiments, for long-term recordings, we applied urethane anesthesia, which is used for non-recovery procedures of exceptionally long duration where the preservation of autonomic reflexes is essential and thus does not need any additional euthanasia procedure.
4.2. EEG Recording
EEGs were recorded using the same electrode that was placed into the CA1 area (see Stereotaxic surgery and electrophysiology), with low-pass and high-pass filters of 1 kHz and 5 Hz, respectively. Seven sessions of 80 s took place during the experimental procedure. Each 80 s session was conducted after recording of the fEPSP amplitude in the CA1 field evoked by paired MS stimulation.
4.3. RNA Isolation and Reverse Transcription
Tissue samples were collected in 1 h after the start of stimulation, placed in 1.5 mL tubes, and frozen in liquid nitrogen. RNA isolation was performed using an ExtractRNA reagent (Evrogen, Moscow, Russia) in accordance with the manufacturer’s recommendations. To remove traces of genomic DNA, RNA samples were treated with DNase I (Thermo Scientific, Vilnius, Lithuania). Reverse transcription was performed using the MMLV RT reagent kit (Evrogen, Moscow, Russia) and murine RNase Inhibitor (New England Biolabs, Ipswich, MA, USA) as recommended by the manufacturer. An equimolar mixture of random decaprimer (Evrogen, Moscow, Russia) and oligo(dT)15 primer (Evrogen, Moscow, Russia) was used; the concentration of each primer in the reaction was 1 μM. After reverse transcription, the reaction mixture was diluted 8-fold with deionized water.
4.4. qPCR
Relative quantities of mRNAs for the genes of interest were evaluated with a BioRad CFX384 real-time PCR station (BioRad, Singapore) using a qPCRmixHS SYBR + LowROX mix for PCR (Evrogen, Moscow, Russia) according to the manufacturer’s recommendations. Relative quantities of mRNAs were normalized to the to the geometric mean of the mRNA expression levels for the ywhaz and osbp genes. The quality of the DNase treatment was evaluated in all the samples and genes by performing a negative control qPCR with the product of DNase I treatment. Primers for qPCR were designed for the mRNA sequences (Table 1) from the NCBI database using the PrimerSelect software package (DNASTAR Lasergene). Gene expression was analyzed by the E−ΔΔCt method.

Table 1.
Primer sequences for qPCR.
4.5. Data Analysis
The data are presented as mean ± SD. All groups were examined for the presence of outliers using the interquartile range method; the outlying values were automatically excluded from analysis by software. Since data distribution did not pass the normality Shapiro–Wilk test, the significance of differences between groups was evaluated using the Kruskal–Wallis ANOVA (the level of significance was p < 0.017 for comparison of three groups) followed by Dunn’s post hoc test (the level of significance was p < 0.025) using scripts on R with commands kruskal.test() и dunn.test(). In the case of comparison of two groups, the Mann–Whitney test was used (the level of significance was p < 0.05).
Author Contributions
Conceptualization, A.P.B., Y.V.D. and V.A.M.; methodology, A.P.B. and Y.V.D.; formal analysis, A.A.K. and T.M.M.; investigation, Y.S.S., A.A.K. and Y.V.D.; writing—original draft preparation, A.P.B. and Y.V.D.; writing—review and editing, A.P.B., Y.V.D., A.A.K. and V.A.M.; visualization, A.A.K., Y.V.D. and T.M.M.; supervision, A.P.B.; project administration, V.A.M.; funding acquisition, A.P.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation, grant no. 22-25-00479.
Institutional Review Board Statement
All experiments were performed in accordance with the ethical principles stated in the EU Directive 2010/63/EU for animal experiments and were approved by the Ethical Committee of the Institute of Higher Nervous Activity and Neurophysiology of the Russian Academy of Sciences (protocol no. 5 from 8 December 2021).
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw data are available from the authors upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Teles-Grilo Ruivo, L.M.; Mellor, J.R. Cholinergic Modulation of Hippocampal Network Function. Front. Synaptic Neurosci. 2013, 5, 2. [Google Scholar] [CrossRef]
- Petersen, P.C.; Buzsáki, G. Cooling of Medial Septum Reveals Theta Phase Lag Coordination of Hippocampal Cell Assemblies. Neuron 2020, 107, 731–744.e3. [Google Scholar] [CrossRef] [PubMed]
- Bolding, K.A.; Ferbinteanu, J.; Fox, S.E.; Muller, R.U. Place Cell Firing Cannot Support Navigation without Intact Septal Circuits. Hippocampus 2020, 30, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Brandon, M.P.; Bogaard, A.R.; Libby, C.P.; Connerney, M.A.; Gupta, K.; Hasselmo, M.E. Reduction of Theta Rhythm Dissociates Grid Cell Spatial Periodicity from Directional Tuning. Science 2011, 332, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Gold, P.E. Impaired and Spared Cholinergic Functions in the Hippocampus after Lesions of the Medial Septum/Vertical Limb of the Diagonal Band with 192 IgG-Saporin. Hippocampus 2004, 14, 170–179. [Google Scholar] [CrossRef]
- Chrobak, J.J.; Stackman, R.W.; Walsh, T.J. Intraseptal Administration of Muscimol Produces Dose-Dependent Memory Impairments in the Rat. Behav. Neural Biol. 1989, 52, 357–369. [Google Scholar] [CrossRef]
- Givens, B.S.; Olton, D.S. Cholinergic and GABAergic Modulation of Medial Septal Area: Effect on Working Memory. Behav. Neurosci. 1990, 104, 849–855. [Google Scholar] [CrossRef]
- Jeffery, K.J.; Donnett, J.G.; O’keefe, J. Medial Septal Control of Theta-Correlated Unit Firing in the Entorhinal Cortex of Awake Rats. Neuroreport 1995, 6, 2166–2170. [Google Scholar] [CrossRef]
- Leutgeb, S.; Mizumori, S.J.Y. Excitotoxic Septal Lesions Result in Spatial Memory Deficits and Altered Flexibility of Hippocampal Single-Unit Representations. J. Neurosci. 1999, 19, 6661–6672. [Google Scholar] [CrossRef]
- Winson, J. Loss of Hippocampal Theta Rhythm Results in Spatial Memory Deficit in the Rat. Science 1978, 201, 160–163. [Google Scholar] [CrossRef]
- Fantie, B.D.; Goddard, G.V. Septal Modulation of the Population Spike in the Fascia Dentata Produced by Perforant Path Stimulation in the Rat. Brain Res. 1982, 252, 227–237. [Google Scholar] [CrossRef]
- Jeantet, Y.; Jaffard, R. Influence of the Medial Septal Nucleus on the Excitability of the Commissural Path-CA1 Pyramidal Cell Synapse in the Hippocampus of Freely Moving Mice. Neuroscience 1983, 8, 291–297. [Google Scholar] [CrossRef]
- Krnjević, K.; Ropert, N. Electrophysiological and Pharmacological Characteristics of Facilitation of Hippocampal Population Spikes by Stimulation of the Medial Septum. Neuroscience 1982, 7, 2165–2183. [Google Scholar] [CrossRef]
- Robinson, G.B.; Racine, R.J. Interactions between Septal and Entorhinal Inputs to the Rat Dentate Gyrus: Facilitation Effects. Brain Res. 1986, 379, 63–67. [Google Scholar] [CrossRef]
- Albrecht, C.; von der Kammer, H.; Mayhaus, M.; Klaudiny, J.; Schweitzer, M.; Nitsch, R.M. Muscarinic Acetylcholine Receptors Induce the Expression of the Immediate Early Growth Regulatory Gene CYR61. J. Biol. Chem. 2000, 275, 28929–28936. [Google Scholar] [CrossRef]
- Von Der Kammer, H.; Albrecht, C.; Mayhaus, M.; Hoffmann, B.; Stanke, G.; Nitsch, R.M. Identification of Genes Regulated by Muscarinic Acetylcholine Receptors: Application of an Improved and Statistically Comprehensive MRNA Differential Display Technique. Nucleic Acids Res. 1999, 27, 2211–2218. [Google Scholar] [CrossRef][Green Version]
- Hughes, P.; Dragunow, M. Muscarinic Receptor-Mediated Induction of Fos Protein in Rat Brain. Neurosci. Lett. 1993, 150, 122–126. [Google Scholar] [CrossRef]
- Chang, K.T.; Berg, D.K. Voltage-Gated Channels Block Nicotinic Regulation of CREB Phosphorylation and Gene Expression in Neurons. Neuron 2001, 32, 855–865. [Google Scholar] [CrossRef]
- Hu, M.; Liu, Q.S.; Chang, K.T.; Berg, D.K. Nicotinic Regulation of CREB Activation in Hippocampal Neurons by Glutamatergic and Nonglutamatergic Pathways. Mol. Cell. Neurosci. 2002, 21, 616–625. [Google Scholar] [CrossRef]
- Lindefors, N.; Ernfors, P.; Falkenberg, T.; Persson, H. Septal Cholinergic Afferents Regulate Expression of Brain-Derived Neurotrophic Factor and Beta-Nerve Growth Factor MRNA in Rat Hippocampus. Exp. Brain Res. 1992, 88, 78–90. [Google Scholar] [CrossRef]
- Fomenko, A.; Lee, D.J.; McKinnon, C.; Lee, E.J.; de Snoo, M.L.; Gondard, E.; Neudorfer, C.; Hamani, C.; Lozano, A.M.; Kalia, L.V.; et al. Deep Brain Stimulation of the Medial Septal Nucleus Induces Expression of a Virally Delivered Reporter Gene in Dentate Gyrus. Front. Neurosci. 2020, 14, 463. [Google Scholar] [CrossRef]
- McNaughton, N.; Miller, J.J. Medial Septal Projections to the Dentate Gyrus of the Rat: Electrophysiological Analysis of Distribution and Plasticity. Exp. Brain Res. 1984, 56, 243–256. [Google Scholar] [CrossRef]
- Mankin, E.A.; Fried, I. Modulation of Human Memory by Deep Brain Stimulation of the Entorhinal-Hippocampal Circuitry. Neuron 2020, 106, 218–235. [Google Scholar] [CrossRef]
- Jeong, D.U.; Lee, J.E.; Lee, S.E.; Chang, W.S.; Kim, S.J.; Chang, J.W. Improvements in Memory after Medial Septum Stimulation Are Associated with Changes in Hippocampal Cholinergic Activity and Neurogenesis. Biomed Res. Int. 2014, 2014, 568587. [Google Scholar] [CrossRef]
- Mamad, O.; McNamara, H.M.; Reilly, R.B.; Tsanov, M. Medial Septum Regulates the Hippocampal Spatial Representation. Front. Behav. Neurosci. 2015, 9, 166. [Google Scholar] [CrossRef]
- Takata, K.; Kimura, H.; Yanagisawa, D.; Harada, K.; Nishimura, K.; Kitamura, Y.; Shimohama, S.; Tooyama, I. Nicotinic Acetylcholine Receptors and Microglia as Therapeutic and Imaging Targets in Alzheimer’s Disease. Molecules 2022, 27, 2780. [Google Scholar] [CrossRef]
- Gamage, R.; Wagnon, I.; Rossetti, I.; Childs, R.; Niedermayer, G.; Chesworth, R.; Gyengesi, E. Cholinergic Modulation of Glial Function During Aging and Chronic Neuroinflammation. Front. Cell. Neurosci. 2020, 14, 577912. [Google Scholar] [CrossRef]
- Minatohara, K.; Akiyoshi, M.; Okuno, H. Role of Immediate-Early Genes in Synaptic Plasticity and Neuronal Ensembles Underlying the Memory Trace. Front. Mol. Neurosci. 2016, 8, 78. [Google Scholar] [CrossRef]
- Krnjević, K.; Ropert, N.; Casullo, J. Septohippocampal Disinhibition. Brain Res. 1988, 438, 182–192. [Google Scholar] [CrossRef]
- Dudar, J.D. The Effect of Septal Nuclei Stimulation on the Release of Acetylcholine from the Rabbit Hippocampus. Brain Res. 1975, 83, 123–133. [Google Scholar] [CrossRef]
- Somjen, G.G. Mechanisms of Spreading Depression and Hypoxic Spreading Depression-like Depolarization. Physiol. Rev. 2001, 81, 1065–1096. [Google Scholar] [CrossRef] [PubMed]
- Hermann, D.M.; Mies, G.; Hossmann, K.A. Expression of C-Fos, JunB, c-Jun, MKP-1 and Hsp72 Following Traumatic Neocortical Lesions in Rats--Relation to Spreading Depression. Neuroscience 1999, 88, 599–608. [Google Scholar] [CrossRef]
- Faraguna, U.; Nelson, A.; Vyazovskiy, V.V.; Cirelli, C.; Tononi, G. Unilateral Cortical Spreading Depression Affects Sleep Need and Induces Molecular and Electrophysiological Signs of Synaptic Potentiation in Vivo. Cereb. Cortex 2010, 20, 2939–2947. [Google Scholar] [CrossRef] [PubMed]
- Sosthenes, M.C.K.; Diniz, D.G.; Roodselaar, J.; Abadie-Guedes, R.; Mendes, F.d.C.C.d.S.; Fernandes, T.N.; Bittencourt, J.C.; Diniz, C.W.P.; Anthony, D.C.; Guedes, R.C.A. Stereological Analysis of Early Gene Expression Using Egr-1 Immunolabeling after Spreading Depression in the Rat Somatosensory Cortex. Front. Neurosci. 2019, 13, 1020. [Google Scholar] [CrossRef]
- Yoshida, K.; Xu, M.; Natsubori, A.; Mimura, M.; Takata, N.; Tanaka, K.F. Identification of the Extent of Cortical Spreading Depression Propagation by Npas4 MRNA Expression. Neurosci. Res. 2015, 98, 1–8. [Google Scholar] [CrossRef]
- Takizawa, T.; Qin, T.; Lopes de Morais, A.; Sugimoto, K.; Chung, J.Y.; Morsett, L.; Mulder, I.; Fischer, P.; Suzuki, T.; Anzabi, M.; et al. Non-Invasively Triggered Spreading Depolarizations Induce a Rapid pro-Inflammatory Response in Cerebral Cortex. J. Cereb. Blood Flow Metab. 2020, 40, 1117–1131. [Google Scholar] [CrossRef]
- Moghaddam, B.; Schenk, J.O.; Stewart, W.B.; Hansen, A.J. Temporal Relationship between Neurotransmitter Release and Ion Flux during Spreading Depression and Anoxia. Can. J. Physiol. Pharmacol. 1987, 65, 1105–1110. [Google Scholar] [CrossRef]
- Fabricius, M.; Jensen, L.H.; Lauritzen, M. Microdialysis of Interstitial Amino Acids during Spreading Depression and Anoxic Depolarization in Rat Neocortex. Brain Res. 1993, 612, 61–69. [Google Scholar] [CrossRef]
- Obrenovitch, T.P.; Urenjak, J.; Wang, M. Nitric Oxide Formation during Cortical Spreading Depression Is Critical for Rapid Subsequent Recovery of Ionic Homeostasis. J. Cereb. Blood Flow Metab. 2002, 22, 680–688. [Google Scholar] [CrossRef]
- Davies, J.A.; Annels, S.J.; Dickie, B.G.M.; Ellis, Y.; Knott, N.J. A Comparison between the Stimulated and Paroxysmal Release of Endogenous Amino Acids from Rat Cerebellar, Striatal and Hippocampal Slices: A Manifestation of Spreading Depression? J. Neurol. Sci. 1995, 131, 8–14. [Google Scholar] [CrossRef]
- Haghir, H.; Kovac, S.; Speckmann, E.J.; Zilles, K.; Gorji, A. Patterns of Neurotransmitter Receptor Distributions Following Cortical Spreading Depression. Neuroscience 2009, 163, 1340–1352. [Google Scholar] [CrossRef]
- Fanselow, M.; Dong, H.-W. Are the Dorsal and Ventral Hippocampus Functionally Distinct Structures. Neuron 2010, 65, 7–19. [Google Scholar] [CrossRef]
- Cembrowski, M.S.; Bachman, J.L.; Wang, L.; Sugino, K.; Shields, B.C.; Spruston, N. Spatial Gene-Expression Gradients Underlie Prominent Heterogeneity of CA1 Pyramidal Neurons. Neuron 2016, 89, 351–368. [Google Scholar] [CrossRef]
- Dutar, P.; Bassant, M.H.; Senut, M.C.; Lamour, Y. The Septohippocampal Pathway: Structure and Function of a Central Cholinergic System. Physiol. Rev. 1995, 75, 393–427. [Google Scholar] [CrossRef]
- Ebert, D.H.; Greenberg, M.E. Activity-Dependent Neuronal Signalling and Autism Spectrum Disorder. Nature 2013, 493, 327. [Google Scholar] [CrossRef]
- Duclot, F.; Kabbaj, M. The Role of Early Growth Response 1 (EGR1) in Brain Plasticity and Neuropsychiatric Disorders. Front. Behav. Neurosci. 2017, 11, 35. [Google Scholar] [CrossRef]
- Fu, J.; Guo, O.; Zhen, Z.; Zhen, J. Essential Functions of the Transcription Factor Npas4 in Neural Circuit Development, Plasticity, and Diseases. Front. Neurosci. 2020, 14, 1262. [Google Scholar] [CrossRef]
- Kaufer, D.; Friedman, A.; Seidman, S.; Soreq, H. Acute Stress Facilitates Long-Lasting Changes in Cholinergic Gene Expression. Nature 1998, 393, 373–377. [Google Scholar] [CrossRef]
- von der Kammer, H.; Demiralay, C.; Andresen, B.; Albrecht, C.; Mayhaus, M.; Nitsch, R.M. Regulation of Gene Expression by Muscarinic Acetylcholine Receptors. Biochem. Soc. Symp. 2001, 1, 131–140. [Google Scholar] [CrossRef]
- Zeisel, A.; Hochgerner, H.; Lönnerberg, P.; Johnsson, A.; Memic, F.; van der Zwan, J.; Häring, M.; Braun, E.; Borm, L.E.; La Manno, G.; et al. Molecular Architecture of the Mouse Nervous System. Cell 2018, 174, 999–1014.e22. [Google Scholar] [CrossRef]
- Zeisel, A.; Muñoz-Manchado, A.B.; Codeluppi, S.; Lönnerberg, P.; La Manno, G.; Juréus, A.; Marques, S.; Munguba, H.; He, L.; Betsholtz, C.; et al. Cell Types in the Mouse Cortex and Hippocampus Revealed by Single-Cell RNA-Seq. Science 2015, 347, 1138–1142. [Google Scholar] [CrossRef]
- Saunders, A.; Macosko, E.Z.; Wysoker, A.; Goldman, M.; Krienen, F.M.; de Rivera, H.; Bien, E.; Baum, M.; Bortolin, L.; Wang, S.; et al. Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 2018, 174, 1015–1030.e16. [Google Scholar] [CrossRef]
- Tasic, B.; Yao, Z.; Graybuck, L.T.; Smith, K.A.; Nguyen, T.N.; Bertagnolli, D.; Goldy, J.; Garren, E.; Economo, M.N.; Viswanathan, S.; et al. Shared and Distinct Transcriptomic Cell Types across Neocortical Areas. Nature 2018, 563, 72–78. [Google Scholar] [CrossRef]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates; Elsevier: San Diego, CA, USA, 2007; ISBN 9780123741219. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).