How RNases Shape Mitochondrial Transcriptomes
Abstract
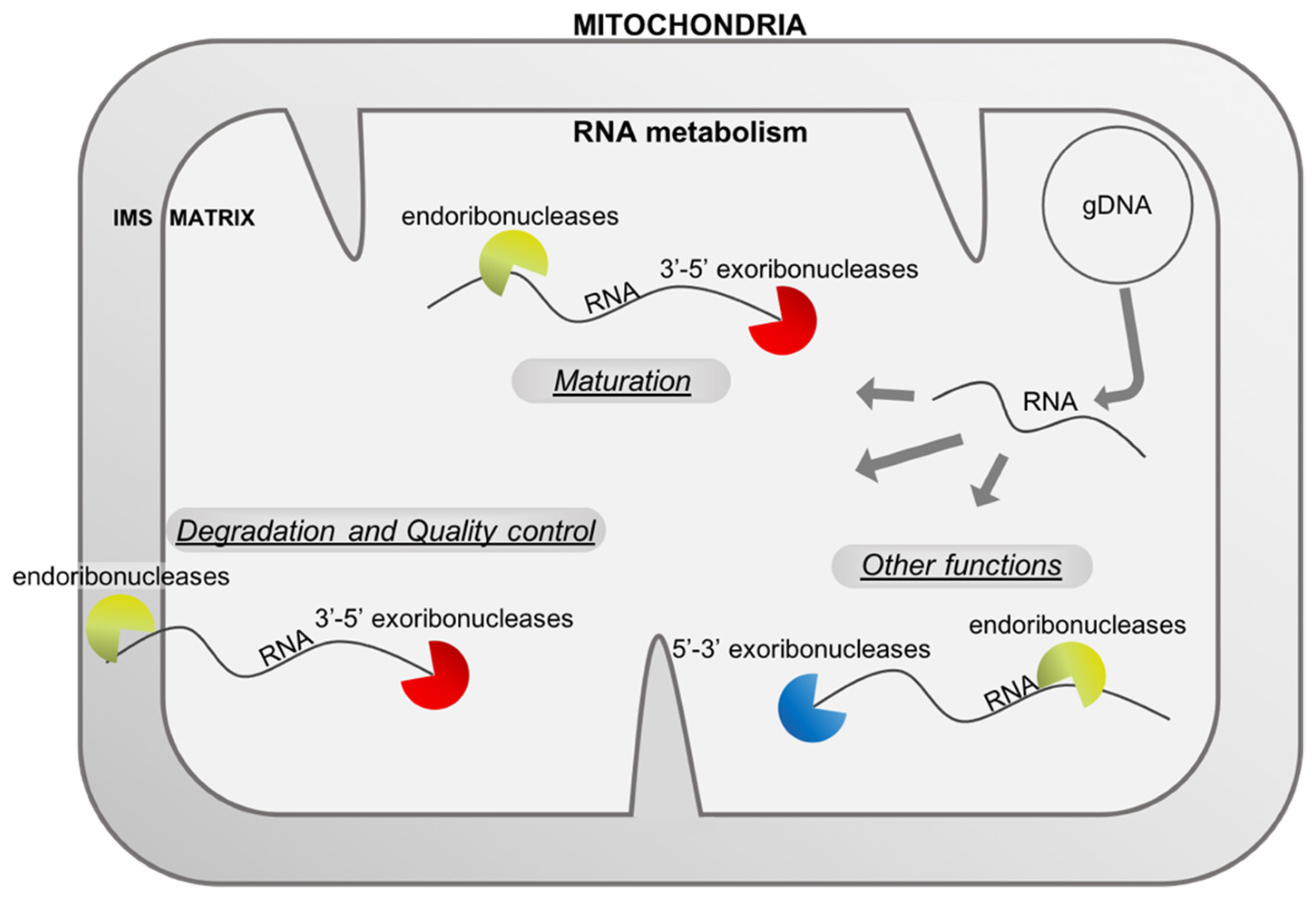
:1. Introduction
2. Mitochondrial RNases Involved in RNA Maturation
2.1. Diversity of RNase P Enzymes
2.2. Diversity of RNase Z Proteins
2.3. MNU Proteins Make a Novel Category of Mitochondrial RNA Maturation Factors
2.4. KREX and KREN Ribonucleases Are Required for RNA Editing in Kinetoplastid Mitochondria
2.5. RAP Domain Nucleases Make a Diverse Family of Putative Mitochondrial RNA Maturation Enzymes
3. Mitochondrial RNases Involved in RNA Quality Control and Degradation
3.1. PNPase Plays a Central Role in Mitochondrial RNA Degradation
3.2. DSS1 Proteins Are Specific Mitochondrial RNA Degradation Enzymes in Fungi and Kinetoplastids
3.3. Participation of Oligoribonucleases in Mitochondrial RNA Degradation
3.4. Other Types of Mitochondrial RNase Enzymes Involved in RNA Degradation
3.5. Functions of RNase T2 Enzymes in Mitochondria
3.6. Mitochondrial RNases That Might Be Involved in RNA Quality Control Pathways
4. Miscellaneous Functions of Mitochondrial Ribonucleases
4.1. RNase H Enzymes Are Involved in Mitochondrial DNA Maintenance
4.2. Putative Mitochondrial Functions of RNase MRP
4.3. The YbeY Ribonuclease Might Participate in the Biogenesis of Mitoribosomes
4.4. Pet127, the Sole 5′ to 3′ Exoribonuclease Identified in Eukaryotes Mitochondria
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zardoya, R. Recent Advances in Understanding Mitochondrial Genome Diversity. F1000Research 2020, 9, 270. [Google Scholar] [CrossRef]
- Aphasizhev, R.; Aphasizheva, I. Mitochondrial RNA Processing in Trypanosomes. Res. Microbiol. 2011, 162, 655–663. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, A.P.; Patel, S.S. Mechanism of Transcription Initiation by the Yeast Mitochondrial RNA Polymerase. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 930–938. [Google Scholar] [CrossRef] [Green Version]
- Gualberto, J.M.; Kühn, K. DNA-Binding Proteins in Plant Mitochondria: Implications for Transcription. Mitochondrion 2014, 19, 323–328. [Google Scholar] [CrossRef]
- Barshad, G.; Marom, S.; Cohen, T.; Mishmar, D. Mitochondrial DNA Transcription and Its Regulation: An Evolutionary Perspective. Trends Genet. 2018, 34, 682–692. [Google Scholar] [CrossRef]
- Basu, U.; Bostwick, A.M.; Das, K.; Dittenhafer-Reed, K.E.; Patel, S.S. Structure, Mechanism, and Regulation of Mitochondrial DNA Transcription Initiation. J. Biol. Chem. 2020, 295, 18406–18425. [Google Scholar] [CrossRef]
- Lechner, M.; Rossmanith, W.; Hartmann, R.K.; Thölken, C.; Gutmann, B.; Giegé, P.; Gobert, A. Distribution of Ribonucleoprotein and Protein-Only RNase P in Eukarya. Mol. Biol. Evol. 2015, 32, 3186–3193. [Google Scholar] [CrossRef] [Green Version]
- Dubrovsky, E.B.; Dubrovskaya, V.A.; Levinger, L.; Schiffer, S.; Marchfelder, A. Drosophila RNase Z Processes Mitochondrial and Nuclear Pre-tRNA 3′ Ends in Vivo. Nucleic Acids Res. 2004, 32, 255–262. [Google Scholar] [CrossRef]
- Canino, G.; Bocian, E.; Barbezier, N.; Echeverría, M.; Forner, J.; Binder, S.; Marchfelder, A. Arabidopsis Encodes Four TRNase Z Enzymes. Plant Physiol. 2009, 150, 1494–1502. [Google Scholar] [CrossRef] [Green Version]
- Siira, S.J.; Rossetti, G.; Richman, T.R.; Perks, K.; Ermer, J.A.; Kuznetsova, I.; Hughes, L.; Shearwood, A.-M.J.; Viola, H.M.; Hool, L.C.; et al. Concerted Regulation of Mitochondrial and Nuclear Non-Coding RNAs by a Dual-Targeted RNase Z. EMBO Rep. 2018, 19, e46198. [Google Scholar] [CrossRef]
- Gan, X.; Yang, J.; Li, J.; Yu, H.; Dai, H.; Liu, J.; Huang, Y. The Fission Yeast Schizosaccharomyces Pombe Has Two Distinct TRNase ZLs Encoded by Two Different Genes and Differentially Targeted to the Nucleus and Mitochondria. Biochem. J. 2011, 435, 103–111. [Google Scholar] [CrossRef] [Green Version]
- Liao, Z.; Schelcher, C.; Smirnov, A. YbeY, Éminence Grise of Ribosome Biogenesis. Biochem. Soc. Trans. 2021, 49, 727–745. [Google Scholar] [CrossRef]
- Bouchoucha, A.; Waltz, F.; Bonnard, G.; Arrivé, M.; Hammann, P.; Kuhn, L.; Schelcher, C.; Zuber, H.; Gobert, A.; Giegé, P. Determination of Protein-Only RNase P Interactome in Arabidopsis Mitochondria and Chloroplasts Identifies a Complex between PRORP1 and Another NYN Domain Nuclease. Plant J. 2019, 100, 549–561. [Google Scholar] [CrossRef]
- Ernst, N.L.; Panicucci, B.; Carnes, J.; Stuart, K. Differential Functions of Two Editosome ExoUases in Trypanosoma brucei. RNA 2009, 15, 947–957. [Google Scholar] [CrossRef] [Green Version]
- Piccinelli, P.; Rosenblad, M.A.; Samuelsson, T. Identification and Analysis of Ribonuclease P and MRP RNA in a Broad Range of Eukaryotes. Nucleic Acids Res. 2005, 33, 4485–4495. [Google Scholar] [CrossRef] [Green Version]
- Rosenblad, M.A.; López, M.D.; Samuelsson, T. The Enigmatic RNase MRP of Kinetoplastids. RNA Biol. 2021, 18, 139–147. [Google Scholar] [CrossRef]
- Cerritelli, S.M.; Crouch, R.J. Ribonuclease H: The Enzymes in Eukaryotes. FEBS J. 2009, 276, 1494–1505. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Huang, J.; Zheng, Q.; Xie, L.; Lu, X.; Jin, J.; Wang, G. Mammalian Mitochondrial RNAs Are Degraded in the Mitochondrial Intermembrane Space by RNASET2. Protein Cell 2017, 8, 735–749. [Google Scholar] [CrossRef] [Green Version]
- Hollin, T.; Jaroszewski, L.; Stajich, J.E.; Godzik, A.; Le Roch, K.G. Identification and Phylogenetic Analysis of RNA Binding Domain Abundant in Apicomplexans or RAP Proteins. Microb. Genom. 2021, 7, 000541. [Google Scholar] [CrossRef]
- Leszczyniecka, M.; DeSalle, R.; Kang, D.; Fisher, P.B. The Origin of Polynucleotide Phosphorylase Domains. Mol. Phylogenet. Evol. 2004, 31, 123–130. [Google Scholar] [CrossRef]
- Razew, M.; Warkocki, Z.; Taube, M.; Kolondra, A.; Czarnocki-Cieciura, M.; Nowak, E.; Labedzka-Dmoch, K.; Kawinska, A.; Piatkowski, J.; Golik, P.; et al. Structural Analysis of MtEXO Mitochondrial RNA Degradosome Reveals Tight Coupling of Nuclease and Helicase Components. Nat. Commun. 2018, 9, 97. [Google Scholar] [CrossRef] [Green Version]
- Mattiacio, J.L.; Read, L.K. Evidence for a Degradosome-like Complex in the Mitochondria of Trypanosoma brucei. FEBS Lett. 2009, 583, 2333–2338. [Google Scholar] [CrossRef] [Green Version]
- Perrin, R.; Meyer, E.H.; Zaepfel, M.; Kim, Y.-J.; Mache, R.; Grienenberger, J.-M.; Gualberto, J.M.; Gagliardi, D. Two Exoribonucleases Act Sequentially to Process Mature 3′-Ends of Atp9 MRNAs in Arabidopsis Mitochondria. J. Biol. Chem. 2004, 279, 25440–25446. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.H.; Erzberger, J.P.; Root, J.; Wilson, D.M. The Human Homolog of Escherichia Coli Orn Degrades Small Single-Stranded RNA and DNA Oligomers. J. Biol. Chem. 2000, 275, 25900–25906. [Google Scholar] [CrossRef] [Green Version]
- Zimmer, S.L.; McEvoy, S.M.; Li, J.; Qu, J.; Read, L.K. A Novel Member of the RNase D Exoribonuclease Family Functions in Mitochondrial Guide RNA Metabolism in Trypanosoma brucei. J. Biol. Chem. 2011, 286, 10329–10340. [Google Scholar] [CrossRef] [Green Version]
- Pearce, S.F.; Rorbach, J.; Van Haute, L.; D’Souza, A.R.; Rebelo-Guiomar, P.; Powell, C.A.; Brierley, I.; Firth, A.E.; Minczuk, M. Maturation of Selected Human Mitochondrial TRNAs Requires Deadenylation. eLife 2017, 6, e27596. [Google Scholar] [CrossRef] [Green Version]
- Carnes, J.; Ernst, N.L.; Wickham, C.; Panicucci, B.; Stuart, K. KREX2 is Not Essential for Either Procyclic or Bloodstream form Trypanosoma brucei. PLoS ONE 2012, 7, e33405. [Google Scholar] [CrossRef] [Green Version]
- Hirayama, T.; Matsuura, T.; Ushiyama, S.; Narusaka, M.; Kurihara, Y.; Yasuda, M.; Ohtani, M.; Seki, M.; Demura, T.; Nakashita, H.; et al. A Poly(A)-Specific Ribonuclease Directly Regulates the Poly(A) Status of Mitochondrial MRNA in Arabidopsis. Nat. Commun. 2013, 4, 2247. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Reuter, M.; Lilie, H.; Liu, Y.; Wahle, E.; Song, H. Structural Insight into Poly(A) Binding and Catalytic Mechanism of Human PARN. EMBO J. 2005, 24, 4082–4093. [Google Scholar] [CrossRef] [Green Version]
- Grover, R.; Burse, S.A.; Shankrit, S.; Aggarwal, A.; Kirty, K.; Narta, K.; Srivastav, R.; Ray, A.K.; Malik, G.; Vats, A.; et al. Myg1 Exonuclease Couples the Nuclear and Mitochondrial Translational Programs through RNA Processing. Nucleic Acids Res. 2019, 47, 5852–5866. [Google Scholar] [CrossRef] [Green Version]
- Faure, E.; Barthélémy, R. True Mitochondrial TRNA Punctuation and Initiation Using Overlapping Stop and Start Codons at Specific and Conserved Positions. In Mitochondrial DNA—New Insights; InTech: London, UK, 2018. [Google Scholar]
- Gobert, A.; Gutmann, B.; Taschner, A.; Göringer, M.; Holzmann, J.; Hartmann, R.K.; Rossmanith, W.; Giegé, P. A Single Arabidopsis Organellar Protein Has RNase P Activity. Nat. Struct. Mol. Biol. 2010, 17, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, B.; Gobert, A.; Giegé, P. PRORP Proteins Support RNase P Activity in Both Organelles and the Nucleus in Arabidopsis. Genes Dev. 2012, 26, 1022–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA Moiety of Ribonuclease P Is the Catalytic Subunit of the Enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Morales, M.J.; Dang, Y.L.; Lou, Y.C.; Sulo, P.; Martin, N.C. A 105-KDa Protein Is Required for Yeast Mitochondrial RNase P Activity. Proc. Natl. Acad. Sci. USA 1992, 89, 9875–9879. [Google Scholar] [CrossRef] [Green Version]
- Lai, L.B.; Bernal-Bayard, P.; Mohannath, G.; Lai, S.M.; Gopalan, V.; Vioque, A. A Functional RNase P Protein Subunit of Bacterial Origin in Some Eukaryotes. Mol. Genet. Genom. 2011, 286, 359–369. [Google Scholar] [CrossRef]
- Seif, E.; Cadieux, A.; Lang, B.F.; Hybrid, E. Coli—Mitochondrial Ribonuclease P RNAs Are Catalytically Active. RNA 2006, 12, 1661–1670. [Google Scholar] [CrossRef] [Green Version]
- Holzmann, J.; Frank, P.; Löffler, E.; Bennett, K.L.; Gerner, C.; Rossmanith, W. RNase P without RNA: Identification and Functional Reconstitution of the Human Mitochondrial TRNA Processing Enzyme. Cell 2008, 135, 462–474. [Google Scholar] [CrossRef] [Green Version]
- Taschner, A.; Weber, C.; Buzet, A.; Hartmann, R.K.; Hartig, A.; Rossmanith, W. Nuclear RNase P of Trypanosoma brucei: A Single Protein in Place of the Multicomponent RNA-Protein Complex. Cell Rep. 2012, 2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Bonnard, G.; Gobert, A.; Arrivé, M.; Pinker, F.; Salinas-Giegé, T.; Giegé, P. Transfer RNA Maturation in Chlamydomonas Mitochondria, Chloroplast and the Nucleus by a Single RNase P Protein. Plant J. 2016, 87, 270–280. [Google Scholar] [CrossRef] [Green Version]
- Reiter, N.J.; Osterman, A.; Torres-Larios, A.; Swinger, K.K.; Pan, T.; Mondragón, A. Structure of a Bacterial Ribonuclease P Holoenzyme in Complex with TRNA. Nature 2010, 468, 784–789. [Google Scholar] [CrossRef]
- Wan, F.; Wang, Q.; Tan, J.; Tan, M.; Chen, J.; Shi, S.; Lan, P.; Wu, J.; Lei, M. Cryo-Electron Microscopy Structure of an Archaeal Ribonuclease P Holoenzyme. Nat. Commun. 2019, 10, 2617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Niu, S.; Tan, M.; Huang, C.; Li, M.; Song, Y.; Wang, Q.; Chen, J.; Shi, S.; Lan, P.; et al. Cryo-EM Structure of the Human Ribonuclease P Holoenzyme. Cell 2018, 175, 1393–1404.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, P.; Tan, M.; Zhang, Y.; Niu, S.; Chen, J.; Shi, S.; Qiu, S.; Wang, X.; Peng, X.; Cai, G.; et al. Structural Insight into Precursor TRNA Processing by Yeast Ribonuclease P. Science 2018, 362, eaat6678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinker, F.; Schelcher, C.; Fernandez-Millan, P.; Gobert, A.; Birck, C.; Thureau, A.; Roblin, P.; Giegé, P.; Sauter, C. Biophysical Analysis of Arabidopsis Protein-Only RNase P Alone and in Complex with TRNA Provides a Refined Model of TRNA Binding. J. Biol. Chem. 2017, 292, 13904–13913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatta, A.; Dienemann, C.; Cramer, P.; Hillen, H.S. Structural Basis of RNA Processing by Human Mitochondrial RNase P. Nat. Struct. Mol. Biol. 2021, 28, 713–723. [Google Scholar] [CrossRef]
- Howard, M.J.; Lim, W.H.; Fierke, C.A.; Koutmos, M. Mitochondrial Ribonuclease P Structure Provides Insight into the Evolution of Catalytic Strategies for Precursor-TRNA 5′ Processing. Proc. Natl. Acad. Sci. USA 2012, 109, 16149–16154. [Google Scholar] [CrossRef] [Green Version]
- Karasik, A.; Shanmuganathan, A.; Howard, M.J.; Fierke, C.A.; Koutmos, M. Nuclear Protein-Only Ribonuclease P2 Structure and Biochemical Characterization Provide Insight into the Conserved Properties of TRNA 5′ End Processing Enzymes. J. Mol. Biol. 2016, 428, 26–40. [Google Scholar] [CrossRef] [Green Version]
- Reinhard, L.; Sridhara, S.; Hallberg, B.M. Structure of the Nuclease Subunit of Human Mitochondrial RNase P. Nucleic Acids Res. 2015, 43, 5664–5672. [Google Scholar] [CrossRef] [Green Version]
- Imai, T.; Nakamura, T.; Maeda, T.; Nakayama, K.; Gao, X.; Nakashima, T.; Kakuta, Y.; Kimura, M. Pentatricopeptide Repeat Motifs in the Processing Enzyme PRORP1 in Arabidopsis Thaliana Play a Crucial Role in Recognition of Nucleotide Bases at TψC Loop in Precursor TRNAs. Biochem. Biophys. Res. Commun. 2014, 450, 1541–1546. [Google Scholar] [CrossRef]
- Chen, T.-H.; Tanimoto, A.; Shkriabai, N.; Kvaratskhelia, M.; Wysocki, V.; Gopalan, V. Use of Chemical Modification and Mass Spectrometry to Identify Substrate-Contacting Sites in Proteinaceous RNase P, a TRNA Processing Enzyme. Nucleic Acids Res. 2016, 44, 5344–5355. [Google Scholar] [CrossRef] [Green Version]
- Gobert, A.; Pinker, F.; Fuchsbauer, O.; Gutmann, B.; Boutin, R.; Roblin, P.; Sauter, C.; Giegé, P. Structural Insights into Protein-Only RNase P Complexed with TRNA. Nat. Commun. 2013, 4, 1353. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, T.; Kaitany, K.J.; Kakuta, Y.; Kimura, M.; Fierke, C.A.; Hall, T.M.T. Pentatricopeptide Repeats of Protein-Only RNase P Use a Distinct Mode to Recognize Conserved Bases and Structural Elements of Pre-TRNA. Nucleic Acids Res. 2020, 48, 11815–11826. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.J.; Klemm, B.P.; Fierke, C.A. Mechanistic Studies Reveal Similar Catalytic Strategies for Phosphodiester Bond Hydrolysis by Protein-Only and RNA-Dependent Ribonuclease P. J. Biol. Chem. 2015, 290, 13454–13464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brillante, N.; Gößringer, M.; Lindenhofer, D.; Toth, U.; Rossmanith, W.; Hartmann, R.K. Substrate Recognition and Cleavage-Site Selection by a Single-Subunit Protein-Only RNase P. Nucleic Acids Res. 2016, 44, 2323–2336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howard, M.J.; Karasik, A.; Klemm, B.P.; Mei, C.; Shanmuganathan, A.; Fierke, C.A.; Koutmos, M. Differential Substrate Recognition by Isozymes of Plant Protein-Only Ribonuclease P. RNA 2016, 22, 782–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, G.; Chen, T.-H.; Srivastava, A.S.; Kosek, D.; Biswas, P.K.; Gopalan, V.; Kirsebom, L.A. Cleavage of Model Substrates by Arabidopsis Thaliana PRORP1 Reveals New Insights into Its Substrate Requirements. PLoS ONE 2016, 11, e0160246. [Google Scholar] [CrossRef] [Green Version]
- Sen, A.; Karasik, A.; Shanmuganathan, A.; Mirkovic, E.; Koutmos, M.; Cox, R.T. Loss of the Mitochondrial Protein-Only Ribonuclease P Complex Causes Aberrant TRNA Processing and Lethality in Drosophila. Nucleic Acids Res. 2016, 44, 6409–6422. [Google Scholar] [CrossRef] [Green Version]
- Vilardo, E.; Rossmanith, W. Molecular Insights into HSD10 Disease: Impact of SDR5C1 Mutations on the Human Mitochondrial RNase P Complex. Nucleic Acids Res. 2015, 43, 5112–5119. [Google Scholar] [CrossRef] [Green Version]
- Hochberg, I.; Demain, L.A.M.; Richer, J.; Thompson, K.; Urquhart, J.E.; Rea, A.; Pagarkar, W.; Rodríguez-Palmero, A.; Schlüter, A.; Verdura, E.; et al. Bi-Allelic Variants in the Mitochondrial RNase P Subunit PRORP Cause Mitochondrial TRNA Processing Defects and Pleiotropic Multisystem Presentations. Am. J. Hum. Genet. 2021, 108, 2195–2204. [Google Scholar] [CrossRef]
- Phizicky, E.M.; Hopper, A.K. TRNA Biology Charges to the Front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [Green Version]
- Ma, M.; Li de la Sierra-Gallay, I.; Lazar, N.; Pellegrini, O.; Durand, D.; Marchfelder, A.; Condon, C.; van Tilbeurgh, H. The Crystal Structure of Trz1, the Long Form RNase Z from Yeast. Nucleic Acids Res. 2017, 45, 6209–6216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yip, M.C.J.; Savickas, S.; Gygi, S.P.; Shao, S. ELAC1 Repairs TRNAs Cleaved during Ribosome-Associated Quality Control. Cell Rep. 2020, 30, 2106–2114.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, A.; Schilling, O.; Späth, B.; Marchfelder, A. The TRNase Z Family of Proteins: Physiological Functions, Substrate Specificity and Structural Properties. Biol. Chem. 2005, 386, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Redko, Y.; de la Sierra-Gallay, I.L.; Condon, C. When All’s Zed and Done: The Structure and Function of RNase Z in Prokaryotes. Nat. Rev. Microbiol. 2007, 5, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; de la Sierra-Gallay, I.L.; Lazar, N.; Pellegrini, O.; Lepault, J.; Condon, C.; Durand, D.; van Tilbeurgh, H. Trz1, the Long Form RNase Z from Yeast, Forms a Stable Heterohexamer with Endonuclease Nuc1 and Mutarotase. Biochem. J. 2017, 474, 3599–3613. [Google Scholar] [CrossRef] [PubMed]
- Daoud, R.; Forget, L.; Lang, B.F. Yeast Mitochondrial RNase P, RNase Z and the RNA Degradosome Are Part of a Stable Supercomplex. Nucleic Acids Res. 2012, 40, 1728–1736. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Su, W.; Yuan, S.; Huang, Y. Functional Conservation of TRNase ZL among Saccharomyces Cerevisiae, Schizosaccharomyces Pombe and Humans. Biochem. J. 2009, 422, 483–492. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Dubrovskaya, V.A.; Dubrovsky, E.B. RNAi Knockdown of DRNaseZ, the Drosophila Homolog of ELAC2, Impairs Growth of Mitotic and Endoreplicating Tissues. Insect Biochem. Mol. Biol. 2011, 41, 167–177. [Google Scholar] [CrossRef]
- Xie, X.; Dubrovsky, E.B. Knockout of Drosophila RNase ZL Impairs Mitochondrial Transcript Processing, Respiration and Cell Cycle Progression. Nucleic Acids Res. 2015, 43, 10364–10375. [Google Scholar] [CrossRef] [Green Version]
- Kopajtich, R.; Mayr, J.A.; Prokisch, H. Analysis of Mitochondrial RNA-Processing Defects in Patient-Derived Tissues by QRT-PCR and RNAseq. In Mitochondria: Practical Protocols; Mokranjac, D., Perocchi, F., Eds.; Springer: New York, NY, USA, 2017; pp. 379–390. ISBN 978-1-4939-6824-4. [Google Scholar]
- Saoura, M.; Powell, C.A.; Kopajtich, R.; Alahmad, A.; AL-Balool, H.H.; Albash, B.; Alfadhel, M.; Alston, C.L.; Bertini, E.; Bonnen, P.E.; et al. Mutations in ELAC2 Associated with Hypertrophic Cardiomyopathy Impair Mitochondrial TRNA 3′-end Processing. Hum. Mutat. 2019, 40, 1731–1748. [Google Scholar] [CrossRef] [Green Version]
- Stoll, B.; Binder, S. Two NYN Domain Containing Putative Nucleases Are Involved in Transcript Maturation in Arabidopsis Mitochondria. Plant J. 2016, 85, 278–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gobert, A.; Bruggeman, M.; Giegé, P. Involvement of PIN-like Domain Nucleases in TRNA Processing and Translation Regulation. IUBMB Life 2019, 71, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Kubíková, J.; Reinig, R.; Salgania, H.K.; Jeske, M. LOTUS-Domain Proteins—Developmental Effectors from a Molecular Perspective. Biol. Chem. 2021, 402, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Schiaffini, M.; Chicois, C.; Pouclet, A.; Chartier, T.; Ubrig, E.; Gobert, A.; Zuber, H.; Mutterer, J.; Chicher, J.; Kuhn, L.; et al. A NYN Domain Protein Directly Interacts with DECAPPING1 and Is Required for Phyllotactic Pattern. Plant Physiol. 2022, 188, 1174–1188. [Google Scholar] [CrossRef]
- Ding, D.; Wei, C.; Dong, K.; Liu, J.; Stanton, A.; Xu, C.; Min, J.; Hu, J.; Chen, C. LOTUS Domain Is a Novel Class of G-Rich and G-Quadruplex RNA Binding Domain. Nucleic Acids Res. 2020, 48, 9262–9272. [Google Scholar] [CrossRef]
- Panigrahi, A.K.; Ernst, N.L.; Domingo, G.J.; Fleck, M.; Salavati, R.; Stuart, K.D. Compositionally and Functionally Distinct Editosomes in Trypanosoma brucei. RNA 2006, 12, 1038–1049. [Google Scholar] [CrossRef] [Green Version]
- McDermott, S.M.; Stuart, K. The Essential Functions of KREPB4 Are Developmentally Distinct and Required for Endonuclease Association with Editosomes. RNA 2017, 23, 1672–1684. [Google Scholar] [CrossRef]
- McDermott, S.M.; Carnes, J.; Stuart, K. Editosome RNase III Domain Interactions Are Essential for Editing and Differ between Life Cycle Stages in Trypanosoma brucei. RNA 2019, 25, 1150–1163. [Google Scholar] [CrossRef]
- Carnes, J.; McDermott, S.M.; Stuart, K. RNase III Domain of KREPB9 and KREPB10 Association with Editosomes in Trypanosoma brucei. mSphere 2018, 3, e00585-17. [Google Scholar] [CrossRef] [Green Version]
- Carnes, J.; Schnaufer, A.; McDermott, S.M.; Domingo, G.; Proff, R.; Steinberg, A.G.; Kurtz, I.; Stuart, K. Mutational Analysis of Trypanosoma brucei Editosome Proteins KREPB4 and KREPB5 Reveals Domains Critical for Function. RNA 2012, 18, 1897–1909. [Google Scholar] [CrossRef] [Green Version]
- Kable, M.L.; Seiwert, S.D.; Heidmann, S.; Stuart, K. RNA Editing: A Mechanism for GRNA-Specified Uridylate Insertion into Precursor MRNA. Science 1996, 273, 1189–1195. [Google Scholar] [CrossRef]
- Seiwert, S.D.; Heidmann, S.; Stuart, K. Direct Visualization of Uridylate Deletion In Vitro Suggests a Mechanism for Kinetoplastid RNA Editing. Cell 1996, 84, 831–841. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, A.; Panigrahi, A.; Cifuentes-Rojas, C.; Sacharidou, A.; Stuart, K.; Cruz-Reyes, J. Determinants for Association and Guide RNA-Directed Endonuclease Cleavage by Purified RNA Editing Complexes from Trypanosoma brucei. J. Mol. Biol. 2008, 381, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.; Hong, W. RAP—A Putative RNA-Binding Domain. Trends Biochem. Sci. 2004, 29, 567–570. [Google Scholar] [CrossRef]
- Boehm, E.; Zaganelli, S.; Maundrell, K.; Jourdain, A.A.; Thore, S.; Martinou, J.-C. FASTKD1 and FASTKD4 Have Opposite Effects on Expression of Specific Mitochondrial RNAs, Depending upon Their Endonuclease-like RAP Domain. Nucleic Acids Res. 2017, 45, 6135–6146. [Google Scholar] [CrossRef]
- Boehm, E.; Zornoza, M.; Jourdain, A.A.; Magdalena, A.D.; García-Consuegra, I.; Merino, R.T.; Orduña, A.; Martín, M.A.; Martinou, J.-C.; De la Fuente, M.A.; et al. Role of FAST Kinase Domains 3 (FASTKD3) in Post-Transcriptional Regulation of Mitochondrial Gene Expression. J. Biol. Chem. 2016, 291, 25877–25887. [Google Scholar] [CrossRef] [Green Version]
- Jourdain, A.A.; Popow, J.; de la Fuente, M.A.; Martinou, J.-C.; Anderson, P.; Simarro, M. The FASTK Family of Proteins: Emerging Regulators of Mitochondrial RNA Biology. Nucleic Acids Res. 2017, 45, 10941–10947. [Google Scholar] [CrossRef] [Green Version]
- Ohkubo, A.; Van Haute, L.; Rudler, D.L.; Stentenbach, M.; Steiner, F.A.; Rackham, O.; Minczuk, M.; Filipovska, A.; Martinou, J.-C. The FASTK Family Proteins Fine-Tune Mitochondrial RNA Processing. PLoS Genet. 2021, 17, e1009873. [Google Scholar] [CrossRef]
- Popow, J.; Alleaume, A.-M.; Curk, T.; Schwarzl, T.; Sauer, S.; Hentze, M.W. FASTKD2 is an RNA-Binding Protein Required for Mitochondrial RNA Processing and Translation. RNA 2015, 21, 1873–1884. [Google Scholar] [CrossRef] [Green Version]
- Kleinknecht, L.; Wang, F.; Stübe, R.; Philippar, K.; Nickelsen, J.; Bohne, A.-V. RAP, the Sole Octotricopeptide Repeat Protein in Arabidopsis, is Required for Chloroplast 16S RRNA Maturation. Plant Cell 2014, 26, 777–787. [Google Scholar] [CrossRef] [Green Version]
- Hollin, T.; Abel, S.; Falla, A.; Pasaje, C.F.A.; Bhatia, A.; Hur, M.; Kirkwood, J.S.; Saraf, A.; Prudhomme, J.; De Souza, A.; et al. Functional Genomics of RAP Proteins and Their Role in Mitoribosome Regulation in Plasmodium Falciparum. Nat. Commun. 2022, 13, 1275. [Google Scholar] [CrossRef] [PubMed]
- Yoo, D.H.; Choi, Y.-C.; Nam, D.E.; Choi, S.S.; Kim, J.W.; Choi, B.-O.; Chung, K.W. Identification of FASTKD2 Compound Heterozygous Mutations as the Underlying Cause of Autosomal Recessive MELAS-like Syndrome. Mitochondrion 2017, 35, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Magraner-Pardo, L.; Gobelli, D.; de la Fuente, M.A.; Pons, T.; Simarro, M. Systematic Analysis of FASTK Gene Family Alterations in Cancer. Int. J. Mol. Sci. 2021, 22, 11337. [Google Scholar] [CrossRef] [PubMed]
- Falchi, F.A.; Pizzoccheri, R.; Briani, F. Activity and Function in Human Cells of the Evolutionary Conserved Exonuclease Polynucleotide Phosphorylase. Int. J. Mol. Sci. 2022, 23, 1652. [Google Scholar] [CrossRef] [PubMed]
- Dendooven, T.; Sinha, D.; Roeselová, A.; Cameron, T.A.; De Lay, N.R.; Luisi, B.F.; Bandyra, K.J. A Cooperative PNPase-Hfq-RNA Carrier Complex Facilitates Bacterial Riboregulation. Mol. Cell 2021, 81, 2901–2913.e5. [Google Scholar] [CrossRef] [PubMed]
- Stoll, B.; Zendler, D.; Binder, S. RNA Processing Factor 7 and Polynucleotide Phosphorylase Are Necessary for Processing and Stability of Nad2 MRNA in Arabidopsis Mitochondria. RNA Biol. 2014, 11, 968–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrin, R.; Heike, L.; Jean-Michel, G.; Dominique, G. AtmtPNPase Is Required for Multiple Aspects of the 18S RRNA Metabolism in Arabidopsis Thaliana Mitochondria. Nucleic Acids Res. 2004, 32, 5174–5182. [Google Scholar] [CrossRef] [Green Version]
- Toompuu, M.; Tuomela, T.; Laine, P.; Paulin, L.; Dufour, E.; Jacobs, H.T. Polyadenylation and Degradation of Structurally Abnormal Mitochondrial TRNAs in Human Cells. Nucleic Acids Res. 2018, 46, 5209–5226. [Google Scholar] [CrossRef] [Green Version]
- Holec, S.; Lange, H.; Kühn, K.; Alioua, M.; Börner, T.; Gagliardi, D. Relaxed Transcription in Arabidopsis Mitochondria is Counterbalanced by RNA Stability Control Mediated by Polyadenylation and Polynucleotide Phosphorylase. Mol. Cell. Biol. 2006, 26, 2869–2876. [Google Scholar] [CrossRef] [Green Version]
- Pietras, Z.; Wojcik, M.A.; Borowski, L.S.; Szewczyk, M.; Kulinski, T.M.; Cysewski, D.; Stepien, P.P.; Dziembowski, A.; Szczesny, R.J. Controlling the Mitochondrial Antisense—Role of the SUV3-PNPase Complex and Its Co-Factor GRSF1 in Mitochondrial RNA Surveillance. Mol. Cell. Oncol. 2018, 5, e1516452. [Google Scholar] [CrossRef] [Green Version]
- Dhir, A.; Dhir, S.; Borowski, L.S.; Jimenez, L.; Teitell, M.; Rötig, A.; Crow, Y.J.; Rice, G.I.; Duffy, D.; Tamby, C.; et al. Mitochondrial Double-Stranded RNA Triggers Antiviral Signalling in Humans. Nature 2018, 560, 238–242. [Google Scholar] [CrossRef]
- Pajak, A.; Laine, I.; Clemente, P.; El-Fissi, N.; Schober, F.A.; Maffezzini, C.; Calvo-Garrido, J.; Wibom, R.; Filograna, R.; Dhir, A.; et al. Defects of Mitochondrial RNA Turnover Lead to the Accumulation of Double-Stranded RNA in vivo. PLoS Genet. 2019, 15, e1008240. [Google Scholar] [CrossRef] [Green Version]
- Shi, Z.; Yang, W.-Z.; Lin-Chao, S.; Chak, K.-F.; Yuan, H.S. Crystal Structure of Escherichia Coli PNPase: Central Channel Residues Are Involved in Processive RNA Degradation. RNA 2008, 14, 2361–2371. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.D.-H.; Shu, Z.; Lieser, S.A.; Chen, P.-L.; Lee, W.-H. Human Mitochondrial SUV3 and Polynucleotide Phosphorylase Form a 330-KDa Heteropentamer to Cooperatively Degrade Double-Stranded RNA with a 3′-to-5′ Directionality. J. Biol. Chem. 2009, 284, 20812–20821. [Google Scholar] [CrossRef] [Green Version]
- Phung, D.K.; Etienne, C.; Batista, M.; Langendijk-Genevaux, P.; Moalic, Y.; Laurent, S.; Liuu, S.; Morales, V.; Jebbar, M.; Fichant, G.; et al. RNA Processing Machineries in Archaea: The 5′-3′ Exoribonuclease ARNase J of the β-CASP Family is Engaged Specifically with the Helicase ASH-Ski2 and the 3′-5′ Exoribonucleolytic RNA Exosome Machinery. Nucleic Acids Res. 2020, 48, 3832–3847. [Google Scholar] [CrossRef]
- Sikorska, N.; Zuber, H.; Gobert, A.; Lange, H.; Gagliardi, D. RNA Degradation by the Plant RNA Exosome Involves Both Phosphorolytic and Hydrolytic Activities. Nat. Commun. 2017, 8, 2162. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.L.; Wang, Y.-T.; Yang, W.-Z.; Hsiao, Y.-Y.; Yuan, H.S. Crystal Structure of Human Polynucleotide Phosphorylase: Insights into Its Domain Function in RNA Binding and Degradation. Nucleic Acids Res. 2012, 40, 4146–4157. [Google Scholar] [CrossRef] [Green Version]
- Rius, R.; Van Bergen, N.J.; Compton, A.G.; Riley, L.G.; Kava, M.P.; Balasubramaniam, S.; Amor, D.J.; Fanjul-Fernandez, M.; Cowley, M.J.; Fahey, M.C.; et al. Clinical Spectrum and Functional Consequences Associated with Bi-Allelic Pathogenic PNPT1 Variants. J. Clin. Med. 2019, 8, 2020. [Google Scholar] [CrossRef] [Green Version]
- Golzarroshan, B.; Lin, C.-L.; Li, C.-L.; Yang, W.-Z.; Chu, L.-Y.; Agrawal, S.; Yuan, H.S. Crystal Structure of Dimeric Human PNPase Reveals Why Disease-Linked Mutants Suffer from Low RNA Import and Degradation Activities. Nucleic Acids Res. 2018, 46, 8630–8640. [Google Scholar] [CrossRef]
- Borowski, L.S.; Dziembowski, A.; Hejnowicz, M.S.; Stepien, P.P.; Szczesny, R.J. Human Mitochondrial RNA Decay Mediated by PNPase–HSuv3 Complex Takes Place in Distinct Foci. Nucleic Acids Res. 2013, 41, 1223–1240. [Google Scholar] [CrossRef]
- Silva, S.; Camino, L.P.; Aguilera, A. Human Mitochondrial Degradosome Prevents Harmful Mitochondrial R Loops and Mitochondrial Genome Instability. Proc. Natl. Acad. Sci. USA 2018, 115, 11024–11029. [Google Scholar] [CrossRef] [Green Version]
- Malla, S.; Li, Z. Functions of Conserved Domains of Human Polynucleotide Phosphorylase on RNA Oxidation. Insights Biomed. Res. 2019, 3, 62–67. [Google Scholar] [CrossRef]
- Khaw, S.-L.; Min-Wen, C.; Koh, C.-G.; Lim, B.; Shyh-Chang, N. Oocyte Factors Suppress Mitochondrial Polynucleotide Phosphorylase to Remodel the Metabolome and Enhance Reprogramming. Cell Rep. 2015, 12, 1080–1088. [Google Scholar] [CrossRef] [Green Version]
- Shimada, E.; Ahsan, F.M.; Nili, M.; Huang, D.; Atamdede, S.; TeSlaa, T.; Case, D.; Yu, X.; Gregory, B.D.; Perrin, B.J.; et al. PNPase Knockout Results in MtDNA Loss and an Altered Metabolic Gene Expression Program. PLoS ONE 2018, 13, e0200925. [Google Scholar] [CrossRef] [Green Version]
- Łabędzka-Dmoch, K.; Kolondra, A.; Karpińska, M.A.; Dębek, S.; Grochowska, J.; Grochowski, M.; Piątkowski, J.; Bui, T.H.D.; Golik, P. Pervasive Transcription of the Mitochondrial Genome in Candida Albicans Is Revealed in Mutants Lacking the MtEXO RNase Complex. RNA Biol. 2021, 18, 303–317. [Google Scholar] [CrossRef]
- Hillen, H.S.; Markov, D.A.; Wojtas, I.D.; Hofmann, K.B.; Lidschreiber, M.; Cowan, A.T.; Jones, J.L.; Temiakov, D.; Cramer, P.; Anikin, M. The Pentatricopeptide Repeat Protein Rmd9 Recognizes the Dodecameric Element in the 3′-UTRs of Yeast Mitochondrial MRNAs. Proc. Natl. Acad. Sci. USA 2021, 118, e2009329118. [Google Scholar] [CrossRef]
- Fujii, S.; Suzuki, T.; Giegé, P.; Higashiyama, T.; Koizuka, N.; Shikanai, T. The Restorer-of-Fertility-like 2 Pentatricopeptide Repeat Protein and RNase P Are Required for the Processing of Mitochondrial Orf291 RNA in Arabidopsis. Plant J. 2016, 86, 504–513. [Google Scholar] [CrossRef] [Green Version]
- Moreno, J.I.; Coleman, I.S.; Johnson, C.L.; Green, D.S.; Piva, M.A. Ccm1p Is a 15S RRNA Primary Transcript Processing Factor as Elucidated by a Novel in Vivo System in Saccharomyces Cerevisiae. Curr. Genet. 2020, 66, 775–789. [Google Scholar] [CrossRef]
- Penschow, J.L.; Sleve, D.A.; Ryan, C.M.; Read, L.K. TbDSS-1, an Essential Trypanosoma brucei Exoribonuclease Homolog That Has Pleiotropic Effects on Mitochondrial RNA Metabolism. Eukaryot. Cell 2004, 3, 1206–1216. [Google Scholar] [CrossRef] [Green Version]
- Aphasizhev, R.; Suematsu, T.; Zhang, L.; Aphasizheva, I. Constructive Edge of Uridylation-Induced RNA Degradation. RNA Biol. 2016, 13, 1078–1083. [Google Scholar] [CrossRef] [Green Version]
- Mattiacio, J.L.; Read, L.K. Roles for TbDSS-1 in RNA Surveillance and Decay of Maturation by-Products from the 12S RRNA Locus. Nucleic Acids Res. 2008, 36, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Muid, K.A.; Kimyon, Ö.; Reza, S.H.; Karakaya, H.C.; Koc, A. Characterization of Long Living Yeast Deletion Mutants That Lack Mitochondrial Metabolism Genes DSS1, PPA2 and AFG3. Gene 2019, 706, 172–180. [Google Scholar] [CrossRef]
- Chu, L.-Y.; Agrawal, S.; Chen, Y.-P.; Yang, W.-Z.; Yuan, H.S. Structural Insights into NanoRNA Degradation by Human Rexo2. RNA 2019, 25, 737–746. [Google Scholar] [CrossRef]
- Nicholls, T.J.; Spåhr, H.; Jiang, S.; Siira, S.J.; Koolmeister, C.; Sharma, S.; Kauppila, J.H.K.; Jiang, M.; Kaever, V.; Rackham, O.; et al. Dinucleotide Degradation by REXO2 Maintains Promoter Specificity in Mammalian Mitochondria. Mol. Cell 2019, 76, 784–796.e6. [Google Scholar] [CrossRef] [Green Version]
- Szewczyk, M.; Malik, D.; Borowski, L.S.; Czarnomska, S.D.; Kotrys, A.V.; Klosowska-Kosicka, K.; Nowotny, M.; Szczesny, R.J. Human REXO2 Controls Short Mitochondrial RNAs Generated by MtRNA Processing and Decay Machinery to Prevent Accumulation of Double-Stranded RNA. Nucleic Acids Res. 2020, 48, 5572–5590. [Google Scholar] [CrossRef]
- Kim, S.-K.; Lormand, J.D.; Weiss, C.A.; Eger, K.A.; Turdiev, H.; Turdiev, A.; Winkler, W.C.; Sondermann, H.; Lee, V.T. A Dedicated Diribonuclease Resolves a Key Bottleneck for the Terminal Step of RNA Degradation. Elife 2019, 8, e46313. [Google Scholar] [CrossRef]
- Bruni, F.; Gramegna, P.; Oliveira, J.M.A.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M.A. REXO2 Is an Oligoribonuclease Active in Human Mitochondria. PLoS ONE 2013, 8, e64670. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Liu, H.; Zhang, C.; Su, S.; Chen, Y.; Chen, X.; Li, Y.; Shao, Z.; Zhang, Y.; Shao, Q.; et al. Structural Basis for Guide RNA Trimming by RNase D Ribonuclease in Trypanosoma brucei. Nucleic Acids Res. 2021, 49, 568–583. [Google Scholar] [CrossRef]
- Kanazawa, M.; Ikeda, Y.; Nishihama, R.; Yamaoka, S.; Lee, N.-H.; Yamato, K.T.; Kohchi, T.; Hirayama, T. Regulation of the Poly(A) Status of Mitochondrial MRNA by Poly(A)-Specific Ribonuclease Is Conserved among Land Plants. Plant Cell Physiol. 2020, 61, 470–480. [Google Scholar] [CrossRef]
- Hirayama, T. PARN-like Proteins Regulate Gene Expression in Land Plant Mitochondria by Modulating MRNA Polyadenylation. Int. J. Mol. Sci. 2021, 22, 10776. [Google Scholar] [CrossRef]
- Monecke, T.; Schell, S.; Dickmanns, A.; Ficner, R. Crystal Structure of the RRM Domain of Poly(A)-Specific Ribonuclease Reveals a Novel M7G-Cap-Binding Mode. J. Mol. Biol. 2008, 382, 827–834. [Google Scholar] [CrossRef]
- Luhtala, N.; Parker, R. T2 Family Ribonucleases: Ancient Enzymes with Diverse Roles. Trends Biochem. Sci. 2010, 35, 253–259. [Google Scholar] [CrossRef] [Green Version]
- Thorn, A.; Steinfeld, R.; Ziegenbein, M.; Grapp, M.; Hsiao, H.-H.; Urlaub, H.; Sheldrick, G.M.; Gärtner, J.; Krätzner, R. Structure and Activity of the Only Human RNase T2. Nucleic Acids Res. 2012, 40, 8733–8742. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Shimada, E.; Koehler, C.M.; Teitell, M.A. PNPASE and RNA Trafficking into Mitochondria. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 998–1007. [Google Scholar] [CrossRef] [Green Version]
- Henneke, M.; Diekmann, S.; Ohlenbusch, A.; Kaiser, J.; Engelbrecht, V.; Kohlschütter, A.; Krätzner, R.; Madruga-Garrido, M.; Mayer, M.; Opitz, L.; et al. RNASET2-Deficient Cystic Leukoencephalopathy Resembles Congenital Cytomegalovirus Brain Infection. Nat. Genet. 2009, 41, 773–775. [Google Scholar] [CrossRef]
- Poulsen, J.B.; Andersen, K.R.; Kjær, K.H.; Durand, F.; Faou, P.; Vestergaard, A.L.; Talbo, G.H.; Hoogenraad, N.; Brodersen, D.E.; Justesen, J.; et al. Human 2′-Phosphodiesterase Localizes to the Mitochondrial Matrix with a Putative Function in Mitochondrial RNA Turnover. Nucleic Acids Res. 2011, 39, 3754–3770. [Google Scholar] [CrossRef]
- Wood, E.R.; Bledsoe, R.; Chai, J.; Daka, P.; Deng, H.; Ding, Y.; Harris-Gurley, S.; Kryn, L.H.; Nartey, E.; Nichols, J.; et al. The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors. J. Biol. Chem. 2015, 290, 19681–19696. [Google Scholar] [CrossRef] [Green Version]
- Rorbach, J.; Nicholls, T.J.J.; Minczuk, M. PDE12 Removes Mitochondrial RNA Poly(A) Tails and Controls Translation in Human Mitochondria. Nucleic Acids Res. 2011, 39, 7750–7763. [Google Scholar] [CrossRef] [Green Version]
- Abshire, E.T.; Hughes, K.L.; Diao, R.; Pearce, S.; Gopalakrishna, S.; Trievel, R.C.; Rorbach, J.; Freddolino, P.L.; Goldstrohm, A.C. Differential Processing and Localization of Human Nocturnin Controls Metabolism of MRNA and Nicotinamide Adenine Dinucleotide Cofactors. J. Biol. Chem. 2020, 295, 15112–15133. [Google Scholar] [CrossRef]
- Estrella, M.A.; Du, J.; Chen, L.; Rath, S.; Prangley, E.; Chitrakar, A.; Aoki, T.; Schedl, P.; Rabinowitz, J.; Korennykh, A. The Metabolites NADP+ and NADPH Are the Targets of the Circadian Protein Nocturnin (Curled). Nat. Commun. 2019, 10, 2367. [Google Scholar] [CrossRef] [Green Version]
- Le, P.T.; Bornstein, S.A.; Motyl, K.J.; Tian, L.; Stubblefield, J.J.; Hong, H.; Takahashi, J.S.; Green, C.B.; Rosen, C.J.; Guntur, A.R. A Novel Mouse Model Overexpressing Nocturnin Results in Decreased Fat Mass in Male Mice. J. Cell. Physiol. 2019, 234, 20228–20239. [Google Scholar] [CrossRef]
- Kojima, S.; Gendreau, K.L.; Sher-Chen, E.L.; Gao, P.; Green, C.B. Changes in Poly(A) Tail Length Dynamics from the Loss of the Circadian Deadenylase Nocturnin. Sci. Rep. 2015, 5, 17059. [Google Scholar] [CrossRef] [Green Version]
- Baggs, J.E.; Green, C.B. Nocturnin, a Deadenylase in Xenopus Laevis Retina: A Mechanism for Posttranscriptional Control of Circadian-Related MRNA. Curr. Biol. 2003, 13, 189–198. [Google Scholar] [CrossRef] [Green Version]
- Garbarino-Pico, E.; Niu, S.; Rollag, M.D.; Strayer, C.A.; Besharse, J.C.; Green, C.B. Immediate Early Response of the Circadian PolyA Ribonuclease Nocturnin to Two Extracellular Stimuli. RNA 2007, 13, 745–755. [Google Scholar] [CrossRef] [Green Version]
- Abshire, E.T.; Chasseur, J.; Bohn, J.A.; Del Rizzo, P.A.; Freddolino, P.L.; Goldstrohm, A.C.; Trievel, R.C. The Structure of Human Nocturnin Reveals a Conserved Ribonuclease Domain That Represses Target Transcript Translation and Abundance in Cells. Nucleic Acids Res. 2018, 46, 6257–6270. [Google Scholar] [CrossRef]
- Estrella, M.A.; Du, J.; Korennykh, A. Crystal Structure of Human Nocturnin Catalytic Domain. Sci. Rep. 2018, 8, 16294. [Google Scholar] [CrossRef] [Green Version]
- Misra, S.; Bennett, J.; Friew, Y.N.; Abdulghani, J.; Irvin-Wilson, C.V.; Tripathi, M.K.; Williams, S.; Chaudhuri, M.; Chaudhuri, G. A Type II Ribonuclease H from Leishmania Mitochondria: An Enzyme Essential for the Growth of the Parasite. Mol. Biochem. Parasitol. 2005, 143, 135–145. [Google Scholar] [CrossRef] [Green Version]
- Cerritelli, S.M.; Frolova, E.G.; Feng, C.; Grinberg, A.; Love, P.E.; Crouch, R.J. Failure to Produce Mitochondrial DNA Results in Embryonic Lethality in Rnaseh1 Null Mice. Mol. Cell 2003, 11, 807–815. [Google Scholar] [CrossRef]
- Engel, M.L.; Hines, J.C.; Ray, D.S. The Crithidia FasciculataRNH1 Gene Encodes Both Nuclear and Mitochondrial Isoforms of RNase H. Nucleic Acids Res. 2001, 29, 725–731. [Google Scholar] [CrossRef] [Green Version]
- De Cózar, J.M.G.; Gerards, M.; Teeri, E.; George, J.; Dufour, E.; Jacobs, H.T.; Jõers, P. RNase H1 Promotes Replication Fork Progression through Oppositely Transcribed Regions of Drosophila Mitochondrial DNA. J. Biol. Chem. 2019, 294, 4331–4344. [Google Scholar] [CrossRef] [Green Version]
- Kuciński, J.; Chamera, S.; Kmera, A.; Rowley, M.J.; Fujii, S.; Khurana, P.; Nowotny, M.; Wierzbicki, A.T. Evolutionary History and Activity of RNase H1-Like Proteins in Arabidopsis thaliana. Plant Cell Physiol. 2020, 61, 1107–1119. [Google Scholar] [CrossRef] [PubMed]
- Holt, I.J. The Jekyll and Hyde Character of RNase H1 and Its Multiple Roles in Mitochondrial DNA Metabolism. DNA Repair 2019, 84, 102630. [Google Scholar] [CrossRef] [PubMed]
- Holmes, J.B.; Akman, G.; Wood, S.R.; Sakhuja, K.; Cerritelli, S.M.; Moss, C.; Bowmaker, M.R.; Jacobs, H.T.; Crouch, R.J.; Holt, I.J. Primer Retention Owing to the Absence of RNase H1 Is Catastrophic for Mitochondrial DNA Replication. Proc. Natl. Acad. Sci. USA 2015, 112, 9334–9339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Behadili, A.; Uhler, J.P.; Berglund, A.-K.; Peter, B.; Doimo, M.; Reyes, A.; Wanrooij, S.; Zeviani, M.; Falkenberg, M. A Two-Nuclease Pathway Involving RNase H1 Is Required for Primer Removal at Human Mitochondrial OriL. Nucleic Acids Res. 2018, 46, 9471–9483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Hage, A.; Webb, S.; Kerr, A.; Tollervey, D. Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in TRNA Genes, Retrotransposons and Mitochondria. PLoS Genet. 2014, 10, e1004716. [Google Scholar] [CrossRef]
- Lima, W.F.; Murray, H.M.; Damle, S.S.; Hart, C.E.; Hung, G.; De Hoyos, C.L.; Liang, X.-H.; Crooke, S.T. Viable RNaseH1 Knockout Mice Show RNaseH1 Is Essential for R Loop Processing, Mitochondrial and Liver Function. Nucleic Acids Res. 2016, 44, 5299–5312. [Google Scholar] [CrossRef] [Green Version]
- Posse, V.; Al-Behadili, A.; Uhler, J.P.; Clausen, A.R.; Reyes, A.; Zeviani, M.; Falkenberg, M.; Gustafsson, C.M. RNase H1 Directs Origin-Specific Initiation of DNA Replication in Human Mitochondria. PLoS Genet. 2019, 15, e1007781. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Wang, W.; Yao, Y.; Sun, Q. Mitochondrial RNase H1 Activity Regulates R-Loop Homeostasis to Maintain Genome Integrity and Enable Early Embryogenesis in Arabidopsis. PLoS Biol. 2021, 19, e3001357. [Google Scholar] [CrossRef]
- Reyes, A.; Melchionda, L.; Nasca, A.; Carrara, F.; Lamantea, E.; Zanolini, A.; Lamperti, C.; Fang, M.; Zhang, J.; Ronchi, D.; et al. RNASEH1 Mutations Impair MtDNA Replication and Cause Adult-Onset Mitochondrial Encephalomyopathy. Am. J. Hum. Genet. 2015, 97, 186–193. [Google Scholar] [CrossRef] [Green Version]
- Young, M.J.; Copeland, W.C. Human Mitochondrial DNA Replication Machinery and Disease. Curr. Opin. Genet. Dev. 2016, 38, 52–62. [Google Scholar] [CrossRef] [Green Version]
- Chang, D.D.; Clayton, D.A. A Novel Endoribonuclease Cleaves at a Priming Site of Mouse Mitochondrial DNA Replication. EMBO J. 1987, 6, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Kiss, T.; Filipowicz, W. Evidence against a Mitochondrial Location of the 7-2/MRP RNA in Mammalian Cells. Cell 1992, 70, 11–16. [Google Scholar] [CrossRef]
- Lu, Q.; Wierzbicki, S.; Krasilnikov, A.S.; Schmitt, M.E. Comparison of Mitochondrial and Nucleolar RNase MRP Reveals Identical RNA Components with Distinct Enzymatic Activities and Protein Components. RNA 2010, 16, 529–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noh, J.H.; Kim, K.M.; Abdelmohsen, K.; Yoon, J.-H.; Panda, A.C.; Munk, R.; Kim, J.; Curtis, J.; Moad, C.A.; Wohler, C.M.; et al. HuR and GRSF1 Modulate the Nuclear Export and Mitochondrial Localization of the LncRNA RMRP. Genes Dev. 2016, 30, 1224–1239. [Google Scholar] [CrossRef] [Green Version]
- Lan, P.; Zhou, B.; Tan, M.; Li, S.; Cao, M.; Wu, J.; Lei, M. Structural Insight into Precursor Ribosomal RNA Processing by Ribonuclease MRP. Science 2020, 369, 656–663. [Google Scholar] [CrossRef]
- Perederina, A.; Li, D.; Lee, H.; Bator, C.; Berezin, I.; Hafenstein, S.L.; Krasilnikov, A.S. Cryo-EM Structure of Catalytic Ribonucleoprotein Complex RNase MRP. Nat. Commun. 2020, 11, 3474. [Google Scholar] [CrossRef]
- Hussen, B.M.; Azimi, T.; Hidayat, H.J.; Taheri, M.; Ghafouri-Fard, S. Long Non-Coding RNA RMRP in the Pathogenesis of Human Disorders. Front. Cell Dev. Biol. 2021, 9, 1130. [Google Scholar] [CrossRef]
- Robertson, N.; Shchepachev, V.; Wright, D.; Turowski, T.W.; Spanos, C.; Helwak, A.; Zamoyska, R.; Tollervey, D. A Disease-Linked LncRNA Mutation in RNase MRP Inhibits Ribosome Synthesis. Nat. Commun. 2022, 13, 649. [Google Scholar] [CrossRef]
- Summer, S.; Smirnova, A.; Gabriele, A.; Toth, U.; Fasemore, A.M.; Förstner, K.U.; Kuhn, L.; Chicher, J.; Hammann, P.; Mitulović, G.; et al. YBEY Is an Essential Biogenesis Factor for Mitochondrial Ribosomes. Nucleic Acids Res. 2020, 48, 9762–9786. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhou, W.; Liu, G.; Yang, C.; Sun, Y.; Wu, W.; Cao, S.; Wang, C.; Hai, G.; Wang, Z.; et al. The Conserved Endoribonuclease YbeY is Required for Chloroplast Ribosomal RNA Processing in Arabidopsis. Plant Physiol. 2015, 168, 205–221. [Google Scholar] [CrossRef] [Green Version]
- Ghosal, A.; Köhrer, C.; Babu, V.M.P.; Yamanaka, K.; Davies, B.W.; Jacob, A.I.; Ferullo, D.J.; Gruber, C.C.; Vercruysse, M.; Walker, G.C. C21orf57 is a Human Homologue of Bacterial YbeY Proteins. Biochem. Biophys. Res. Commun. 2017, 484, 612–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Souza, A.R.; Van Haute, L.; Powell, C.A.; Mutti, C.D.; Páleníková, P.; Rebelo-Guiomar, P.; Rorbach, J.; Minczuk, M. YbeY is Required for Ribosome Small Subunit Assembly and TRNA Processing in Human Mitochondria. Nucleic Acids Res. 2021, 49, 5798–5812. [Google Scholar] [CrossRef] [PubMed]
- Horpaopan, S.; Spier, I.; Zink, A.M.; Altmüller, J.; Holzapfel, S.; Laner, A.; Vogt, S.; Uhlhaas, S.; Heilmann, S.; Stienen, D.; et al. Genome-Wide CNV Analysis in 221 Unrelated Patients and Targeted High-Throughput Sequencing Reveal Novel Causative Candidate Genes for Colorectal Adenomatous Polyposis. Int. J. Cancer 2015, 136, E578–E589. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, M.-Y.; Shen, Y.-J.; Zhuo, X.-B.; Gao, P.; Zhou, F.-S.; Liang, B.; Zu, J.; Zhang, Q.; Suleman, S.; et al. A Large-Scale, Exome-Wide Association Study of Han Chinese Women Identifies Three Novel Loci Predisposing to Breast Cancer. Cancer Res. 2018, 78, 3087–3097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fekete, Z.; Ellis, T.P.; Schonauer, M.S.; Dieckmann, C.L. Pet127 Governs a 5′ → 3′-Exonuclease Important in Maturation of Apocytochrome b MRNA in Saccharomyces Cerevisiae. J. Biol. Chem. 2008, 283, 3767–3772. [Google Scholar] [CrossRef] [Green Version]
- Wegierski, T.; Dmochowska, A.; Jabłonowska, A.; Dziembowski, A.; Bartnik, E.; Stepień, P.P. Yeast Nuclear PET127 Gene Can Suppress Deletions of the SUV3 or DSS1 Genes: An Indication of a Functional Interaction between 3′ and 5′ Ends of Mitochondrial MRNAs. Acta Biochim. Pol. 1998, 45, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Wiesenberger, G.; Fox, T.D. Pet127p, a Membrane-Associated Protein Involved in Stability and Processing of Saccharomyces Cerevisiae Mitochondrial RNAs. Mol. Cell. Biol. 1997, 17, 2816–2824. [Google Scholar] [CrossRef] [Green Version]
- Łabędzka-Dmoch, K.; Razew, M.; Gapinska, M.; Piatkowski, J.; Kolondra, A.; Salmonowicz, H.; Wenda, J.M.; Nowotny, M.; Golik, P. The Pet127 Protein is a Mitochondrial 5′-to-3′ Exoribonuclease from the PD-(D/E)XK Superfamily Involved in RNA Maturation and Intron Degradation in Yeasts. RNA 2022, 28, 711–728. [Google Scholar] [CrossRef]

| Enzymes | RBD | Amoebozoa | Fungi | Animals | Chlorophyta | Streptophyta | Euglenozoa | Stramenopiles | Alveolata | Ref. nb | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Endoribonucleases | RNase P | RNP | RNA | + | - | +/- | - | - | + | + | [7] | |
| PRORP | PPR | - | - | + | + | + | + | + | + | [7] | ||
| RNase Z | ZS | - | - | - | - | [8,9,10,11] | ||||||
| ZL | exosite | + | + | + | + | [8,9,10,11] | ||||||
| YbeY | + | + | [12] | |||||||||
| MNU1/2 | Lotus | - | - | - | - | spermatophta | - | - | - | [13] | ||
| KREN | ZnFD PUF | + | [14] | |||||||||
| RNase MRP | +/- | + | + | + | + | + | + | + | [15,16] | |||
| RNase H1 | HBD | (+) | + | + | + | [17] | ||||||
| RNase T2 | + | [18] | ||||||||||
| RAP | HPR/OPR | + | + | [19] | ||||||||
| 3′-5′ | PNPase | KH-S1 | - | + | + | - | [20] | |||||
| RNase II/R | DSS1 | WH-HTH S1 or CSD1-CSD2 S1 | + | + | [21,22] | |||||||
| RNR1 | CSD1-CSD2 S1 | + | [23] | |||||||||
| REXO2 | + | [24] | ||||||||||
| RNase D | + | [25] | ||||||||||
| PDE12 | + | [26] | ||||||||||
| KREX | + | [14,27] | ||||||||||
| PARN | + | [28,29] | ||||||||||
| Myg1 | + | + | [30] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cartalas, J.; Coudray, L.; Gobert, A. How RNases Shape Mitochondrial Transcriptomes. Int. J. Mol. Sci. 2022, 23, 6141. https://doi.org/10.3390/ijms23116141
Cartalas J, Coudray L, Gobert A. How RNases Shape Mitochondrial Transcriptomes. International Journal of Molecular Sciences. 2022; 23(11):6141. https://doi.org/10.3390/ijms23116141
Chicago/Turabian StyleCartalas, Jérémy, Léna Coudray, and Anthony Gobert. 2022. "How RNases Shape Mitochondrial Transcriptomes" International Journal of Molecular Sciences 23, no. 11: 6141. https://doi.org/10.3390/ijms23116141
APA StyleCartalas, J., Coudray, L., & Gobert, A. (2022). How RNases Shape Mitochondrial Transcriptomes. International Journal of Molecular Sciences, 23(11), 6141. https://doi.org/10.3390/ijms23116141






