Adaptation and Changes in Actin Dynamics and Cell Motility as Early Responses of Cultured Mammalian Cells to Altered Gravitational Vector
Abstract
:1. Introduction
2. Results
2.1. Proteomic Characterization of Cells Exposed to Simulated Microgravity
2.2. Proteomic Events Related to Cell Motility
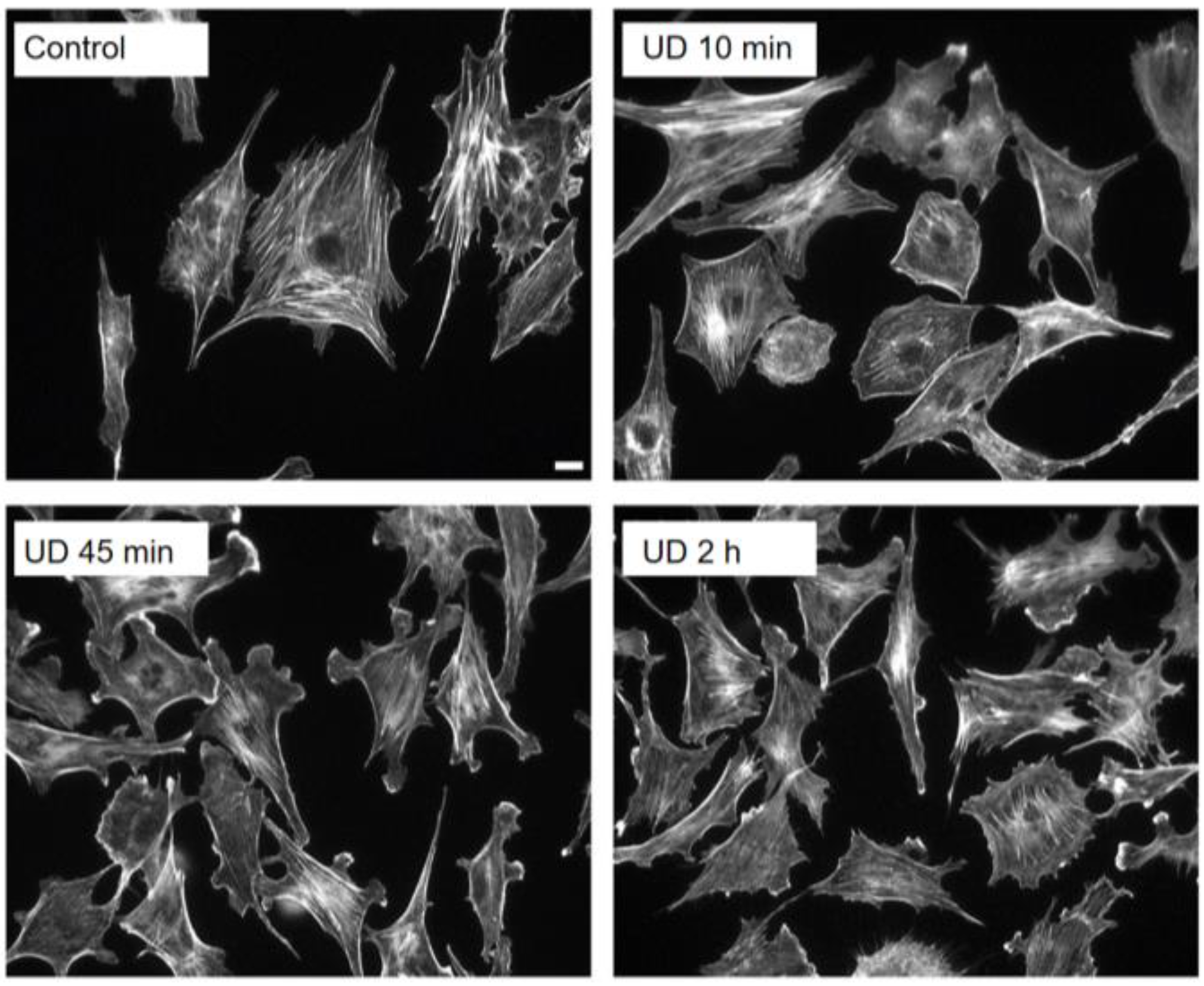
2.3. Live Recording of Cells Exposed to Altered Gravitational Vector: Upside-down Cell Cultures
2.4. Development of Lamellipodia
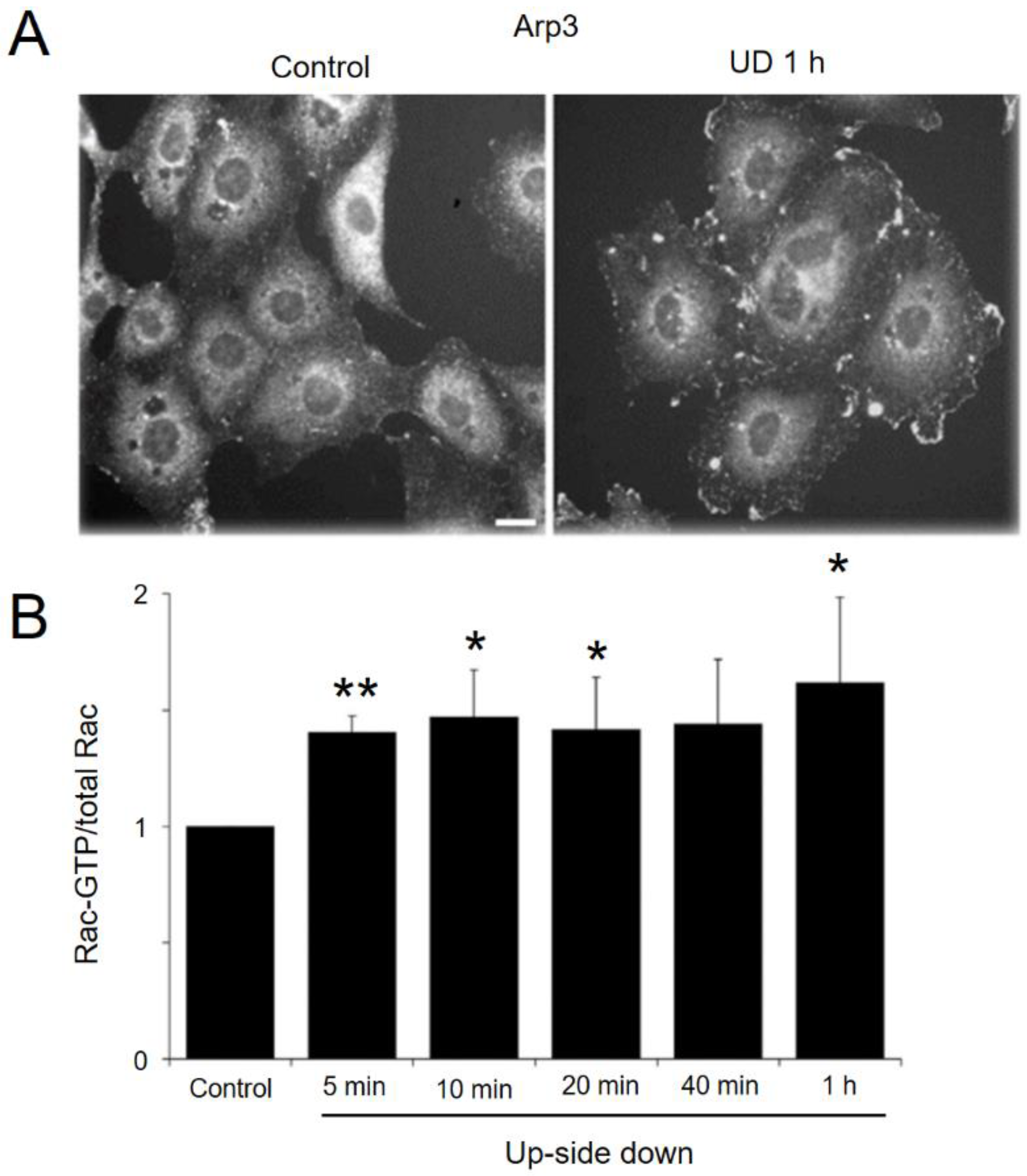
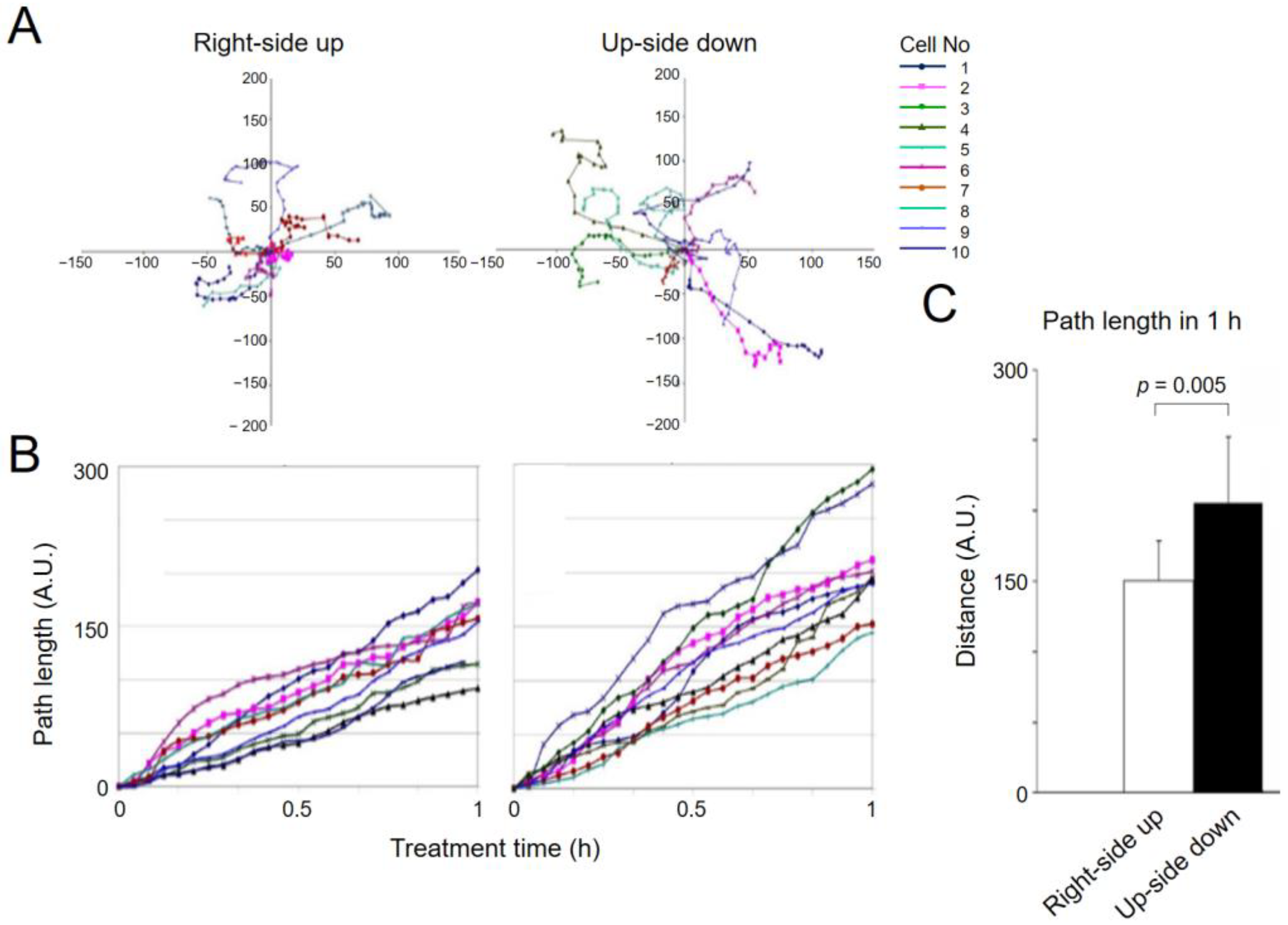
2.5. Increased Migration of Sparsely Cultured Upside-down Cells
2.6. Gravity Effects on Cells in Confluent Culture
3. Discussion
4. Materials and Methods
4.1. Cells
4.2. 3-D Clinostat
4.3. Sample Preparation for RPPA
4.4. RPPA Experiment and Data Analysis
4.5. Upside-down Experiments
4.6. Fluorescence Microscopy
4.7. Cell Motility Analysis
4.8. Rac1 Activity Assay
4.9. Wound Healing Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 1BR-hTERT cells | immortalized human fibroblasts |
| 3D | three dimensional |
| A.U. | arbitrary unit |
| AKT | Ak strain transforming |
| Arp3 | actin related protein 3 |
| ATCC | American Type Culture Collection |
| BAEC | bovine arterial endothelial cell |
| CCD | charge-coupled device |
| DAB | 3,3′-diaminobenzidine |
| DCC | disposable closed cell culture chamber |
| DMEM | Dulbecco’s modified Eagle’s medium |
| ECGS | endothelial cell growth supplement |
| ECM | endothelial cell medium |
| EGTA | ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid |
| FBS | fetal bovine serum |
| FDR | false discovery rate |
| GST | glutathione S-transferase |
| GTP | guanosine-5′-triphosphate |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| HSP27 | heat shock protein 27 |
| HUVEC | human umbilical vein endothelial cell |
| IgG | immunoglobulin G |
| LIMMA | linear models for microarray data |
| MAPK | mitogen-activated protein kinase |
| N.A. | numerical aperture |
| PAK1 | p21-activated protein kinase 1 |
| PBD | protein-binding domain |
| PBS | phosphate buffered saline |
| PKC | protein kinase C |
| QC | quality control |
| Rac1 | Ras-related C3 botulinum toxin substrate 1 |
| Rho | Ras homologous |
| RPPA | reverse phase protein array |
| RT | room temperature |
| S6 | ribosomal protein S6 |
| SDS | sodium dodecyl sulfate |
| SHP2 | Src homology-2 domain-containing protein tyrosine phosphatase-2 |
| SμG | simulated microgravity |
| TTBS | Tween-Tris-buffered saline |
| UD | upside down |
| μG | microgravity |
| YB1 | Y box binding protein 1 |
References
- Liu, Y.; Wang, E. Transcriptional analysis of normal human fibroblast responses to microgravity stress. Genom. Proteom. Bioinform. 2008, 6, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Hader, D.-P.; Braun, M.; Grimm, D.; Hemmersbach, R. Gravireceptors in eukaryotes—A comparison of case studies on the cellular level. NPJ Microgravity 2017, 3, 13. [Google Scholar] [CrossRef] [Green Version]
- Buken, C.; Sahana, J.; Corydon, T.J.; Melnik, D.; Bauer, J.; Wehland, M.; Kruger, M.; Balk, S.; Abuagela, N.; Infanger, M.; et al. Morphological and molecular changes in juvenile normal human fibroblasts exposed to simulated microgravity. Sci. Rep. 2019, 9, 11882. [Google Scholar] [CrossRef]
- Aventaggiato, M.; Barreca, F.; Vernucci, E.; Bizzarri, M.; Ferretti, E.; Russo, M.A.; Tafani, M. Putative receptors for gravity sensing in mammalian cells: The effects of microgravity. Appl. Sci. 2020, 10, 2028. [Google Scholar] [CrossRef] [Green Version]
- El Gindi, M.; Sapudom, J.; Ibrahim, I.H.; Al-Sayegh, M.; Chen, W.; Garcia-Sabaté, A.; Teo, J.C.M. May the force be with you (or not): The immune system under microgravity. Cells 2021, 10, 1941. [Google Scholar] [CrossRef]
- Genchi, G.G.; Cialdai, F.; Monici, M.; Mazzolai, B.; Mattoli, V.; Ciofani, G. Hypergravity stimulation enhances PC12 neuron-like cell differentiation. Biomed. Res. Int. 2015, 2015, 748121. [Google Scholar] [CrossRef] [Green Version]
- Eckert, J.; van Loon, J.J.W.A.; Eng, L.M.; Schmidt, T.E. Hypergravity affects cell traction forces of fibroblasts. Biophy. J. 2021, 120, 773–780. [Google Scholar] [CrossRef]
- Woodcock, E.M.; Girvan, P.; Eckert, J.; Lopez-Duarte, I.; Kubánková, M.; van Loon, J.J.W.A.; Brooks, N.J.; Kuimova, M.K. Measuring intracellular viscosity in conditions of hypergravity. Biophy. J. 2019, 116, 1984–1993. [Google Scholar] [CrossRef]
- De Cesari, C.; Barravecchia, I.; Pyankova, O.V.; Vezza, M.; Germani, M.M.; Scebba, F.; van Loon, J.J.W.A.; Angeloni, D. Hypergravity activates a pro-angiogenic homeostatic response by human capillary endothelial cells. Int. J. Mol. Sci. 2020, 21, 2354. [Google Scholar] [CrossRef] [Green Version]
- Herranz, R.; Anken, R.; Boonstra, J.; Braun, M.; Christianen, P.C.M.; de Geest, M.; Hauslage, J.; Hilbig, R.; Hill, R.J.A.; Lebert, M.; et al. Ground-based facilities for simulation of microgravity: Organism-specific recommendations for their use, and recommended terminology. Astrobiology 2013, 13, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Amselem, S. Remote controlled autonomous microgravity lab platforms for drug research in space. Pharm. Res. 2019, 36, 183. [Google Scholar] [CrossRef] [PubMed]
- Ferranti, F.; Del Bianco, M.; Pacelli, C. Advantages and limitations of current microgravity platforms for space biology research. Appl. Sci. 2021, 11, 68. [Google Scholar] [CrossRef]
- Morita, T.M. Directional gravity sensing in gravitropism. Annu. Rev. Plant Biol. 2010, 61, 705–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, D.; Tocquard, K.; Venisse, J.-S.; Legué, V.; Roeckel-Drevet, P. Gravity sensing, a largely misunderstood trigger of plant orientated growth. Front. Plant Sci. 2014, 5, 610. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Z.; Du, H.; Chen, S.; Huang, W.; Ge, L. LAZY gene family in plant gravitropism. Front. Plant Sci. 2021, 11, 606241. [Google Scholar] [CrossRef]
- Takahashi, K.; Takahashi, H.; Furuichi, T.; Toyota, M.; Furutani-Seiki, M.; Kobayashi, T.; Watanabe-Takano, H.; Shinohara, M.; Numaga-Tomita, T.; Sakaue-Sawano, A.; et al. Gravity sensing in plant and animal cells. NPJ Microgravity 2021, 7, 2. [Google Scholar] [CrossRef]
- Bradbury, P.; Wu, H.; Choi, J.U.; Rowan, A.E.; Zhang, H.; Poole, K.; Lauko, J.; Chou, J. Modeling the impact of microgravity at the cellular level: Implications for human disease. Front. Cell Dev. Biol. 2020, 8, 96. [Google Scholar] [CrossRef] [Green Version]
- Hada, M.; Ikeda, H.; Rhone, J.R.; Beitman, A.J.; Plante, I.; Souda, H.; Yoshida, Y.; Held, K.D.; Fujiwara, K.; Saganti, P.B.; et al. Increased chromosome aberrations in cells exposed simultaneously to simulated microgravity and radiation. Int. J. Mol. Sci. 2019, 20, 43. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, H.; Muratani, M.; Hidema, J.; Hada, M.; Fujiwara, K.; Souda, H.; Yoshida, Y.; Takahashi, A. Expression profile of cell cycle-related genes in human fibroblasts exposed simultaneously to radiation and simulated microgravity. Int. J. Mol. Sci. 2019, 20, 4791. [Google Scholar] [CrossRef] [Green Version]
- Enomoto, A.; Murakami, H.; Asai, N.; Morone, N.; Watanabe, T.; Kawai, K.; Murakumo, Y.; Usukura, J.; Kaibuchi, K.; Takahashi, M. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev. Cell 2005, 9, 389–402. [Google Scholar] [CrossRef] [Green Version]
- Xue, G.; Hemmings, B.A. PKB/Akt–dependent regulation of cell motility. J. Natl. Cancer Inst. 2013, 105, 393–404. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Yamada, N.; Tanaka, T.; Hori, T.; Yokoyama, S.; Hayakawa, Y.; Yano, S.; Fukuoka, J.; Koizumi, K.; Saiki, I.; et al. Crucial roles of RSK in cell motility by catalysing serine phosphorylation of EphA2. Nat. Commun. 2015, 6, 7679. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Oh, D.; Biswas, K.H.; Yu, C.-H.; Zaidel-Bar, R.; Groves, J.T. Spatially modulated ephrinA1:EphA2 signaling increases local contractility and global focal adhesion dynamics to promote cell motility. Proc. Nat. Acad. Sci. USA 2017, 115, E5705. [Google Scholar] [CrossRef] [Green Version]
- Piotrowicz, R.S.; Hickey, E.; Levin, E.G. Heat shock protein 27 kDa expression and phosphorylation regulates endothelial cell migration. FASEB J. 1998, 12, 1481–1490. [Google Scholar] [CrossRef]
- Hirano, S.; Shelden, E.A.; Gilmont, R.R. HSP27 regulates fibroblast adhesion, motility, and matrix contraction. Cell Stress Chaperones 2004, 9, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Xie, F.; Aziz, A.R.; Shao, S.; Li, W.; Deng, S.; Liao, X.; Liu, B. Heat shock protein 27 phosphorylation regulates tumor cell migration under shear stress. Biomolecules 2019, 9, 50. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.-S.; Wu, J.-R.; Hu, C.-T. Signal cross talks for sustained MAPK activation and cell migration: The potential role of reactive oxygen species. Cancer Metastasis Rev. 2008, 27, 303–314. [Google Scholar] [CrossRef]
- Tanimura, S.; Takeda, K. ERK signalling as a regulator of cell motility. J. Biochem. 2017, 162, 145–154. [Google Scholar] [CrossRef] [Green Version]
- Lavoie, H.; Gagnon, J.; Therrien, M. ERK signalling: A master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 2020, 21, 607. [Google Scholar] [CrossRef]
- Ng, T.; Parsons, M.; Hughes, W.E.; Monypenny, J.; Zicha, D.; Gautreau, A.; Arpin, M.; Gschmeissner, S.; Verveer, P.J.; Bastiaens, P.I.H.; et al. Ezrin is a downstream effector of trafficking PKC–integrin complexes involved in the control of cell motility. EMBO J. 2001, 20, 2723–2741. [Google Scholar] [CrossRef] [Green Version]
- Sumagin, R.; Robin, A.Z.; Nusrat, A.; Parkos, C.A. Activation of PKCβII by PMA facilitates enhanced epithelial wound repair through increased cell spreading and migration. PLoS ONE 2013, 8, e55775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fogh, B.S.; Multhaupt, H.A.B.; Couchman, J.R. Protein kinase C, focal adhesions and the regulation of cell migration. J. Histochem. Cytochemv. 2014, 62, 172–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bevven, L.A.; Crouch, M.F. Cellular function of p70S6K: A role in regulating cell motility. Immunol. Cell Biol. 2000, 78, 447–451. [Google Scholar] [CrossRef]
- Yang, X.; Xu, L.; Yang, Y.-E.; Xiong, C.; Yu, J.; Wang, Y.; Lin, Y. Knockdown of ribosomal protein S6 suppresses proliferation, migration, and invasion in epithelial ovarian cancer. J. Ovarian Res. 2020, 13, 100. [Google Scholar] [CrossRef]
- Qi, J.H.; Ito, N.; Claesson-Welsh, L. Tyrosine phosphatase SHP-2 is involved in regulation of platelet-derived growth factor-induced migration. J. Biol. Chem. 1999, 274, 14455–14463. [Google Scholar] [CrossRef] [Green Version]
- Howe, A.K. Regulation of actin-based cell migration by cAMP/PKA. Biochim. Biophys. Acta Mol. Cell Res. 2004, 1692, 159–174. [Google Scholar] [CrossRef]
- Zhang, X.; Ding, Z.; Mo, J.; Sang, B.; Shi, Q.; Hu, J.; Xie, S.; Zhan, W.; Lu, D.; Yang, M.; et al. GOLPH3 promotes glioblastoma cell migration and invasion via the mTOR-YB1 pathway in vitro. Mol. Carcinog. 2015, 54, 1252–1263. [Google Scholar] [CrossRef]
- Liu, Q.; Tao, T.; Liu, F.; Ni, R.; Lu, C.; Shen, A. Hyper-O-GlcNAcylation of YB-1 affects Ser102 phosphorylation and promotes cell proliferation in hepatocellular carcinoma. Exp. Cell Res. 2016, 349, 230–238. [Google Scholar] [CrossRef]
- Kim, A.; Shim, S.; Kim, Y.-H.; Kim, M.-J.; Park, S.; Myung, J.K. Inhibition of Y box binding protein 1 suppresses cell growth and motility in colorectal cancer. Mol. Cancer Ther. 2020, 19, 479–489. [Google Scholar] [CrossRef] [Green Version]
- Carlsson, S.I.M.; Bertilaccio, M.T.S.; Ballabio, E.; Maier, J.A.M. Endothelial stress by gravitational unloading: Effects on cell growth and cytoskeletal organization. Biochim. Biophys. Acta. 2003, 1642, 173–179. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhang, S.; Chen, J.; Du, T.; Wang, Y.; Wang, Z. Modeled microgravity causes changes in the cytoskeleton and focal adhesions, and decreases in migration in malignant human MCF-7 cells. Protoplasma 2009, 238, 23. [Google Scholar] [CrossRef] [PubMed]
- Meyers, V.E.; Zayzafoon, M.; Douglas, J.T.; McDonald, J.M. RhoA and cytoskeletal disruption mediate reduced osteoblastogenesis and enhanced adipogenesis of human mesenchymal stem cells in modeled microgravity. J. Bone Miner. Res. 2005, 10, 1858–1866. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Dai, Z.-Q.; Ling, S.-K.; Zhang, H.-Y.; Wan, Y.-M.; Li, Y.-H. Gravity, a regulation factor in the differentiation of rat bone marrow mesenchymal stem cells. J. Biomed. Sci. 2009, 16, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higashibata, A.; Imamizo-Sato, M.; Seki, M.; Yamazaki, T.; Ishioka, N. Influence of simulated microgravity on the activation of the small GTPase Rho involved in cytoskeletal formation—Molecular cloning and sequencing of bovine leukemia-associated guanine nucleotide exchange factor. BMC Biochem. 2006, 7, 19. [Google Scholar] [CrossRef] [Green Version]
- Pache, C.; Kühn, J.; Westphal, K.; Toy, M.F.; Parent, J.; Buchi, O.; Franco-Obregon, A.; Depeursinge, C.D.; Egli, M. Digital holographic microscopy real-time monitoring of cytoarchitectural alterations during simulated microgravity. J. Biomed. Opt. 2010, 15, 026021. [Google Scholar] [CrossRef] [Green Version]
- Vorselen, D.; Roos, W.H.; MacKintosh, F.C.; Wuite, G.J.L.; van Loon, J.J.W.A. The role of the cytoskeleton in sensing changes in gravity by nonspecialized cells. FASEB J. 2014, 28, 536–547. [Google Scholar] [CrossRef] [Green Version]
- Thiel, C.S.; Tauber, S.; Lauber, B.; Polzer, J.; Seebacher, C.; Uhl, R.; Neelam, S.; Zhang, Y.; Levine, H.; Ullrich, O. Rapid morphological and cytoskeletal response to microgravity in human primary macrophages. Int. J. Mol. Sci. 2019, 20, 2402. [Google Scholar] [CrossRef] [Green Version]
- Osawa, M.; Masuda, M.; Kusano, K.; Fujiwara, K. Evidence for a role of platelet endothelial cell adhesion molecule-1 in endothelial cell mechanosignal transduction: Is it a mechanoresponsive molecule? J. Cell Biol. 2002, 158, 773–785. [Google Scholar] [CrossRef] [Green Version]
- Chiu, Y.J.; McBeath, E.; Fujiwara, K. Mechanotransduction in an extracted cell model: Fyn drives stretch- and flow-elicited PECAM-1 phosphorylation. J. Cell Biol. 2008, 182, 753–763. [Google Scholar] [CrossRef] [Green Version]
- Davies, P.F. Hemodynamic shear stress and the endothelium in cardiovascular pathophysiology. Nat. Clin. Pract. Cardiovasc. Med. 2009, 6, 16–26. [Google Scholar] [CrossRef] [Green Version]
- Shi, F.; Wang, Y.-C.; Zhao, T.-Z.; Zhang, S.; Du, T.-Y.; Yang, C.-B.; Li, Y.-H.; Sun, X.-Q. Effects of simulated microgravity on human umbilical vein endothelial cell angiogenesis and role of the PI3K-Akt-eNOS signal pathway. PLoS ONE 2012, 7, e40365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maier, J.A.M.; Cialdai, F.; Monici, M.; Morbidelli, L. The impact of microgravity and hypergravity on endothelial cells. Biomed. Res. Int. 2015, 2015, 434803. [Google Scholar] [CrossRef] [PubMed]
- Yamao, M.; Naoki, H.; Kunida, K.; Aoki, K.; Matsuda, M.; Ishii, S. Distinct predictive performance of Rac1 and Cdc42 in cell migration. Sci. Rep. 2015, 5, 17527. [Google Scholar] [CrossRef] [Green Version]
- Lawson, C.D.; Ridley, A.J. Rho GTPase signaling complexes in cell migration and invasion. J. Cell Biol. 2018, 217, 447–457. [Google Scholar] [CrossRef]
- Hausmann, N.; Fengler, S.; Hennig, A.; Franz-Wachtel, M.; Hampp, R.; Neef, M. Cytosolic calcium, hydrogen peroxide and related gene expression and protein modulation in Arabidopsis thaliana cell cultures respond immediately to altered gravitation: Parabolic flight data. Plant Biol. 2014, 16, 120–128. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, M.; Liu, Y.; Wang, C.; Takahashi, K.; Naruse, K. Gravity-sensitive genes in human vascular endothelial cells. Front. Cell Dev. Biol. 2021, 9, 689662. [Google Scholar] [CrossRef]
- Neelam, S.; Richardson, B.; Barker, R.; Udave, C.; Gilroy, S.; Cameron, M.J.; Levine, H.G.; Zhang, Y. Changes in nuclear shape and gene expression in response to simulated microgravity are LINC complex-dependent. Int. J. Mol. Sci. 2020, 21, 6762. [Google Scholar] [CrossRef]
- Tauber, S.; Hauschild, S.; Paulsen, K.; Gutewort, A.; Raig, C.; Hürlimann, E.; Biskup, J.; Philpot, C.; Lier, H.; Engelmann, F.; et al. Signal transduction in primary human T lymphocytes in altered gravity during parabolic flight and clinostat experiments. Cell. Physiol. Biochem. 2015, 35, 1034–1051. [Google Scholar] [CrossRef]
- Neelam, S.; Lee, A.; Lane, M.A.; Udave, C.; Levine, H.G.; Zhang, Y. Module to support real-time microscopic imaging of living organisms on ground-based microgravity analogs. Appl. Sci. 2021, 11, 3122. [Google Scholar] [CrossRef]
- Wang, S.; Yin, Z.; Zhao, B.; Qi, Y.; Liu, J.; Rahimi, S.A.; Lee, L.Y.; Li, S. Microgravity simulation activates Cdc42 via Rap1GDS1 to promote vascular branch morphogenesis during vasculogenesis. Stem Cell Res. 2017, 25, 157–165. [Google Scholar] [CrossRef]
- Cialdai, F.; Vignali, L.; Morbidelli, L.; Colciago, A.; Celotti, F.; Santi, A.; Caselli, A.; Cirri, P.; Monici, M. Modeled microgravity affects fibroblast functions related to wound healing. Micrograv. Sci. Technol. 2017, 29, 121–132. [Google Scholar] [CrossRef]
- Tanigawa, N.; Yano, S.; Higashibata, A.; Tsuchiya, Y.; Tanii, H.; Ando, N.; Kubota, K.; Nagase, M. Development and validation of a closed chamber for cell culture experiments in space. Biol. Sci. Space 2014, 28, 1–5. [Google Scholar] [CrossRef]
- Crampton, S.P.; Davis, J.; Hughes, C.C.W. Isolation of human umbilical vein endothelial cells (HUVEC). J. Vis. Exp. 2007, 3, 183. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, H.; Souda, H.; Puspitasari, A.; Held, K.D.; Hidema, J.; Nikawa, T.; Yoshida, Y.; Kanai, T.; Takahashi, A. Development and performance evaluation of a three-dimensional clinostat synchronized heavy-ion irradiation system. Life Sci. Space Res. 2017, 12, 51–60. [Google Scholar] [CrossRef]
- Yamanouchi, S.; Rhone, J.R.; Mao, J.-H.; Fujiwara, K.; Saganti, P.B.; Takahashi, A.; Hada, M. Simultaneous exposure of cultured human lymphoblastic cells to simulated microgravity and radiation increases chromosome aberrations. Life 2020, 10, 187. [Google Scholar] [CrossRef]
- Yamanouchi, S.; Adachi, T.; Yoshida, Y.; Rhone, J.R.; Mao, J.-H.; Fujiwara, K.; Saganti, P.B.; Takahashi, A.; Hada, M. The combined effect of simulated microgravity and radiation on chromosome aberrations in human peripheral blood lymphocytes. Biol. Sci. Space 2021, 35, 15–23. [Google Scholar] [CrossRef]
- Hennessy, B.T.; Lu, Y.; Poradosu, E.; Yu, Q.; Yu, S.; Hall, H.; Carey, M.S.; Ravoori, M.; Gonzalez-Angulo, A.M.; Birch, R.; et al. Pharmacodynamic markers of perifosine efficacy. Clin. Cancer Res. 2007, 13, 7421–7431. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Angulo, A.M.; Hennessy, B.T.; Meric-Bernstam, F.; Sahin, A.; Liu, W.; Ju, Z.; Carey, M.S.; Myhre, S.; Speers, C.; Deng, L.; et al. Functional proteomics can define prognosis and predict pathologic complete response in patients with breast cancer. Clin. Proteom. 2011, 8, 11. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; He, X.; Baggerly, K.A.; Coombes, K.R.; Hennessy, B.T.; Mills, G.B. Non-parametric quantification of protein lysate arrays. Bioinformatics 2007, 23, 1986–1994. [Google Scholar] [CrossRef]
- Neeley, E.S.; Kornblau, S.M.; Coombes, K.R.; Baggerly, K.A. Variable slope normalization of reverse phase protein arrays. Bioinformatics 2009, 25, 1384–1389. [Google Scholar] [CrossRef] [Green Version]
- Ju, Z.; Liu, W.; Roebuck, P.L.; Siwak, D.R.; Zhang, N.; Lu, Y.; Davies, M.A.; Akbani, R.; Weinstein, J.N.; Mills, G.B.; et al. Development of a robust classifier for quality control of reverse-phase protein arrays. Bioinformatics 2015, 31, 912–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl. Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
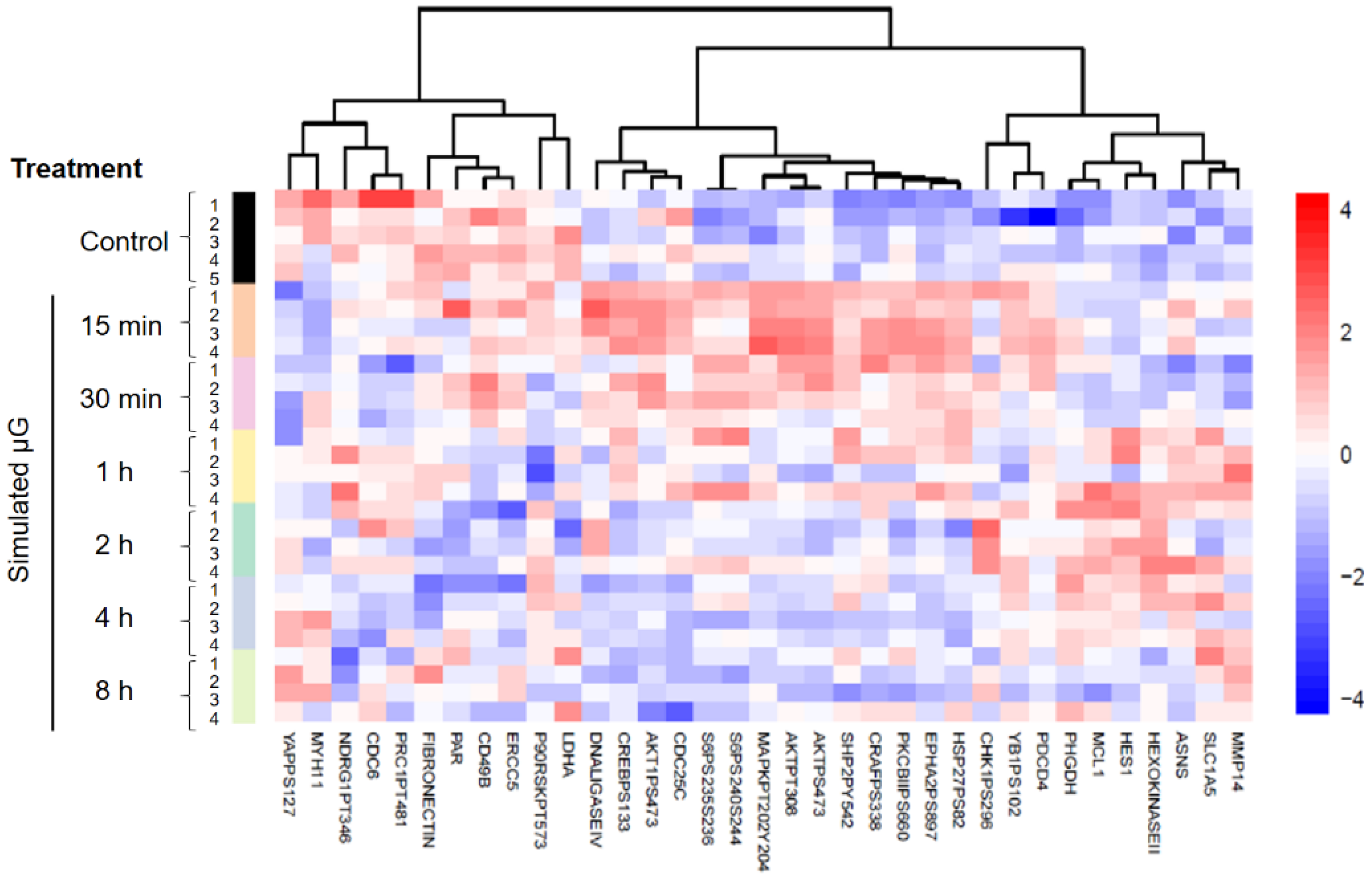
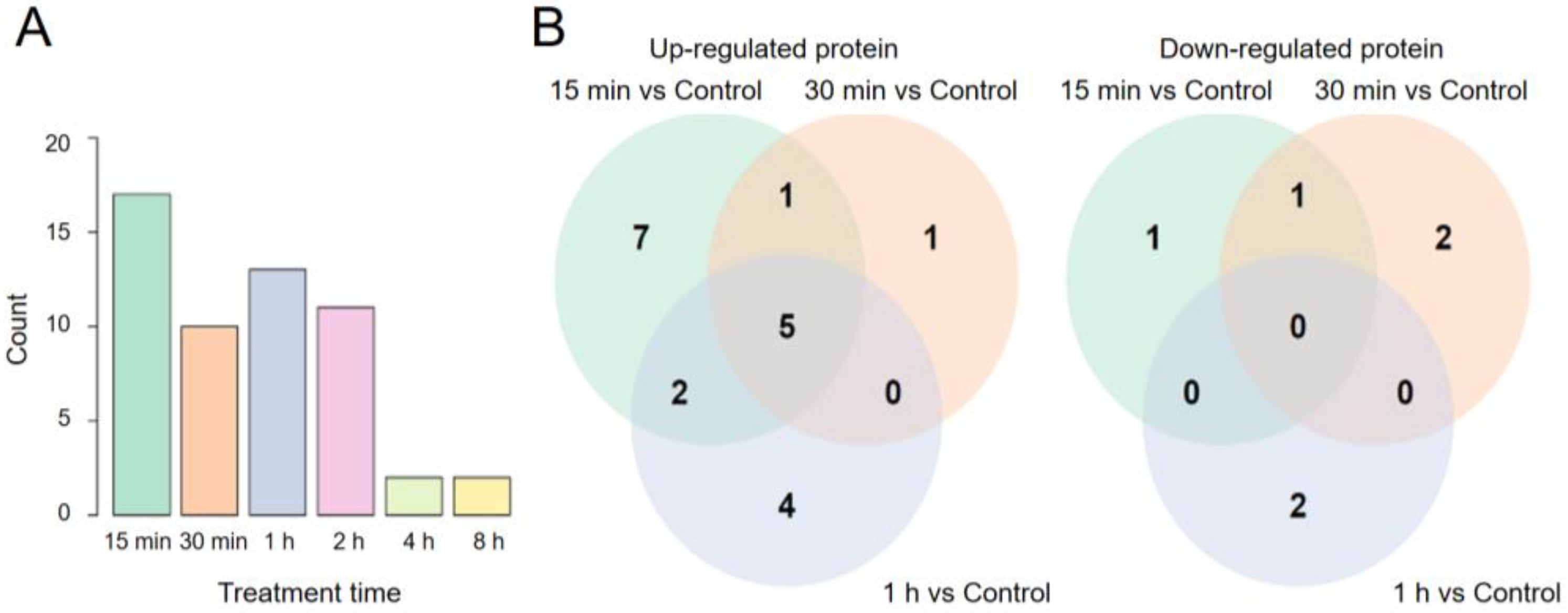

| Protein | Treatment Time (Minutes) | |||||
|---|---|---|---|---|---|---|
| 15 | 30 | 60 | 120 | 240 | 480 | |
| AKT1PS473 * | 0.27 | |||||
| AKTPS473 * | 0.45 | |||||
| AKTPT308 * | 0.52 | 0.29 | ||||
| ASNA | 0.36 | 0.41 | 0.36 | |||
| CRAFPS338 | 0.24 | |||||
| CD49B | −0.18 | −0.20 | ||||
| CDC35C | −0.26 | |||||
| CDC6 | −0.16 | |||||
| CHK1PS296 | 0.15 | |||||
| CREBPS133 | 0.29 | 0.18 | ||||
| DNALIGASEIV | 0.25 | |||||
| EPHA2PS897 * | 0.99 | 0.83 | 0.70 | |||
| ERCC5 | −0.16 | |||||
| FIBRONECTIN | −0.54 | −0.75 | ||||
| HES1 | 0.52 | 0.58 | ||||
| HEXOKINASEII | 0.37 | 0.61 | ||||
| HSP27PS82 * | 0.44 | 0.37 | 0.36 | |||
| LDHA | −0.23 | |||||
| MAPKPT202Y204 * | 1.15 | 0.64 | 0.42 | |||
| MCL1 | 0.30 | |||||
| MMP14 | 0.19 | |||||
| MYH11 | −0.23 | |||||
| NDRG1PT346 | −0.28 | |||||
| P90RSKPT573 | −0.63 | |||||
| PAR | −0.90 | |||||
| PDCD4 | 0.35 | |||||
| PHGDH | 0.30 | 0.32 | ||||
| PKCBIIPS660 * | 0.36 | |||||
| PRC1PT481 | −0.15 | |||||
| S6PS235S236 * | 0.58 | 0.66 | 0.57 | |||
| S6PS240S244 * | 0.61 | |||||
| SHP2PY542 * | 0.22 | 0.20 | 0.24 | |||
| SLC1A5 | 0.38 | |||||
| YAPPS127 | −0.21 | −0.23 | ||||
| YB1PS102 * | 0.26 | |||||
 value > 1;
value > 1;  , 0.5 ≦ value < 1;
, 0.5 ≦ value < 1;  , 0 ≦ value < 0.5;
, 0 ≦ value < 0.5;  , −0.5 ≦ value < 0;
, −0.5 ≦ value < 0;  , −0.5 < value.
, −0.5 < value.Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ju, Z.; Thomas, T.N.; Chiu, Y.-J.; Yamanouchi, S.; Yoshida, Y.; Abe, J.-i.; Takahashi, A.; Wang, J.; Fujiwara, K.; Hada, M. Adaptation and Changes in Actin Dynamics and Cell Motility as Early Responses of Cultured Mammalian Cells to Altered Gravitational Vector. Int. J. Mol. Sci. 2022, 23, 6127. https://doi.org/10.3390/ijms23116127
Ju Z, Thomas TN, Chiu Y-J, Yamanouchi S, Yoshida Y, Abe J-i, Takahashi A, Wang J, Fujiwara K, Hada M. Adaptation and Changes in Actin Dynamics and Cell Motility as Early Responses of Cultured Mammalian Cells to Altered Gravitational Vector. International Journal of Molecular Sciences. 2022; 23(11):6127. https://doi.org/10.3390/ijms23116127
Chicago/Turabian StyleJu, Zhenlin, Tamlyn N. Thomas, Yi-Jen Chiu, Sakuya Yamanouchi, Yukari Yoshida, Jun-ichi Abe, Akihisa Takahashi, Jing Wang, Keigi Fujiwara, and Megumi Hada. 2022. "Adaptation and Changes in Actin Dynamics and Cell Motility as Early Responses of Cultured Mammalian Cells to Altered Gravitational Vector" International Journal of Molecular Sciences 23, no. 11: 6127. https://doi.org/10.3390/ijms23116127
APA StyleJu, Z., Thomas, T. N., Chiu, Y.-J., Yamanouchi, S., Yoshida, Y., Abe, J.-i., Takahashi, A., Wang, J., Fujiwara, K., & Hada, M. (2022). Adaptation and Changes in Actin Dynamics and Cell Motility as Early Responses of Cultured Mammalian Cells to Altered Gravitational Vector. International Journal of Molecular Sciences, 23(11), 6127. https://doi.org/10.3390/ijms23116127









