Endometrial Epithelial ARID1A Is Required for Uterine Immune Homeostasis during Early Pregnancy
Abstract
1. Introduction
2. Results
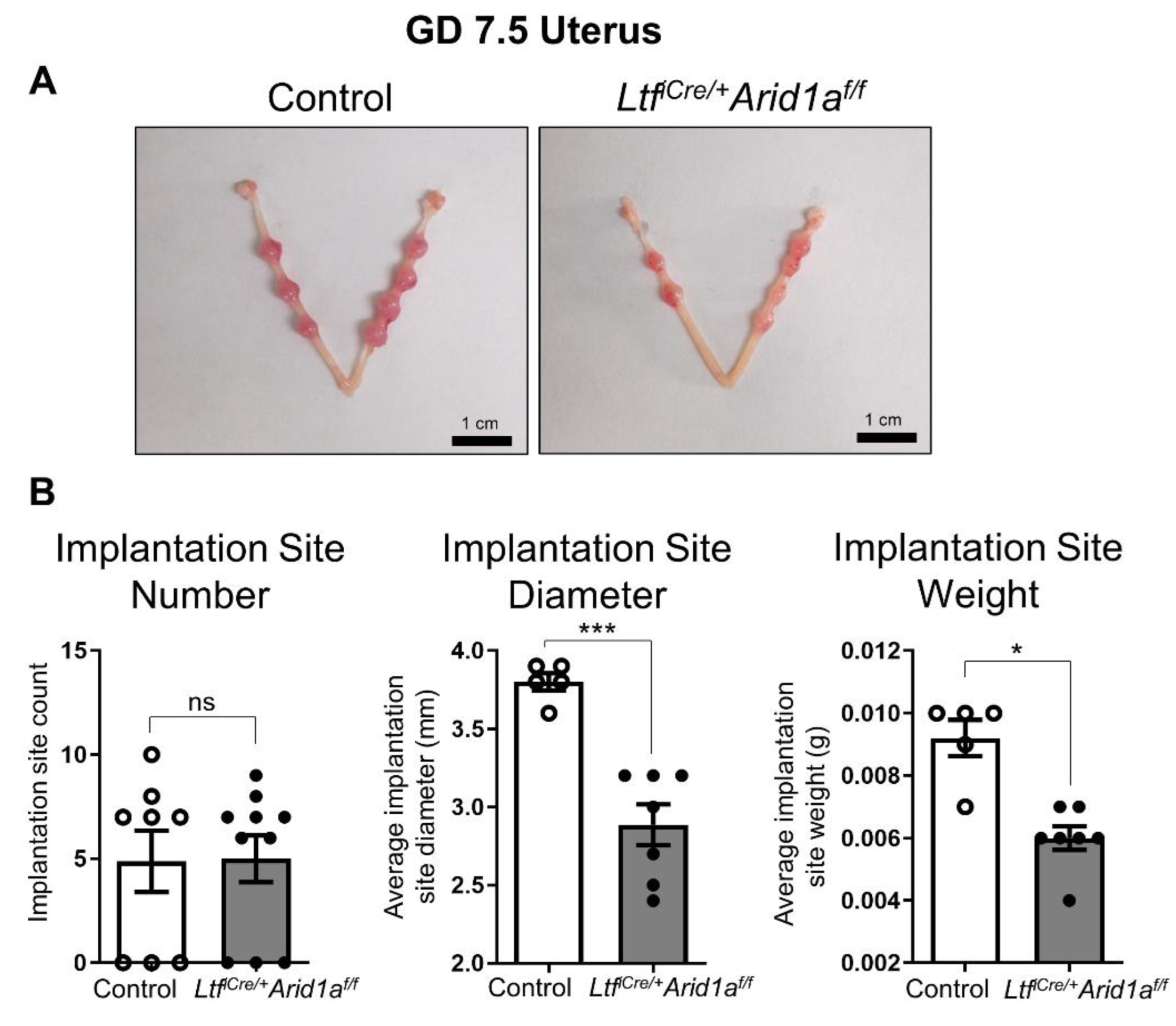
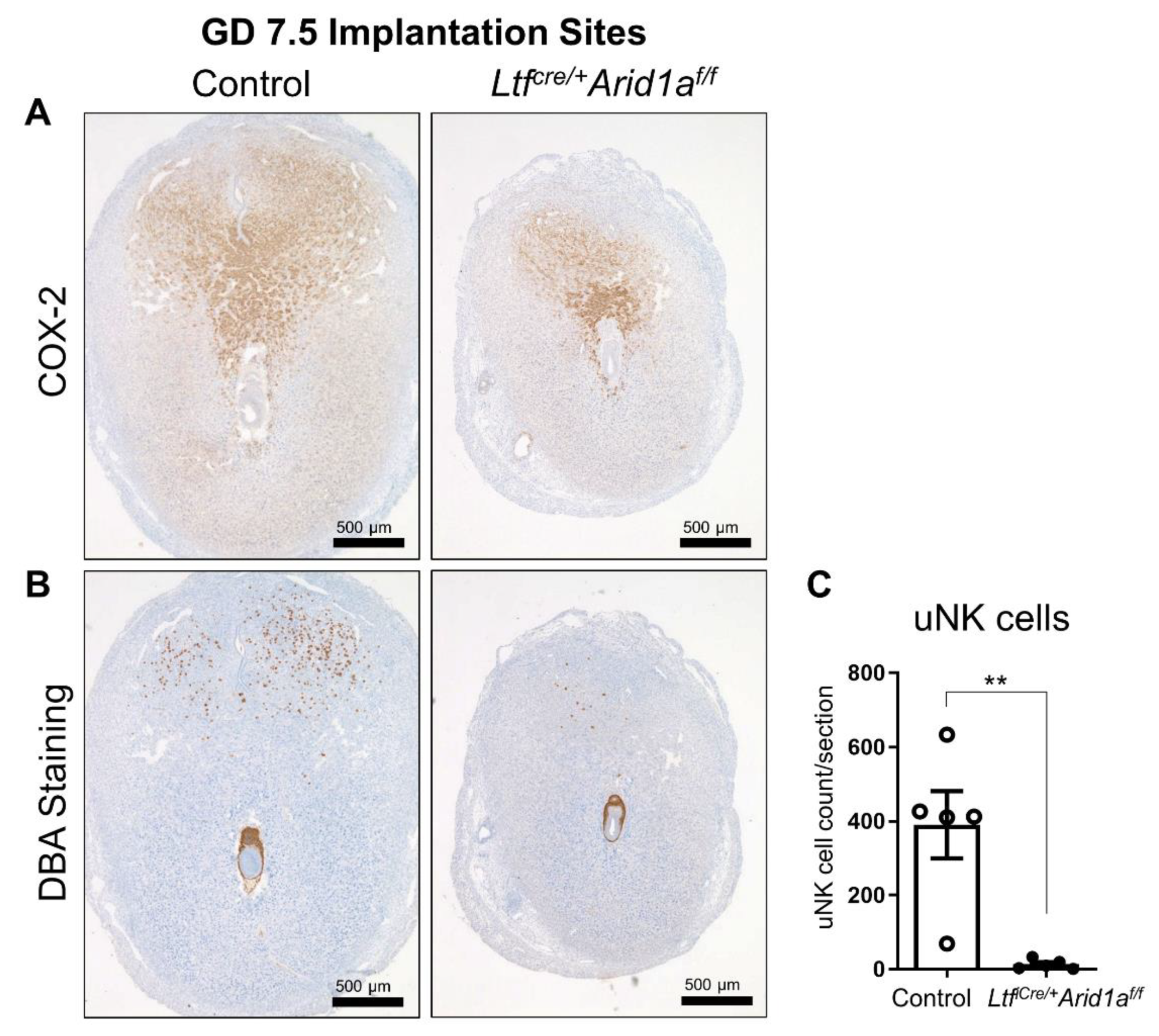
2.1. Deletion of Endometrial Epithelial Arid1a in Mice Causes Diminished Implantation Site Size and uNK Cell Numbers at GD 7.5
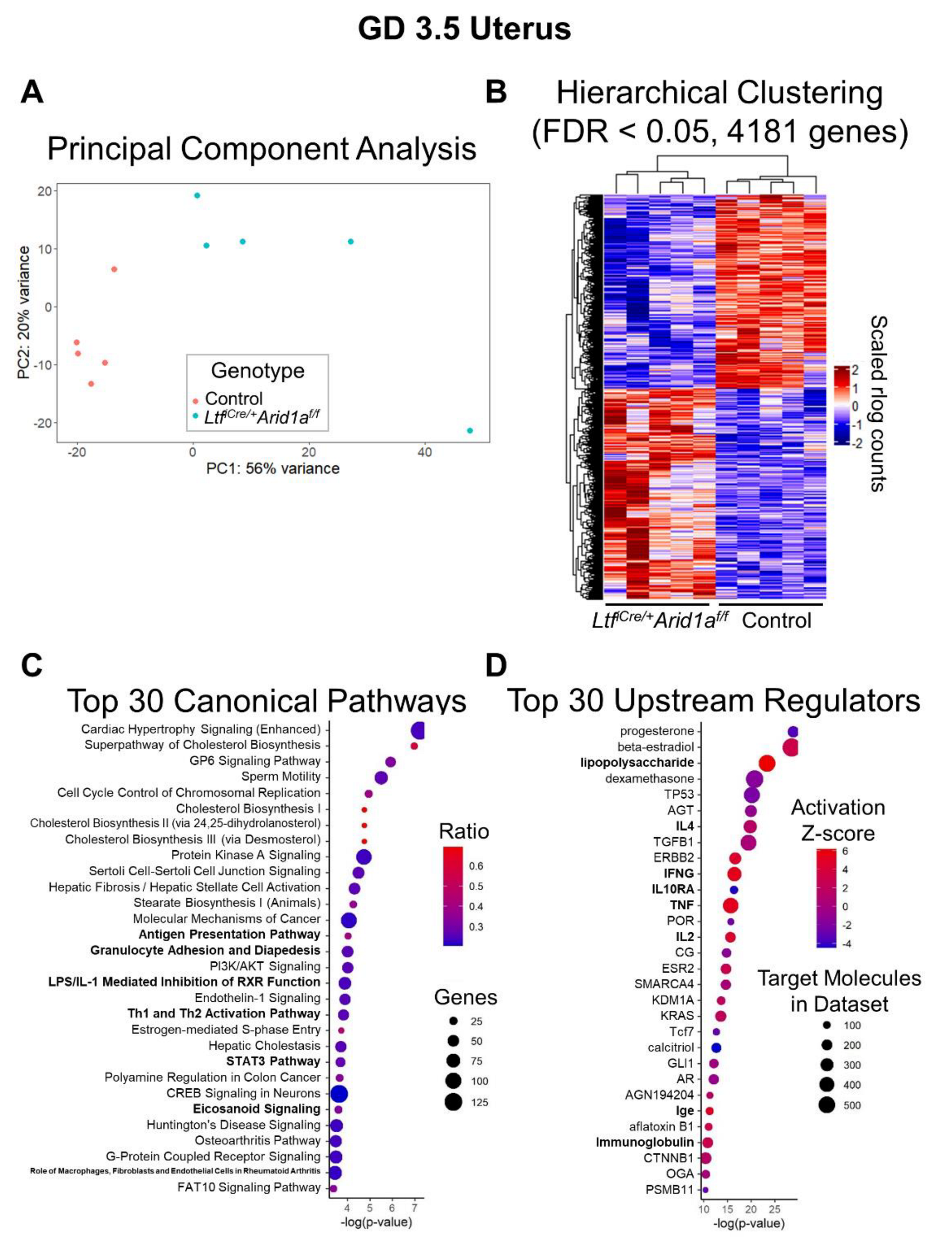
2.2. RNA-Sequencing Analysis of LtfiCre/+Arid1af/f Uteri at GD 3.5 Reveals Altered Immune Pathways
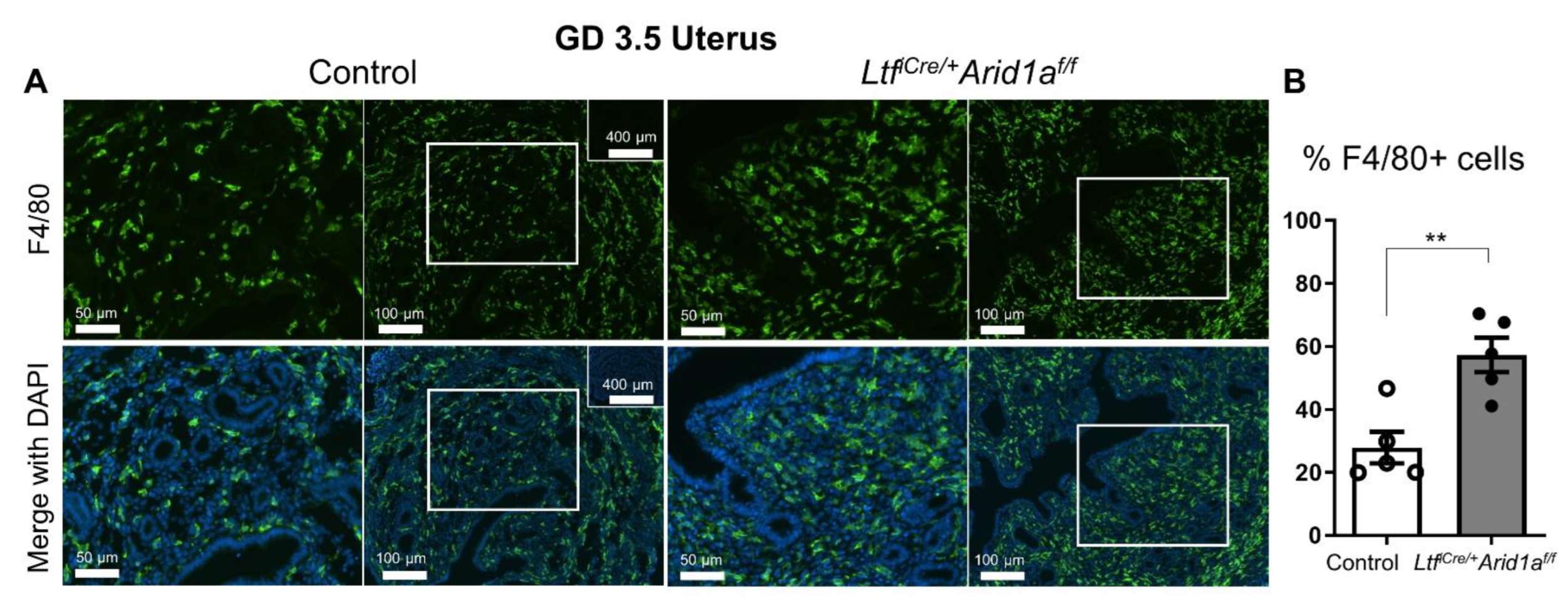
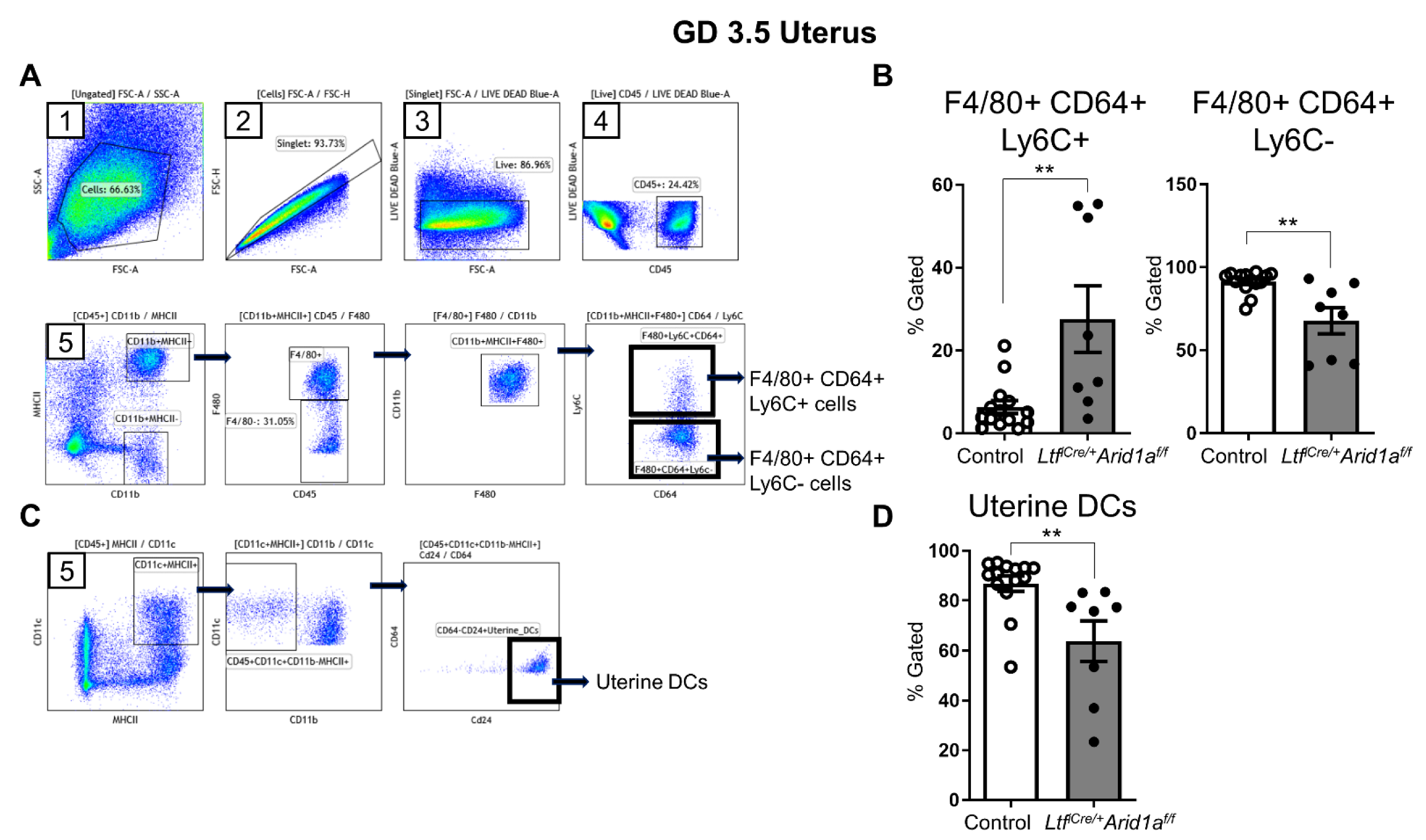
2.3. Uterine Macrophage Numbers Are Elevated in LtfiCre/+Arid1af/f Mice at GD 3.5
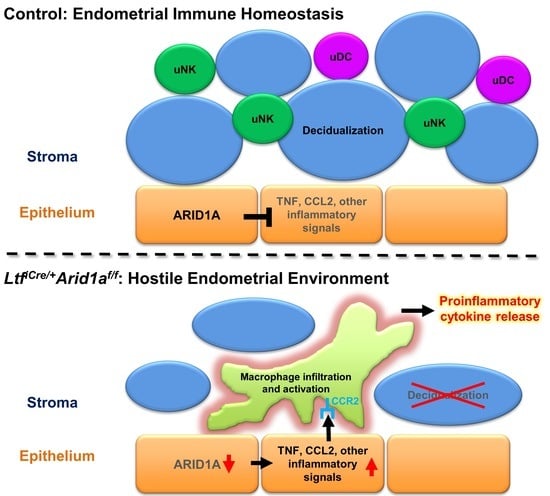
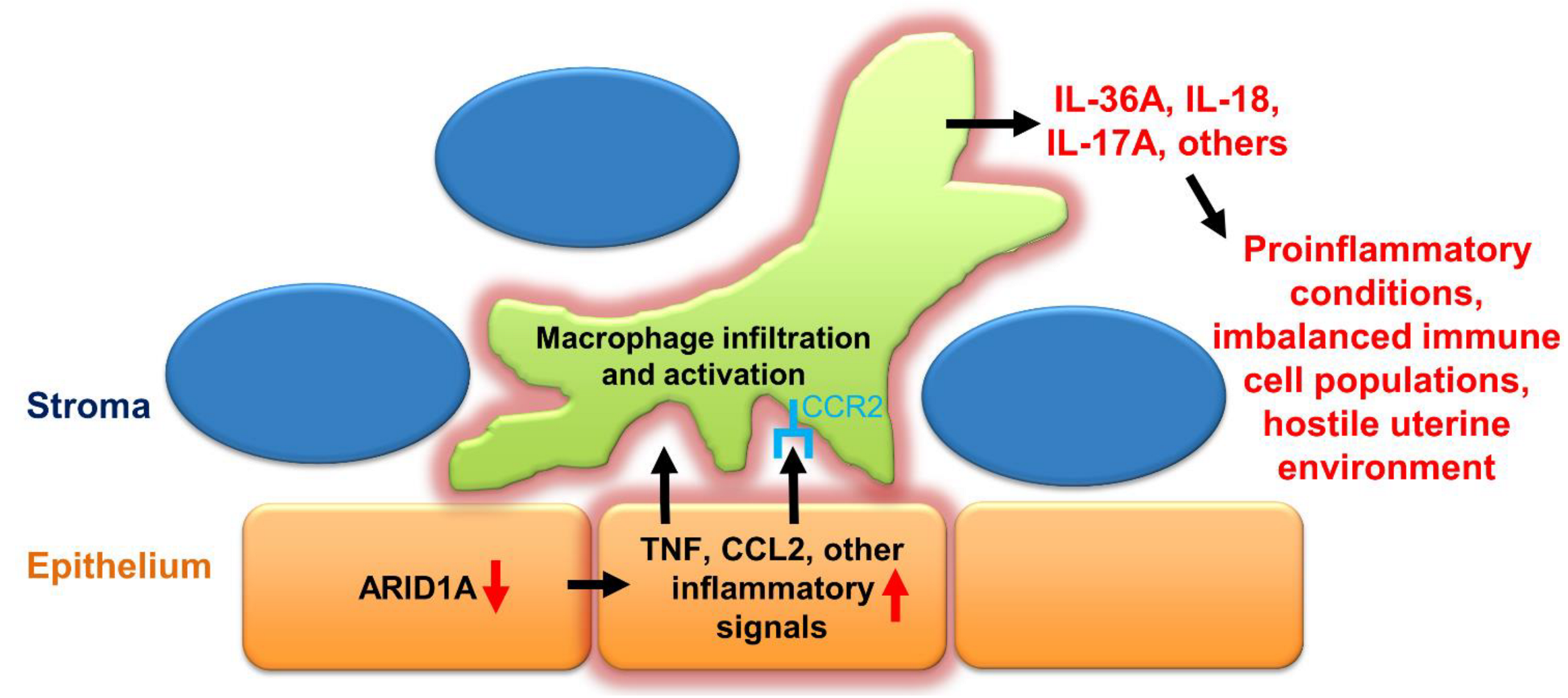
3. Discussion
4. Materials and Methods
4.1. Mouse Models
4.2. Mouse Procedures and Tissue Collection
4.3. Histology and Immunostaining
4.4. RNA Isolation and RT-qPCR
4.5. RNA-Sequencing
4.6. RNA-Sequencing Analysis
4.7. Flow Cytometry
4.8. Statistical Analysis
4.9. Data and Code Availability
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kelleher, A.M.; DeMayo, F.J.; Spencer, T.E. Uterine Glands: Developmental Biology and Functional Roles in Pregnancy. Endocr. Rev. 2019, 40, 1424–1445. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.P.; Li, R.; DeMayo, F.J. Progesterone Receptor Regulation of Uterine Adaptation for Pregnancy. Trends Endocrinol. Metab. 2018, 29, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, X.; Huang, Z.; Balaji, J.; Kim, T.H.; Wang, T.; Zhou, L.; Deleon, A.; Cook, M.E.; Marbrey, M.W.; et al. The role of epithelial progesterone receptor isoforms in embryo implantation. iScience 2021, 24, 103487. [Google Scholar] [CrossRef]
- Gellersen, B.; Brosens, J.J. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr. Rev. 2014, 35, 851–905. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.; Sun, X.; Dey, S.K. Mechanisms of implantation: Strategies for successful pregnancy. Nat. Med. 2012, 18, 1754–1767. [Google Scholar] [CrossRef] [PubMed]
- Ticconi, C.; Di Simone, N.; Campagnolo, L.; Fazleabas, A. Clinical consequences of defective decidualization. Tissue Cell 2021, 72, 101586. [Google Scholar] [CrossRef]
- Mori, M.; Bogdan, A.; Balassa, T.; Csabai, T.; Szekeres-Bartho, J. The decidua-the maternal bed embracing the embryo-maintains the pregnancy. Semin. Immunopathol. 2016, 38, 635–649. [Google Scholar] [CrossRef]
- Koopman, L.A.; Kopcow, H.D.; Rybalov, B.; Boyson, J.E.; Orange, J.S.; Schatz, F.; Masch, R.; Lockwood, C.J.; Schachter, A.D.; Park, P.J.; et al. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J. Exp. Med. 2003, 198, 1201–1212. [Google Scholar] [CrossRef]
- Brighton, P.J.; Maruyama, Y.; Fishwick, K.; Vrljicak, P.; Tewary, S.; Fujihara, R.; Muter, J.; Lucas, E.S.; Yamada, T.; Woods, L.; et al. Clearance of senescent decidual cells by uterine natural killer cells in cycling human endometrium. eLife 2017, 6, e31274. [Google Scholar] [CrossRef]
- Lucas, E.S.; Vrljicak, P.; Muter, J.; Diniz-da-Costa, M.M.; Brighton, P.J.; Kong, C.S.; Lipecki, J.; Fishwick, K.J.; Odendaal, J.; Ewington, L.J.; et al. Recurrent pregnancy loss is associated with a pro-senescent decidual response during the peri-implantation window. Commun. Biol. 2020, 3, 37. [Google Scholar] [CrossRef]
- Croy, B.A.; Zhang, J.; Tayade, C.; Colucci, F.; Yadi, H.; Yamada, A.T. Analysis of uterine natural killer cells in mice. Methods Mol. Biol. 2010, 612, 465–503. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.; Rees, A.; Cronin, J.G.; Nair, M.; Jones, N.; Thornton, C.A. Macrophage Plasticity in Reproduction and Environmental Influences on Their Function. Front. Immunol. 2020, 11, 607328. [Google Scholar] [CrossRef] [PubMed]
- Plaks, V.; Birnberg, T.; Berkutzki, T.; Sela, S.; BenYashar, A.; Kalchenko, V.; Mor, G.; Keshet, E.; Dekel, N.; Neeman, M.; et al. Uterine DCs are crucial for decidua formation during embryo implantation in mice. J. Clin. Investig. 2008, 118, 3954–3965. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.K.; Tay, C.S.; Erlebacher, A. Dendritic cell entrapment within the pregnant uterus inhibits immune surveillance of the maternal/fetal interface in mice. J. Clin. Investig. 2009, 119, 2062–2073. [Google Scholar] [CrossRef] [PubMed]
- Vallve-Juanico, J.; Houshdaran, S.; Giudice, L.C. The endometrial immune environment of women with endometriosis. Hum. Reprod. Updat. 2019, 25, 564–591. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, R.M.; Kim, T.H.; Shin, J.H.; Jeong, J.W. Progesterone and Estrogen Signaling in the Endometrium: What Goes Wrong in Endometriosis? Int. J. Mol. Sci. 2019, 20, 3822. [Google Scholar] [CrossRef]
- Saunders, P.T.K.; Horne, A.W. Endometriosis: Etiology, pathobiology, and therapeutic prospects. Cell 2021, 184, 2807–2824. [Google Scholar] [CrossRef]
- Zondervan, K.T.; Becker, C.M.; Missmer, S.A. Endometriosis. N. Engl. J. Med. 2020, 382, 1244–1256. [Google Scholar] [CrossRef]
- Bulun, S.E.; Yilmaz, B.D.; Sison, C.; Miyazaki, K.; Bernardi, L.; Liu, S.; Kohlmeier, A.; Yin, P.; Milad, M.; Wei, J. Endometriosis. Endocr. Rev. 2019, 40, 1048–1079. [Google Scholar] [CrossRef]
- Kim, T.H.; Young, S.L.; Sasaki, T.; Deaton, J.L.; Schammel, D.P.; Palomino, A.W.; Jeong, J.W.; Lessey, B.A. Role of SIRT1 and Progesterone Resistance in Normal and Abnormal Endometrium. J. Clin. Endocrinol. Metab. 2021, 107, 788–800. [Google Scholar] [CrossRef]
- Yoo, J.Y.; Kim, T.H.; Fazleabas, A.T.; Palomino, W.A.; Ahn, S.H.; Tayade, C.; Schammel, D.P.; Young, S.L.; Jeong, J.W.; Lessey, B.A. KRAS Activation and over-expression of SIRT1/BCL6 Contributes to the Pathogenesis of Endometriosis and Progesterone Resistance. Sci. Rep. 2017, 7, 6765. [Google Scholar] [CrossRef] [PubMed]
- Mai, H.; Liao, Y.; Luo, S.; Wei, K.; Yang, F.; Shi, H. Histone deacetylase HDAC2 silencing prevents endometriosis by activating the HNF4A/ARID1A axis. J. Cell. Mol. Med. 2021, 25, 9972–9982. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Yoo, J.Y.; Choi, K.C.; Shin, J.H.; Leach, R.E.; Fazleabas, A.T.; Young, S.L.; Lessey, B.A.; Yoon, H.G.; Jeong, J.W. Loss of HDAC3 results in nonreceptive endometrium and female infertility. Sci. Transl. Med. 2019, 11, eaaf7533. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Yoo, J.Y.; Wang, Z.; Lydon, J.P.; Khatri, S.; Hawkins, S.M.; Leach, R.E.; Fazleabas, A.T.; Young, S.L.; Lessey, B.A.; et al. ARID1A Is Essential for Endometrial Function during Early Pregnancy. PLoS Genet. 2015, 11, e1005537. [Google Scholar] [CrossRef]
- Zhang, Q.; Dong, P.; Liu, X.; Sakuragi, N.; Guo, S.W. Enhancer of Zeste homolog 2 (EZH2) induces epithelial-mesenchymal transition in endometriosis. Sci. Rep. 2017, 7, 6804. [Google Scholar] [CrossRef]
- Maeda, D.; Shih Ie, M. Pathogenesis and the role of ARID1A mutation in endometriosis-related ovarian neoplasms. Adv. Anat. Pathol. 2013, 20, 45–52. [Google Scholar] [CrossRef]
- Anglesio, M.S.; Papadopoulos, N.; Ayhan, A.; Nazeran, T.M.; Noe, M.; Horlings, H.M.; Lum, A.; Jones, S.; Senz, J.; Seckin, T.; et al. Cancer-Associated Mutations in Endometriosis without Cancer. N. Engl. J. Med. 2017, 376, 1835–1848. [Google Scholar] [CrossRef]
- Kim, H.I.; Kim, T.H.; Yoo, J.Y.; Young, S.L.; Lessey, B.A.; Ku, B.J.; Jeong, J.W. ARID1A and PGR proteins interact in the endometrium and reveal a positive correlation in endometriosis. Biochem. Biophys. Res. Commun. 2021, 550, 151–157. [Google Scholar] [CrossRef]
- Wilson, M.R.; Reske, J.J.; Holladay, J.; Wilber, G.E.; Rhodes, M.; Koeman, J.; Adams, M.; Johnson, B.; Su, R.W.; Joshi, N.R.; et al. ARID1A and PI3-kinase pathway mutations in the endometrium drive epithelial transdifferentiation and collective invasion. Nat. Commun. 2019, 10, 3554. [Google Scholar] [CrossRef]
- Reske, J.J.; Wilson, M.R.; Holladay, J.; Siwicki, R.A.; Skalski, H.; Harkins, S.; Adams, M.; Risinger, J.I.; Hostetter, G.; Lin, K.; et al. Co-existing TP53 and ARID1A mutations promote aggressive endometrial tumorigenesis. PLoS Genet. 2021, 17, e1009986. [Google Scholar] [CrossRef]
- Marquardt, R.M.; Kim, T.H.; Yoo, J.Y.; Teasley, H.E.; Fazleabas, A.T.; Young, S.L.; Lessey, B.A.; Arora, R.; Jeong, J.W. Endometrial epithelial ARID1A is critical for uterine gland function in early pregnancy establishment. FASEB J. 2021, 35, e21209. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.A. GM-CSF-Dependent Inflammatory Pathways. Front. Immunol. 2019, 10, 2055. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Kalish, F.; Schulz, S.; Yang, Y.; Wong, R.J.; Stevenson, D.K. Unique roles of infiltrating myeloid cells in the murine uterus during early to midpregnancy. J. Immunol. 2015, 194, 3713–3722. [Google Scholar] [CrossRef] [PubMed]
- Akinrinmade, O.A.; Chetty, S.; Daramola, A.K.; Islam, M.U.; Thepen, T.; Barth, S. CD64: An Attractive Immunotherapeutic Target for M1-type Macrophage Mediated Chronic Inflammatory Diseases. Biomedicines 2017, 5, 56. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, Y.; Pei, G.; Deng, X.; Jiang, H.; Wu, J.; Zhou, C.; Guo, Y.; Yao, Y.; Zeng, R.; et al. Bone marrow-derived Ly6C(-) macrophages promote ischemia-induced chronic kidney disease. Cell Death Dis. 2019, 10, 291. [Google Scholar] [CrossRef]
- Groves, A.M.; Johnston, C.J.; Williams, J.P.; Finkelstein, J.N. Role of Infiltrating Monocytes in the Development of Radiation-Induced Pulmonary Fibrosis. Radiat. Res. 2018, 189, 300–311. [Google Scholar] [CrossRef]
- Krishnarajah, S.; Ingelfinger, F.; Friebel, E.; Cansever, D.; Amorim, A.; Andreadou, M.; Bamert, D.; Litscher, G.; Lutz, M.; Mayoux, M.; et al. Single-cell profiling of immune system alterations in lymphoid, barrier and solid tissues in aged mice. Nat. Aging 2022, 2, 74–89. [Google Scholar] [CrossRef]
- Yu, Y.R.; O’Koren, E.G.; Hotten, D.F.; Kan, M.J.; Kopin, D.; Nelson, E.R.; Que, L.; Gunn, M.D. A Protocol for the Comprehensive Flow Cytometric Analysis of Immune Cells in Normal and Inflamed Murine Non-Lymphoid Tissues. PLoS ONE 2016, 11, e0150606. [Google Scholar] [CrossRef]
- Hogg, C.; Panir, K.; Dhami, P.; Rosser, M.; Mack, M.; Soong, D.; Pollard, J.W.; Jenkins, S.J.; Horne, A.W.; Greaves, E. Macrophages inhibit and enhance endometriosis depending on their origin. Proc. Natl. Acad. Sci. USA 2021, 118, e2013776118. [Google Scholar] [CrossRef]
- Zondervan, K.T.; Becker, C.M.; Koga, K.; Missmer, S.A.; Taylor, R.N.; Vigano, P. Endometriosis. Nat. Rev. Dis. Primers 2018, 4, 9. [Google Scholar] [CrossRef]
- Ahn, S.H.; Khalaj, K.; Young, S.L.; Lessey, B.A.; Koti, M.; Tayade, C. Immune-inflammation gene signatures in endometriosis patients. Fertil. Steril. 2016, 106, 1420–1431 e1427. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.H.; Edwards, A.K.; Singh, S.S.; Young, S.L.; Lessey, B.A.; Tayade, C. IL-17A Contributes to the Pathogenesis of Endometriosis by Triggering Proinflammatory Cytokines and Angiogenic Growth Factors. J. Immunol. 2015, 195, 2591–2600. [Google Scholar] [CrossRef] [PubMed]
- Richter, O.N.; Dorn, C.; Rosing, B.; Flaskamp, C.; Ulrich, U. Tumor necrosis factor alpha secretion by peritoneal macrophages in patients with endometriosis. Arch. Gynecol. Obs. 2005, 271, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Ledee-Bataille, N.; Olivennes, F.; Kadoch, J.; Dubanchet, S.; Frydman, N.; Chaouat, G.; Frydman, R. Detectable levels of interleukin-18 in uterine luminal secretions at oocyte retrieval predict failure of the embryo transfer. Hum. Reprod. 2004, 19, 1968–1973. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.; Moor, J.; Jenkins, C.; Miller, H.; Walker, J.J.; McLean, M.A.; Norman, J.; McInnes, I.B. Abnormal first trimester serum interleukin 18 levels are associated with a poor outcome in women with a history of recurrent miscarriage. Am. J. Reprod. Immunol. 2004, 51, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.; Knofler, M. Human tumour necrosis factor: Physiological and pathological roles in placenta and endometrium. Placenta 2009, 30, 111–123. [Google Scholar] [CrossRef]
- Winger, E.E.; Reed, J.L. Treatment with tumor necrosis factor inhibitors and intravenous immunoglobulin improves live birth rates in women with recurrent spontaneous abortion. Am. J. Reprod. Immunol. 2008, 60, 8–16. [Google Scholar] [CrossRef]
- Murrieta-Coxca, J.M.; Gomez-Chavez, F.; Baeza-Martinez, D.A.; Cancino-Diaz, M.E.; Cancino-Diaz, J.C.; Perez-Tapia, S.M.; Reyes-Maldonado, E.; Rodriguez-Martinez, S. Estrous Cycle and Gestational Age-Dependent Expression of Members of the Interleukin-36 Subfamily in a Semi-Allogeneic Model of Infected and Non-Infected Murine Pregnancy. Front. Immunol. 2016, 7, 376. [Google Scholar] [CrossRef]
- Dosiou, C.; Giudice, L.C. Natural killer cells in pregnancy and recurrent pregnancy loss: Endocrine and immunologic perspectives. Endocr. Rev. 2005, 26, 44–62. [Google Scholar] [CrossRef]
- Renaud, S.J.; Scott, R.L.; Chakraborty, D.; Rumi, M.A.; Soares, M.J. Natural killer-cell deficiency alters placental development in rats. Biol. Reprod. 2017, 96, 145–158. [Google Scholar] [CrossRef]
- Ashkar, A.A.; Black, G.P.; Wei, Q.; He, H.; Liang, L.; Head, J.R.; Croy, B.A. Assessment of requirements for IL-15 and IFN regulatory factors in uterine NK cell differentiation and function during pregnancy. J. Immunol. 2003, 171, 2937–2944. [Google Scholar] [CrossRef] [PubMed]
- Ratsep, M.T.; Felker, A.M.; Kay, V.R.; Tolusso, L.; Hofmann, A.P.; Croy, B.A. Uterine natural killer cells: Supervisors of vasculature construction in early decidua basalis. Reproduction 2015, 149, R91-102. [Google Scholar] [CrossRef] [PubMed]
- Schulke, L.; Berbic, M.; Manconi, F.; Tokushige, N.; Markham, R.; Fraser, I.S. Dendritic cell populations in the eutopic and ectopic endometrium of women with endometriosis. Hum. Reprod. 2009, 24, 1695–1703. [Google Scholar] [CrossRef] [PubMed]
- Maridas, D.E.; Hey-Cunningham, A.J.; Ng, C.H.M.; Markham, R.; Fraser, I.S.; Berbic, M. Peripheral and endometrial dendritic cell populations during the normal cycle and in the presence of endometriosis. J. Endometr. Pelvic Pain Disord. 2014, 6, 67–119. [Google Scholar] [CrossRef]
- Parameswaran, N.; Patial, S. Tumor necrosis factor-alpha signaling in macrophages. Crit. Rev. Eukaryot. Gene Expr. 2010, 20, 87–103. [Google Scholar] [CrossRef]
- Straub, R.H. The complex role of estrogens in inflammation. Endocr. Rev. 2007, 28, 521–574. [Google Scholar] [CrossRef]
- Schofield, G.; Kimber, S.J. Leukocyte subpopulations in the uteri of leukemia inhibitory factor knockout mice during early pregnancy. Biol. Reprod. 2005, 72, 872–878. [Google Scholar] [CrossRef][Green Version]
- Laird, S.M.; Tuckerman, E.M.; Saravelos, H.; Li, T.C. The production of tumour necrosis factor alpha (TNF-alpha) by human endometrial cells in culture. Hum. Reprod. 1996, 11, 1318–1323. [Google Scholar] [CrossRef][Green Version]
- Roulis, M.; Armaka, M.; Manoloukos, M.; Apostolaki, M.; Kollias, G. Intestinal epithelial cells as producers but not targets of chronic TNF suffice to cause murine Crohn-like pathology. Proc. Natl. Acad. Sci. USA 2011, 108, 5396–5401. [Google Scholar] [CrossRef]
- Robertson, S.A.; Mayrhofer, G.; Seamark, R.F. Uterine epithelial cells synthesize granulocyte-macrophage colony-stimulating factor and interleukin-6 in pregnant and nonpregnant mice. Biol. Reprod. 1992, 46, 1069–1079. [Google Scholar] [CrossRef]
- Zhao, Y.; Chegini, N. The expression of granulocyte macrophage-colony stimulating factor (GM-CSF) and receptors in human endometrium. Am. J. Reprod. Immunol. 1999, 42, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Deshmane, S.L.; Kremlev, S.; Amini, S.; Sawaya, B.E. Monocyte chemoattractant protein-1 (MCP-1): An overview. J. Interferon. Cytokine Res. 2009, 29, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Jolicoeur, C.; Boutouil, M.; Drouin, R.; Paradis, I.; Lemay, A.; Akoum, A. Increased expression of monocyte chemotactic protein-1 in the endometrium of women with endometriosis. Am. J. Pathol. 1998, 152, 125–133. [Google Scholar]
- Daikoku, T.; Ogawa, Y.; Terakawa, J.; Ogawa, A.; DeFalco, T.; Dey, S.K. Lactoferrin-iCre: A new mouse line to study uterine epithelial gene function. Endocrinology 2014, 155, 2718–2724. [Google Scholar] [CrossRef]
- Kovacic, B.; Hoelbl-Kovacic, A.; Fischhuber, K.M.; Leitner, N.R.; Gotthardt, D.; Casanova, E.; Sexl, V.; Muller, M. Lactotransferrin-Cre reporter mice trace neutrophils, monocytes/macrophages and distinct subtypes of dendritic cells. Haematologica 2014, 99, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Cunha, G.R.; Sinclair, A.; Ricke, W.A.; Robboy, S.J.; Cao, M.; Baskin, L.S. Reproductive tract biology: Of mice and men. Differentiation 2019, 110, 49–63. [Google Scholar] [CrossRef]
- Wang, H.; Dey, S.K. Roadmap to embryo implantation: Clues from mouse models. Nat. Rev. Genet. 2006, 7, 185–199. [Google Scholar] [CrossRef]
- Gao, X.; Tate, P.; Hu, P.; Tjian, R.; Skarnes, W.C.; Wang, Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl. Acad. Sci. USA 2008, 105, 6656–6661. [Google Scholar] [CrossRef]
- Mendes-Jorge, L.; Ramos, D.; Valenca, A.; Lopez-Luppo, M.; Pires, V.M.R.; Catita, J.; Nacher, V.; Navarro, M.; Carretero, A.; Rodriguez-Baeza, A.; et al. Correction: L-Ferritin Binding to Scara5: A New Iron Traffic Pathway Potentially Implicated in Retinopathy. PLoS ONE 2017, 12, e0180288. [Google Scholar] [CrossRef]
- Nittoli, V.; Colella, M.; Porciello, A.; Reale, C.; Roberto, L.; Russo, F.; Russo, N.A.; Porreca, I.; De Felice, M.; Mallardo, M.; et al. Multi Species Analyses Reveal Testicular T3 Metabolism and Signalling as a Target of Environmental Pesticides. Cells 2021, 10, 2187. [Google Scholar] [CrossRef]
- Allam, R.; Maillard, M.H.; Tardivel, A.; Chennupati, V.; Bega, H.; Yu, C.W.; Velin, D.; Schneider, P.; Maslowski, K.M. Epithelial NAIPs protect against colonic tumorigenesis. J. Exp. Med. 2015, 212, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, T.; Tomari, H.; Onoyama, I.; Araki, H.; Yasunaga, M.; Lin, C.; Kawamura, K.; Yokota, N.; Yoshida, S.; Yagi, H.; et al. Identification of genes associated with endometrial cell ageing. Mol. Hum. Reprod. 2021, 27, gaaa078. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Zhuang, F.; Liu, J.T.; Li, N.; Chen, Y.X.; Su, X.B.; Yao, A.H.; Yao, Q.P.; Han, Y.; Li, S.S.; et al. Single-cell analyses reveal functional classification of dendritic cells and their potential roles in inflammatory disease. FASEB J. 2019, 33, 3784–3794. [Google Scholar] [CrossRef] [PubMed]
- Schokrpur, S.; Hu, J.; Moughon, D.L.; Liu, P.; Lin, L.C.; Hermann, K.; Mangul, S.; Guan, W.; Pellegrini, M.; Xu, H.; et al. CRISPR-Mediated VHL Knockout Generates an Improved Model for Metastatic Renal Cell Carcinoma. Sci. Rep. 2016, 6, 29032. [Google Scholar] [CrossRef]
- Chan, H.Y.; Moldenhauer, L.M.; Groome, H.M.; Schjenken, J.E.; Robertson, S.A. Toll-like receptor-4 null mutation causes fetal loss and fetal growth restriction associated with impaired maternal immune tolerance in mice. Sci. Rep. 2021, 11, 16569. [Google Scholar] [CrossRef]
- Sand, J.; Haertel, E.; Biedermann, T.; Contassot, E.; Reichmann, E.; French, L.E.; Werner, S.; Beer, H.D. Expression of inflammasome proteins and inflammasome activation occurs in human, but not in murine keratinocytes. Cell Death Dis. 2018, 9, 24. [Google Scholar] [CrossRef]
- Murayama, M.A.; Kakuta, S.; Maruhashi, T.; Shimizu, K.; Seno, A.; Kubo, S.; Sato, N.; Saijo, S.; Hattori, M.; Iwakura, Y. CTRP3 plays an important role in the development of collagen-induced arthritis in mice. Biochem. Biophys. Res. Commun. 2014, 443, 42–48. [Google Scholar] [CrossRef]
- Sneezum, L.; Eislmayr, K.; Dworak, H.; Sedlyarov, V.; Le Heron, A.; Ebner, F.; Fischer, I.; Iwakura, Y.; Kovarik, P. Context-Dependent IL-1 mRNA-Destabilization by TTP Prevents Dysregulation of Immune Homeostasis Under Steady State Conditions. Front. Immunol. 2020, 11, 1398. [Google Scholar] [CrossRef]
- Shi, M.; Sekulovski, N.; Whorton, A.E.; MacLean, J.A., 2nd; Greaves, E.; Hayashi, K. Efficacy of niclosamide on the intra-abdominal inflammatory environment in endometriosis. FASEB J. 2021, 35, e21584. [Google Scholar] [CrossRef]
- Hashiguchi, Y.; Yabe, R.; Chung, S.H.; Murayama, M.A.; Yoshida, K.; Matsuo, K.; Kubo, S.; Saijo, S.; Nakamura, Y.; Matsue, H.; et al. IL-36alpha from Skin-Resident Cells Plays an Important Role in the Pathogenesis of Imiquimod-Induced Psoriasiform Dermatitis by Forming a Local Autoamplification Loop. J. Immunol. 2018, 201, 167–182. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Anders, S.; Kim, V.; Huber, W. RNA-Seq workflow: Gene-level exploratory analysis and differential expression. F1000Research 2015, 4, 1070. [Google Scholar] [CrossRef]
- Ignatiadis, N.; Klaus, B.; Zaugg, J.B.; Huber, W. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 2016, 13, 577–580. [Google Scholar] [CrossRef]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. In Use R! 2nd ed.; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]

| Gene Name | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Source |
|---|---|---|---|
| Klra7 | TCACAGCACACAGGTAGAGG | AGCTGGAAATCTGGCAGGTC | Self-designed |
| Scara5 | AGGAGGGAAAGCCAGGTAGC | CCCCTAGCTTCCCATCATCA | [69] |
| Dio2 | CCTCTTCCTGGCGCTCTATG | TTCAGGATTGGAGACGTGCA | [70] |
| Myd88 | CCCACTCGCAGTTTGTTGGA | TAGGGGGTCATCAAGGGTGG | Self-designed |
| Naip1 | CAGCCACCTAAAATAAGCTCTGG | GGACCCATGTTGGTCACTCC | [71] |
| Il17rb | CCATCCCTCCAGATGACAAC | TGCTCCTTCCTTGCCTCCAAGTTA | [72] |
| Ccr2 | GACAAGCACTTAGACCAGGC | ACCTTCGGAACTTCTCTCCA | [73] |
| Ccr4 | CCATTCTGGGGCTACTACGC | ACCAGGTACATCCATGAAACGA | [73] |
| Tnfsf13b | ACACTGCCCAACAATTCCTG | TCGTCTCCGTTGCGTGAAATC | [74] |
| Csf3 | GCAGACACAGTGCCTAAGCCA | CATCCAGCTGAAGCAAGTCCA | [75] |
| Il18 | GACTCTTGCGTCAACTTCAAGG | CAGGCTGTCTTTTGTCAACGA | [76] |
| Tnf | GCCTCCCTCTCATCAGTTCT | CACTTGGTGGTTTGCTACGA | [77] |
| Il1a | CCATCCAACCCAGATCAGCA | GTTTCTGGCAACTCCTTCAGC | [78] |
| Csf2 | CCTGGGCATTGTGGTCTACAG | GGCATGTCATCCAGGAGGTT | [75] |
| Il17a | GGAGAGCTTCATCTGTGTCTCTG | TTGGCCTCAGTGTTTGGACA | [79] |
| Il36a | CTACAGCTTGGGGAAGGGAACATA | CCCTTTAGAGCAGACAGCGATGAA | [80] |
| Antibody | Conjugate | Clone | Company | Cat. # |
|---|---|---|---|---|
| CD45 | PE Cy5 | 30-F11 | BioLegend, San Diego, CA, USA | 103110 |
| CD11c | PE | N418 | BioLegend, San Diego, CA, USA | 117307 |
| CD11b | Percp Cy5.5 | M1/70 | BioLegend, San Diego, CA, USA | 101228 |
| Ly6C | BV510 | HK1.4 | BioLegend, San Diego, CA, USA | 128033 |
| Ly6G | BV711 | 1A8 | BioLegend, San Diego, CA, USA | 127643 |
| CD64 | FITC | X54-5/7.1 | BioLegend, San Diego, CA, USA | 136316 |
| CD24 | APC | M1/69 | BioLegend, San Diego, CA, USA | 101813 |
| F4/80 | APC Cy7 | BM8 | BioLegend, San Diego, CA, USA | 127117 |
| MHCII | EF450 | M5/114.15.2 | Invitrogen, Waltham, MA, USA | 48-5321-82 |
| LD | Blue | - | Thermo Fisher, Waltham, MA, USA | L23105 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marquardt, R.M.; Ahn, S.H.; Reske, J.J.; Chandler, R.L.; Petroff, M.G.; Kim, T.H.; Jeong, J.-W. Endometrial Epithelial ARID1A Is Required for Uterine Immune Homeostasis during Early Pregnancy. Int. J. Mol. Sci. 2022, 23, 6067. https://doi.org/10.3390/ijms23116067
Marquardt RM, Ahn SH, Reske JJ, Chandler RL, Petroff MG, Kim TH, Jeong J-W. Endometrial Epithelial ARID1A Is Required for Uterine Immune Homeostasis during Early Pregnancy. International Journal of Molecular Sciences. 2022; 23(11):6067. https://doi.org/10.3390/ijms23116067
Chicago/Turabian StyleMarquardt, Ryan M., Soo Hyun Ahn, Jake J. Reske, Ronald L. Chandler, Margaret G. Petroff, Tae Hoon Kim, and Jae-Wook Jeong. 2022. "Endometrial Epithelial ARID1A Is Required for Uterine Immune Homeostasis during Early Pregnancy" International Journal of Molecular Sciences 23, no. 11: 6067. https://doi.org/10.3390/ijms23116067
APA StyleMarquardt, R. M., Ahn, S. H., Reske, J. J., Chandler, R. L., Petroff, M. G., Kim, T. H., & Jeong, J.-W. (2022). Endometrial Epithelial ARID1A Is Required for Uterine Immune Homeostasis during Early Pregnancy. International Journal of Molecular Sciences, 23(11), 6067. https://doi.org/10.3390/ijms23116067