RNAi Efficiency through dsRNA Injection Is Enhanced by Knockdown of dsRNA Nucleases in the Fall Webworm, Hyphantria cunea (Lepidoptera: Arctiidae)
Abstract
:1. Introduction
2. Results
2.1. Both Hemolymph and Gut Content of H. cunea Rapidly Degrade dsRNA
2.2. Identification and Characterization of Four HcdsRNase Genes from H. cunea
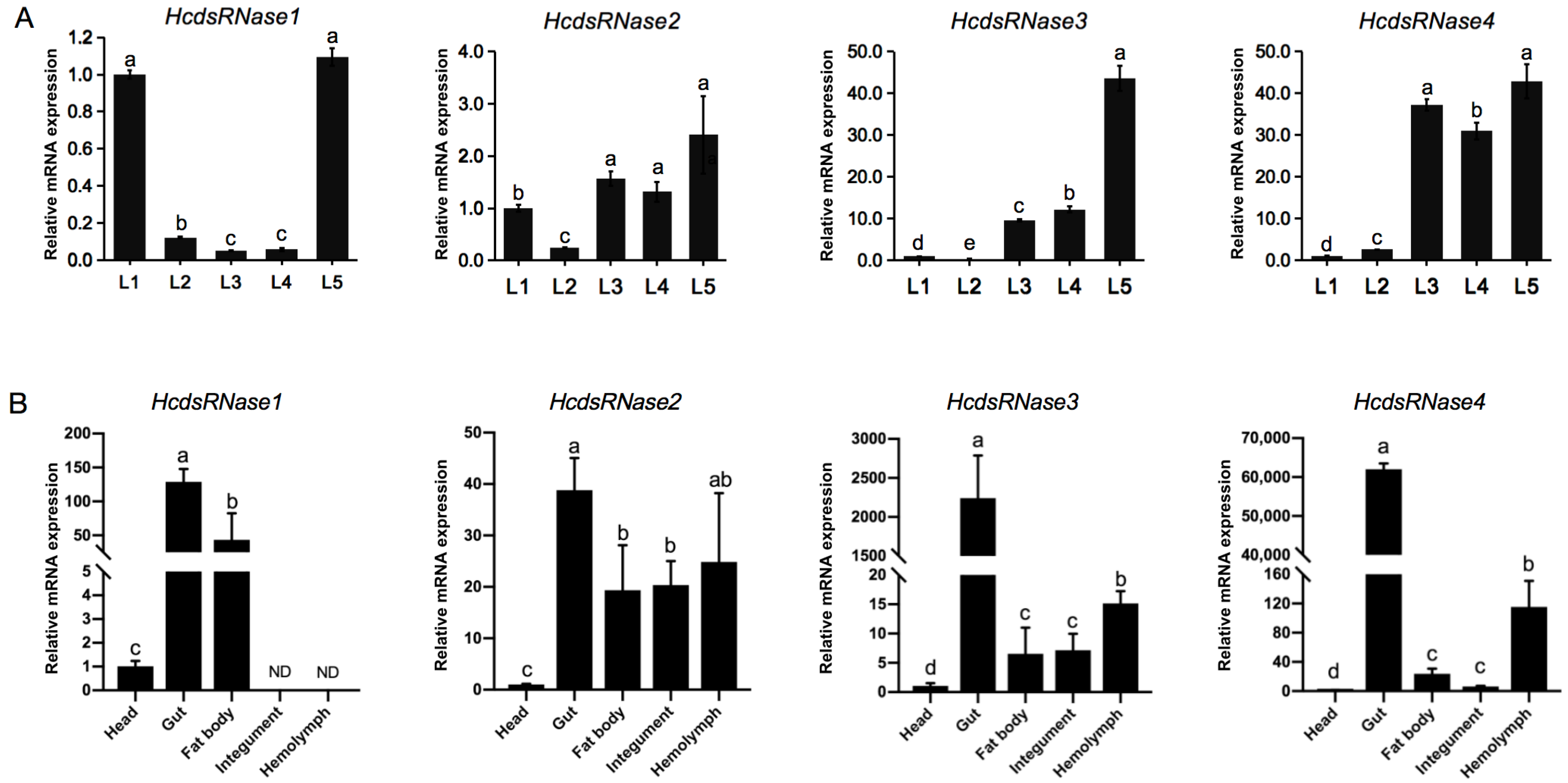
2.3. HcdsRNase Genes Are Expressed in all the Instars of H. cunea Larvae and Are Mainly Functional in Gut and Hemolymph Tissues
2.4. dsRNA Injection can Induce Expression of HcdsRNase3 and HcdsRNase4
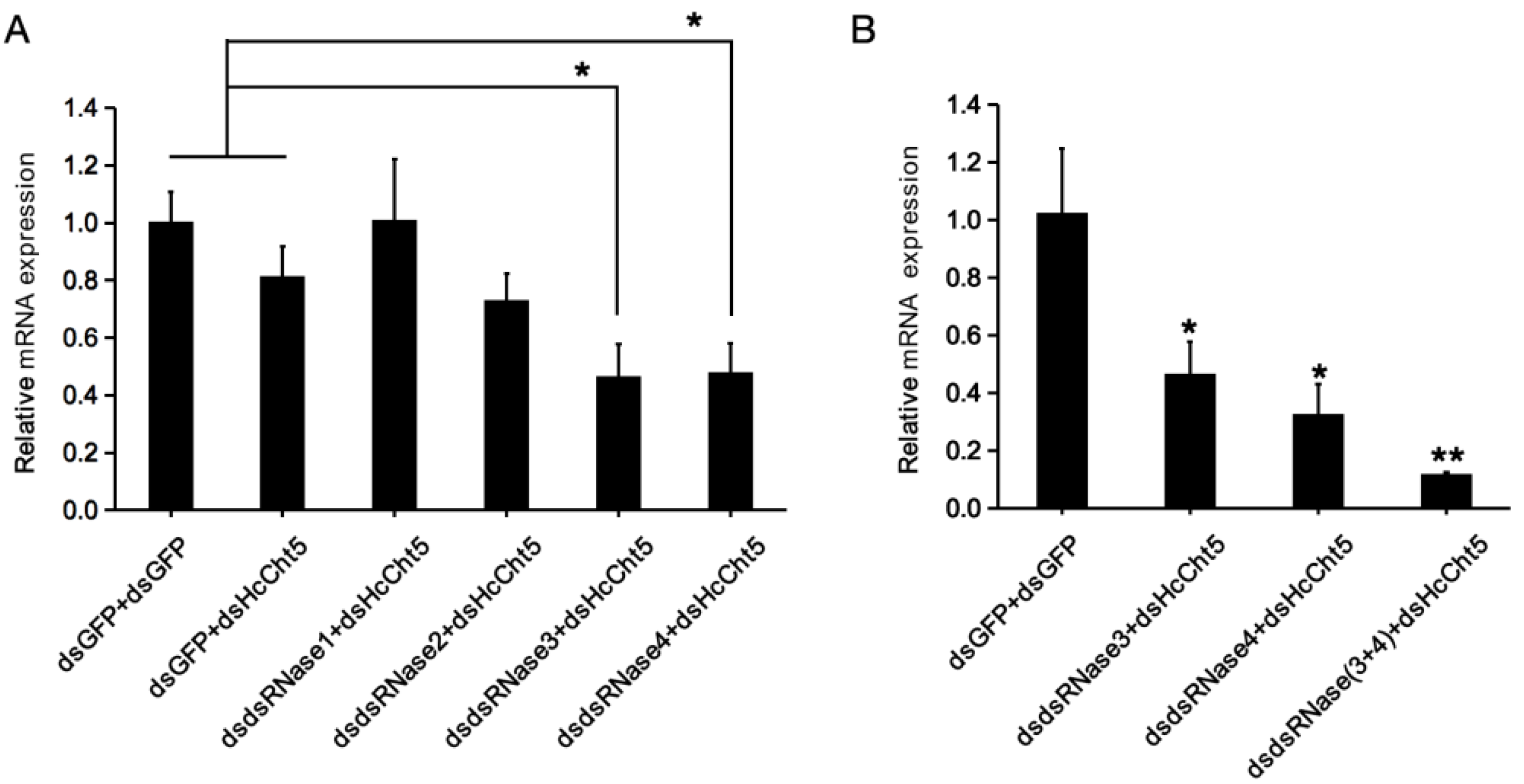
2.5. HcdsRNase Genes can Be Effectively Silenced by Injecting the Corresponding dsRNAs
2.6. HcdsRNase3 and HcdsRNase4 Inhibits RNAi Efficacy
3. Discussion
4. Materials and Methods
4.1. Insect Rearing
4.2. Identification and Characterization of dsRNases from H. cunea
4.3. Tissue-Specific and Developmental Expression Analysis
4.4. RNA Isolation, cDNA Synthesis, and RT-qPCR
4.5. Synthesis of dsRNA
4.6. Incubations of dsRNA in Insect Tissue Extracts
4.7. Transcriptional Responses of HcdsRNases after dsRNA Injection
4.8. Knockdown of HcdsRNases and RNAi Efficacy Assessment by RNAi-of-RNAi Assays
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gomi, T. Seasonal adaptations of the fall webworm Hyphantria cunea (Drury) (Lepidoptera: Arctiidae) following its invasion of Japan. Ecol. Res. 2007, 22, 855–861. [Google Scholar] [CrossRef]
- Ji, R.; Xie, B.; Li, X.; Gao, Z.; Li, D. Research progress on the invasive species, Hyphantria cunea. Entomol. Knowl. 2003, 40, 13–18. [Google Scholar]
- Sun, L.; Ma, H.; Gao, Y.; Wang, Z.; Cao, C. Functional Identification and Characterization of Leucokinin and Its Receptor in the Fall Webworm, Hyphantria cunea. Front. Physiol. 2021, 12, 741362. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; He, S.; Zhu, C.; Wang, T.; Xu, Z.; Zong, S. Projecting the current and future potential global distribution of Hyphantria cunea (Lepidoptera: Arctiidae) using CLIMEX. Pest. Manag. Sci. 2019, 75, 160–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Feng, K.; Tang, F.; Xu, M. Activation of the Host Immune Response in Hyphantria cunea (Drury) (Lepidoptera: Noctuidae) Induced by Serratia marcescens Bizio. Insects 2021, 12, 983. [Google Scholar] [CrossRef]
- Sun, L.; Liu, P.; Sun, S.; Yan, S.; Cao, C. Transcriptomic analysis of interactions between Hyphantria cunea larvae and nucleopolyhedrovirus. Pest. Manag. Sci. 2019, 75, 1024–1033. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.Q.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Baum, J.A.; Roberts, J.K. Progress Towards RNAi-Mediated Insect Pest Management. Adv. Insect Physiol. 2014, 47, 249–295. [Google Scholar]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi Efficiency, Systemic Properties, and Novel Delivery Methods for Pest Insect Control: What We Know So Far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef] [Green Version]
- Zhu, K.Y.; Palli, S.R. Mechanisms, Applications, and Challenges of Insect RNA Interference. Annu. Rev. Entomol. 2020, 65, 293–311. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Khan, S.A.; Hasse, C.; Ruf, S.; Heckel, D.G.; Bock, R. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 2015, 347, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.M.W.; Silver, K.; Zhang, J.; Park, Y.; Zhu, K.Y. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest. Manag. Sci. 2019, 75, 18–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yen, S.; Ren, B.; Zeng, B.; Shen, J. Improving RNAi efficiency for pest control in crop species. Biotechniques 2020, 68, 283–290. [Google Scholar] [CrossRef] [Green Version]
- Silver, K.; Cooper, A.M.W.; Zhu, K.Y. Strategies for enhancing the efficiency of RNA interference in insects. Pest. Manag. Sci. 2021, 77, 2645–2658. [Google Scholar] [CrossRef] [PubMed]
- Prentice, K.; Smagghe, G.; Gheysen, G.; Christians, O. Nuclease activity decreases the RNAi response in the sweetpotato weevil Cylas puncticollis. Insect Biochem. Mol. Biol. 2019, 110, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Song, H.F.; Fan, Y.H.; Zhang, J.Q.; Cooper, A.M.W.; Silver, K.; Li, D.Q.; Li, T.; Ma, E.B.; Zhu, K.Y.; Zhang, J.Z. Contributions of dsRNases to differential RNAi efficiencies between the injection and oral delivery of dsRNA in Locusta migratoria. Pest. Manag. Sci. 2019, 75, 1707–1717. [Google Scholar] [CrossRef]
- Al Baki, A.; Jung, J.K.; Kim, Y. Alteration of insulin signaling to control insect pest by using transformed bacteria expressing dsRNA. Pest. Manag. Sci. 2020, 76, 1020–1030. [Google Scholar] [CrossRef]
- Chen, J.-X.; Lyu, Z.-H.; Wang, C.-Y.; Cheng, J.; Lin, T. RNA interference of a trehalose-6-phosphate synthase gene reveals its roles in the biosynthesis of chitin and lipids in Heortia vitessoides (Lepidoptera: Crambidae). Insect Sci. 2020, 27, 212–223. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.-Z.; Li, N.-Y.; Xie, Y.-X.; Zhang, Q.; Wang, Y.; Lu, Z.-J. Identification and Functional Analysis of Two Chitin Synthase Genes in the Common Cutworm, Spodoptera litura. Insects 2020, 11, 253. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Zhao, D.; Zhang, Y.; Guo, W.; Wang, W.; Zhao, K.; Gao, Y.; Wang, X. Identification and characterization of chitin deacetylase2 from the American white moth, Hyphantria cunea (Drury). Gene 2018, 670, 98–105. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Zhang, S.; Kong, X.; Liu, F.; Zhang, Z. RNAi-Mediated Silencing of the Chitinase 5 Gene for Fall Webworm (Hyphantria cunea) Can Inhibit Larval Molting Depending on the Timing of dsRNA Injection. Insects 2021, 12, 406. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Whyard, S.; Velez, A.M.; Smagghe, G. Double-Stranded RNA Technology to Control Insect Pests: Current Status and Challenges. Front. Plant Sci. 2020, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.H.; Song, H.F.; Abbas, M.; Wang, Y.L.; Li, T.; Ma, E.B.; Cooper, A.M.W.; Silver, K.; Zhu, K.Y.; Zhang, J.Z. A dsRNA-degrading nuclease (dsRNase2) limits RNAi efficiency in the Asian corn borer (Ostrinia furnacalis). Insect Sci. 2021, 28, 1677–1689. [Google Scholar] [CrossRef] [PubMed]
- Gurusamy, D.; Howell, J.L.; Chereddy, S.C.R.R.; Mogilicherla, K.; Palli, S.R. Improving RNA interference in the southern green stink bug, Nezara viridula. J. Pest Sci. 2021, 94, 1461–1472. [Google Scholar] [CrossRef]
- Peng, Y.C.; Zhu, G.H.; Wang, K.X.; Chen, J.S.; Liu, X.L.; Wu, M.; Zhao, C.Q.; Xiao, H.J.; Palli, S.R.; Han, Z.J. Knockout of SldsRNase1 and SldsRNase2 revealed their function in dsRNA degradation and contribution to RNAi efficiency in the tobacco cutworm, Spodoptera litura. J. Pest Sci. 2021, 94, 1449–1460. [Google Scholar] [CrossRef]
- Arimatsu, Y.; Kotani, E.; Sugimura, Y.; Furusawa, T. Molecular characterization of a cDNA encoding extracellular dsRNase and its expression in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2007, 37, 176–183. [Google Scholar] [CrossRef]
- Powell, M.E.; Bradish, H.M.; Gatehouse, J.A.; Fitches, E.C. Systemic RNAi in the small hive beetle Aethina tumida Murray (Coleoptera: Nitidulidae), a serious pest of the European honey bee Apis mellifera. Pest. Manag. Sci. 2017, 73, 53–63. [Google Scholar] [CrossRef] [Green Version]
- Guan, R.-B.; Li, H.-C.; Fan, Y.-J.; Hu, S.-R.; Christiaens, O.; Smagghe, G.; Miao, X.-X. A nuclease specific to lepidopteran insects suppresses RNAi. J. Biol. Chem. 2018, 293, 6011–6021. [Google Scholar] [CrossRef] [Green Version]
- Lomate, P.R.; Bonning, B.C. Distinct properties of proteases and nucleases in the gut, salivary gland and saliva of southern green stink bug, Nezara viridula. Sci. Rep. 2016, 6, 27587. [Google Scholar] [CrossRef]
- Allen, M.L.; Walker, W.B., III. Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J. Insect Physiol. 2012, 58, 391–396. [Google Scholar] [CrossRef]
- Shukla, J.N.; Kalsi, M.; Sethi, A.; Narva, K.E.; Fishilevich, E.; Singh, S.; Mogilicherla, K.; Palli, S.R. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 2016, 13, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Mogilicherla, K.; Howell, J.L.; Palli, S.R. Improving RNAi in the Brown Marmorated Stink Bug: Identification of target genes and reference genes for RT-qPCR. Sci. Rep. 2018, 8, 3720. [Google Scholar] [CrossRef] [Green Version]
- Tayler, A.; Heschuk, D.; Giesbrecht, D.; Park, J.Y.; Whyard, S. Efficiency of RNA interference is improved by knockdown of dsRNA nucleases in tephritid fruit flies. Open Biol. 2019, 9, 190198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spit, J.; Philips, A.; Wynant, N.; Santos, D.; Plaetinck, G.; Broeck, J.V. Knockdown of nuclease activity in the gut enhances RNAi efficiency in the Colorado potato beetle, Leptinotarsa decemlineata, but not in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2017, 81, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Taning, C.N.T.; Smagghe, G.; Christiaens, O. Silencing of Double-Stranded Ribonuclease Improves Oral RNAi Efficacy in Southern Green Stinkbug Nezara viridula. Insects 2021, 12, 115. [Google Scholar] [CrossRef]
- Singh, I.K.; Singh, S.; Mogilicherla, K.; Shukla, J.N.; Palli, S.R. Comparative analysis of double-stranded RNA degradation and processing in insects. Sci. Rep. 2017, 7, 17059. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Smagghe, G.; Swevers, L. Transcriptional response of BmTo119-1 and RNAi machinery genes to exogenous dsRNA in the midgut of Bombyx mori. J. Insect Physiol. 2013, 59, 646–654. [Google Scholar] [CrossRef]
- Garbutt, J.S.; Belles, X.; Richards, E.H.; Reynolds, S.E. Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: Evidence from Manduca sexta and Blattella germanica. J. Insect Physiol. 2013, 59, 171–178. [Google Scholar] [CrossRef]
- Wang, K.; Peng, Y.; Pu, J.; Fu, W.; Wang, J.; Han, Z. Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol. 2016, 77, 1–9. [Google Scholar] [CrossRef]
- Friedhoff, P.; Gimadutdinow, O.; Pingoud, A. Identificationof catalytically relevant amino acids of the extracellular Serratia marcescens endonuclease by alignment guided mutagenesis. Nucleic Acids Res. 1994, 22, 3280–3287. [Google Scholar] [CrossRef] [Green Version]
- Cooper, A.M.W.; Song, H.F.; Shi, X.K.; Yu, Z.T.; Lorenzen, M.; Silver, K.; Zhang, J.Z.; Zhu, K.Y. Molecular Characterizations of Double-Stranded RNA Degrading Nuclease Genes from Ostrinia nubilalis. Insects 2020, 11, 652. [Google Scholar] [CrossRef] [PubMed]
- Vatanparast, M.; Kim, Y. Optimization of recombinant bacteria expressing dsRNA to enhance insecticidal activity against a lepidopteran insect, Spodoptera exigua. PLoS ONE 2017, 12, e0183054. [Google Scholar]
- Chen, J.Z.; Jiang, Y.X.; Li, M.W.; Li, J.W.; Zha, B.H.; Yang, G. Double-Stranded RNA-Degrading Enzymes Reduce the Efficiency of RNA Interference in Plutella xylostella. Insects 2021, 12, 712. [Google Scholar] [CrossRef] [PubMed]
- Wynant, N.; Santos, D.; Verdonck, R.; Spit, J.; Van Wielendaele, P.; Vanden Broeck, J. Identification, functional characterization and phylogenetic analysis of double stranded RNA degrading enzymes present in the gut of the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2014, 46, 1–8. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, K.; Chen, J.; Wang, J.; Zhang, H.; Ze, L.; Zhu, G.; Zhao, C.; Xiao, H.; Han, Z. Identification of a double-stranded RNA-degrading nuclease influencing both ingestion and injection RNA interference efficiency in the red flour beetle Tribolium castaneum. Insect Biochem. Mol. Biol. 2020, 125, 103440. [Google Scholar] [CrossRef]
- Lomate, P.R.; Bonning, B.C. Proteases and nucleases involved in the biphasic digestion process of the brown marmorated stink bug, Halyomorpha halys (Hemiptera: Pentatomidae). Arch. Insect Biochem. Physiol. 2018, 98, e21459. [Google Scholar] [CrossRef]
- Christiaens, O.; Swevers, L.; Smagghe, G. DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 2014, 53, 307–314. [Google Scholar] [CrossRef]
- Yasunaga, A.; Hanna, S.L.; Li, J.; Cho, H.; Rose, P.P.; Spiridigliozzi, A.; Gold, B.; Diamond, M.S.; Cherry, S. Genome-Wide RNAi Screen Identifies Broadly-Acting Host Factors That Inhibit Arbovirus Infection. PLoS Pathog. 2014, 10, e1003914. [Google Scholar] [CrossRef] [Green Version]
- Sadler, A.J.; Williams, B.R.G. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008, 8, 559–568. [Google Scholar] [CrossRef]
- Song, H.; Zhang, J.; Li, D.; Cooper, A.M.W.; Silver, K.; Li, T.; Liu, X.; Ma, E.; Zhu, K.Y.; Zhang, J. A double-stranded RNA degrading enzyme reduces the efficiency of oral RNA interference in migratory locust. Insect Biochem. Mol. Biol. 2017, 86, 68–80. [Google Scholar] [CrossRef]
- Peng, Y.C.; Wang, K.X.; Fu, W.X.; Sheng, C.W.; Han, Z.J. Biochemical Comparison of dsRNA Degrading Nucleases in Four Different Insects. Front. Physiol. 2018, 9, 624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.; Ren, B.-Y.; Shen, J. Nanoparticle-mediated double-stranded RNA delivery system: A promising approach for sustainable pest management. Insect Sci. 2021, 28, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-H.; Huang, J.-H.; Liu, Y.; Belles, X.; Lee, H.-J. Oral delivery of dsRNA lipoplexes to German cockroach protects dsRNA from degradation and induces RNAi response. Pest. Manag. Sci. 2017, 73, 960–966. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.-Z.; Zhou, H.; Wei, Y.-L.; Yan, S.; Shen, J. A novel plasmid-Escherichia coli system produces large batch dsRNAs for insect gene silencing. Pest. Manag. Sci. 2020, 76, 2505–2512. [Google Scholar] [CrossRef]
- Kennedy, S.; Wang, D.; Ruvkun, G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C-elegans. Nature 2004, 427, 645–649. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinf. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]

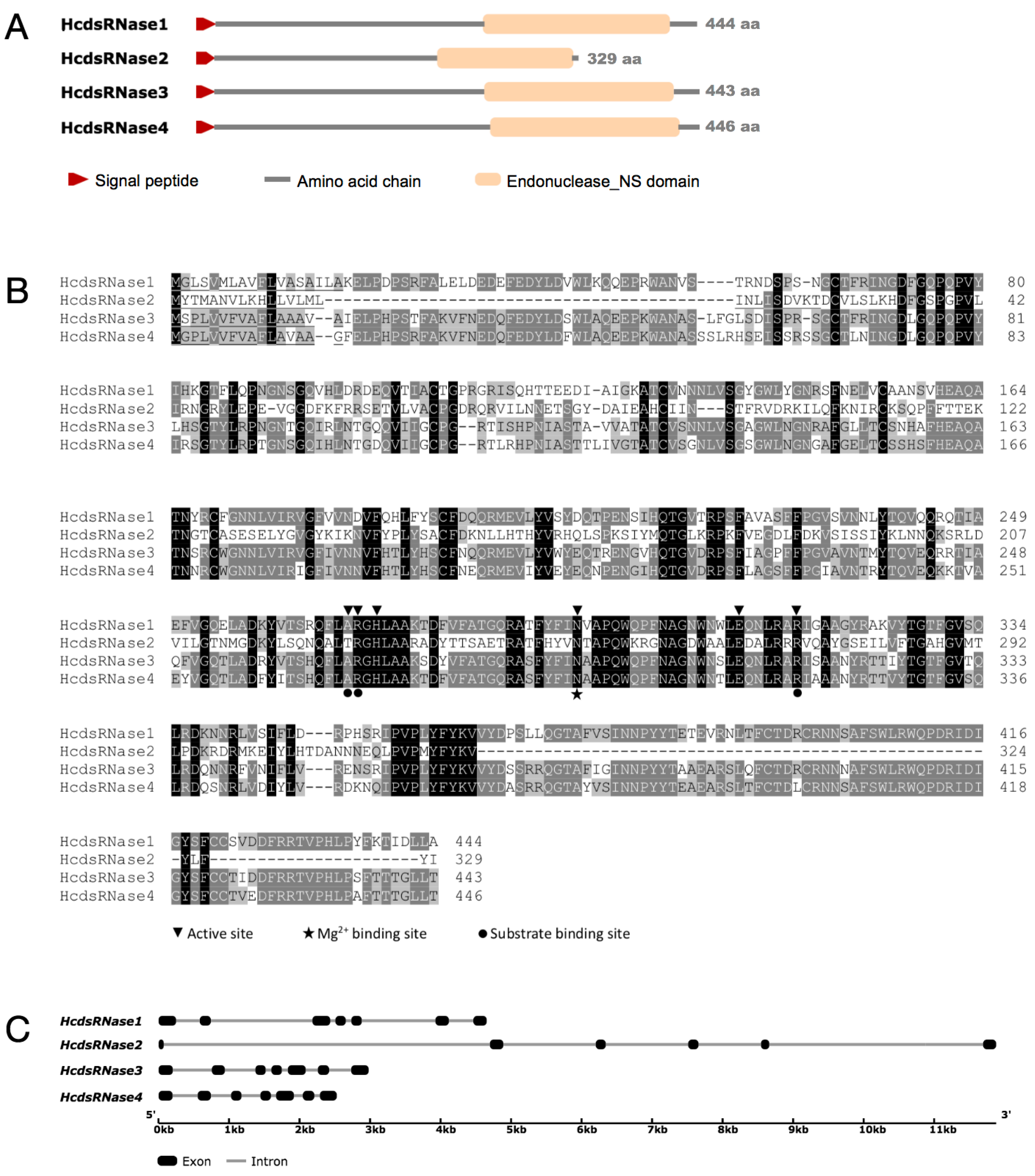


| Amino Acids (a.a.) | Molecular Weight (kD) | Isoelectric Point (pI) | Signal Peptide (Position) | NUC Domain (Position) | |
|---|---|---|---|---|---|
| HcdsRNase1 | 444 | 50.56 | 6.51 | Yes (1–18) | Yes (187–400) |
| HcdsRNase2 | 329 | 37.64 | 9.25 | Yes (1–25) | Yes (145–328) |
| HcdsRNase3 | 443 | 49.87 | 9.00 | Yes (1–16) | Yes (186–399) |
| HcdsRNase4 | 446 | 49.99 | 7.17 | Yes (1–16) | Yes (189–402) |
| HcdsRNase1 | HcdsRNase2 | HcdsRNase3 | HcdsRNase4 | |
|---|---|---|---|---|
| HcdsRNase1 | — | 34% | 70% | 68% |
| HcdsRNase2 | — | 32% | 34% | |
| HcdsRNase3 | — | 82% | ||
| HcdsRNase4 | — |
| Gene Name | Primer Sequence (5′-3′) 1 | Product Size (bp) | Primer Usage | |
|---|---|---|---|---|
| HcdsRNase1 | 5′ RACE | CTGCAAGGTGACCTCGGGCCAAGA | RACE-PCR | |
| 3′ RACE | TCCGTGCCCGTATTGGAGCTGCTG | |||
| HcdsRNase1 | Forward | ATGGGTCTGAGCGTTATG | 1335 | Full length ORF |
| Reverse | TTATGCCAGGAGATCAATAG | |||
| HcdsRNase2 | Forward | ATGTACACAATGGCCAACGTTTTGA | 990 | |
| Reverse | TTATATGTAAAATAAATATACCTTGTAGAA | |||
| HcdsRNase3 | Forward | ATGAGTCCGCTTGTTGTATTCGTAG | 1332 | |
| Reverse | TTATGTCAATAGACCAGTGGTGGTG | |||
| HcdsRNase4 | Forward | ATGGGGCCGCTTGTCGTGTTCGTAG | 1341 | |
| Reverse | TTATGTCAATAGACCAGTGGTGGTGAA | |||
| HcdsRNase1 | Forward | AAGTTCAGCAGAGGCAAACA | 199 | RT-qPCR |
| Reverse | CAGTTCCAGTTACCAGCATTG | |||
| HcdsRNase2 | Forward | TAGCTTGCCCTGGAGATAGA | 147 | |
| Reverse | GCTGTGATTTACACCGAATGT | |||
| HcdsRNase3 | Forward | CAGGACAAATCCGCCTCAAT | 142 | |
| Reverse | CAACCAGCGCCAGACACTAA | |||
| HcdsRNase4 | Forward | CACCTATCTACGACCCACCG | 172 | |
| Reverse | CCAACCAGAGCCAGACACTAA | |||
| β-actin | Forward | GGTTACTCTTTCACCACCACAG | 129 | |
| Reverse | GGACTTCTCAAGGGAACTGC | |||
| HcCht5 | Forward | TCGGTCGTTCACTTTAGCAG | 205 | |
| Reverse | TTTGTAAGCGTAGGGGCAT | |||
| dsdsRNase1 | Forward | TTGGTGTTTCGCAACTGCGT | 280 | dsRNA synthesis |
| Reverse | CTGCAGCAGAAGCTGTATCCA | |||
| dsdsRNase2 | Forward | AACATGCGCAAGCGAAAGTG | 328 | |
| Reverse | TGCACGTGCTGCTAAGTGTCC | |||
| dsdsRNase3 | Forward | GATAGATATGTCACCAGCCACC | 234 | |
| Reverse | CAGTTGAGTTACCCCGAAGG | |||
| dsdsRNase4 | Forward | GTGTCTCAACTGCGTGACCA | 211 | |
| Reverse | TGTTGTTGCGACACAGGTCC | |||
| dsGFP | Forward | TGAGCAAGGGCGAGGAG | 678 | |
| Reverse | CGGCGGTCACGAACTCCAG | |||
| dsHcCht5 | Forward | CACGCATCTCATCTACTCA | 388 | |
| Reverse | CGAACCTTTACCGACCCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Fan, Z.; Wang, Q.; Kong, X.; Liu, F.; Fang, J.; Zhang, S.; Zhang, Z. RNAi Efficiency through dsRNA Injection Is Enhanced by Knockdown of dsRNA Nucleases in the Fall Webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int. J. Mol. Sci. 2022, 23, 6182. https://doi.org/10.3390/ijms23116182
Zhang X, Fan Z, Wang Q, Kong X, Liu F, Fang J, Zhang S, Zhang Z. RNAi Efficiency through dsRNA Injection Is Enhanced by Knockdown of dsRNA Nucleases in the Fall Webworm, Hyphantria cunea (Lepidoptera: Arctiidae). International Journal of Molecular Sciences. 2022; 23(11):6182. https://doi.org/10.3390/ijms23116182
Chicago/Turabian StyleZhang, Xun, Zhizhi Fan, Qinghua Wang, Xiangbo Kong, Fu Liu, Jiaxing Fang, Sufang Zhang, and Zhen Zhang. 2022. "RNAi Efficiency through dsRNA Injection Is Enhanced by Knockdown of dsRNA Nucleases in the Fall Webworm, Hyphantria cunea (Lepidoptera: Arctiidae)" International Journal of Molecular Sciences 23, no. 11: 6182. https://doi.org/10.3390/ijms23116182
APA StyleZhang, X., Fan, Z., Wang, Q., Kong, X., Liu, F., Fang, J., Zhang, S., & Zhang, Z. (2022). RNAi Efficiency through dsRNA Injection Is Enhanced by Knockdown of dsRNA Nucleases in the Fall Webworm, Hyphantria cunea (Lepidoptera: Arctiidae). International Journal of Molecular Sciences, 23(11), 6182. https://doi.org/10.3390/ijms23116182







