Genetic Evidence for a Causal Relationship between Hyperlipidemia and Type 2 Diabetes in Mice
Abstract
1. Introduction
2. Results
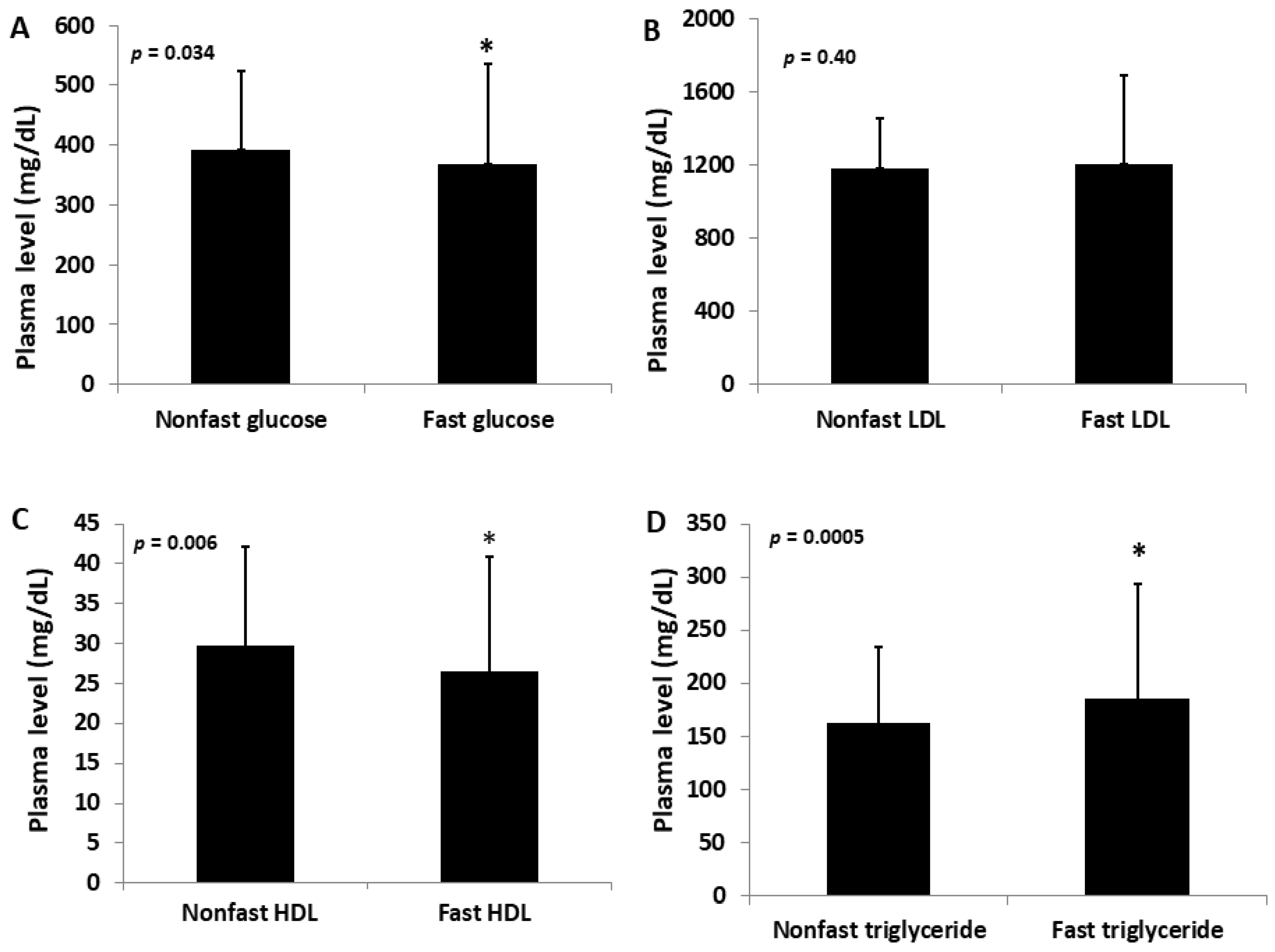
2.1. Fasting versus Non-Fasting Lipid and Glucose Levels
2.2. QTL Analysis of Fasting versus Non-Fasting Plasma Lipid Levels
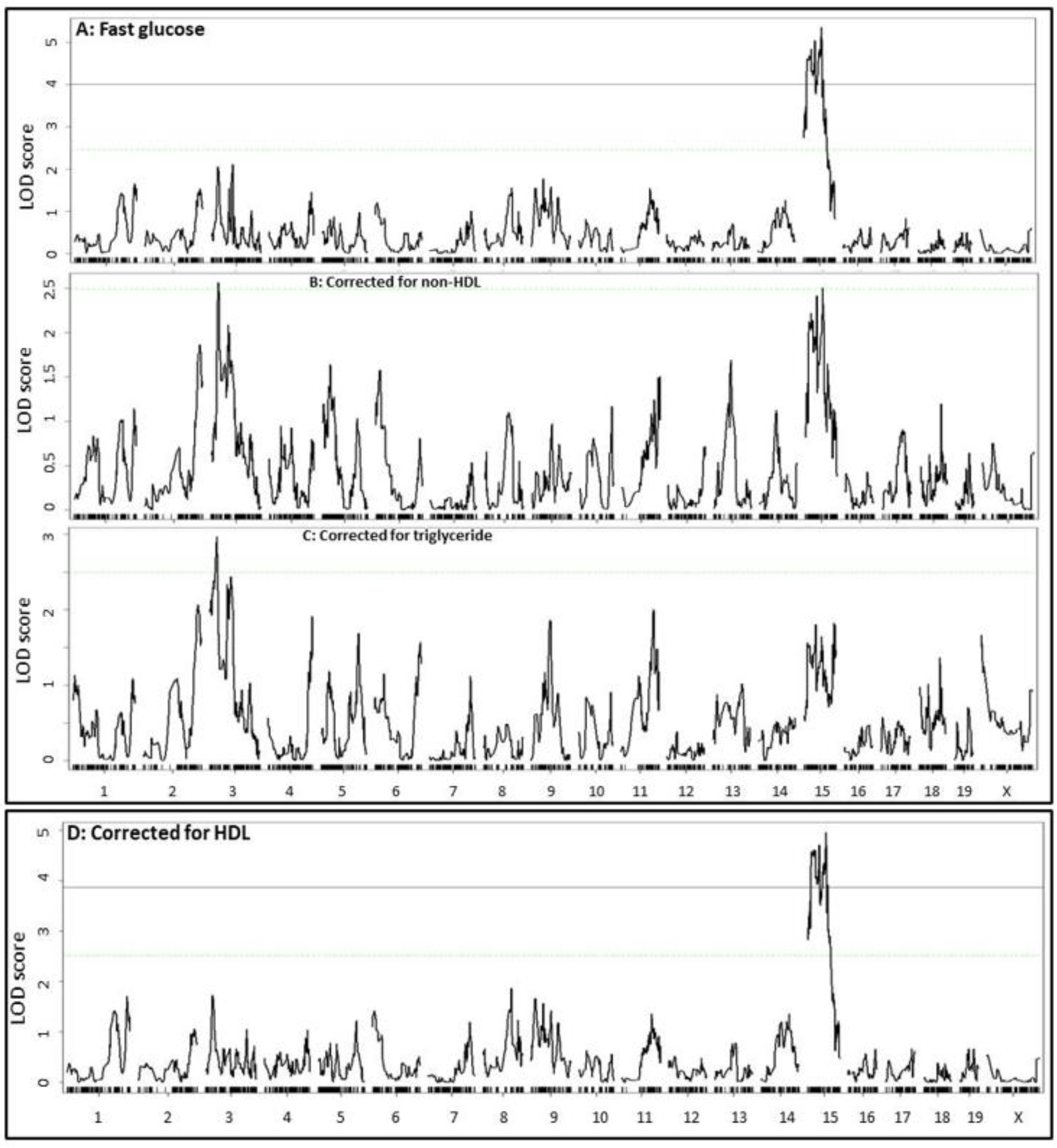
2.3. Coincident QTL for Plasma Lipids and Plasma Glucose
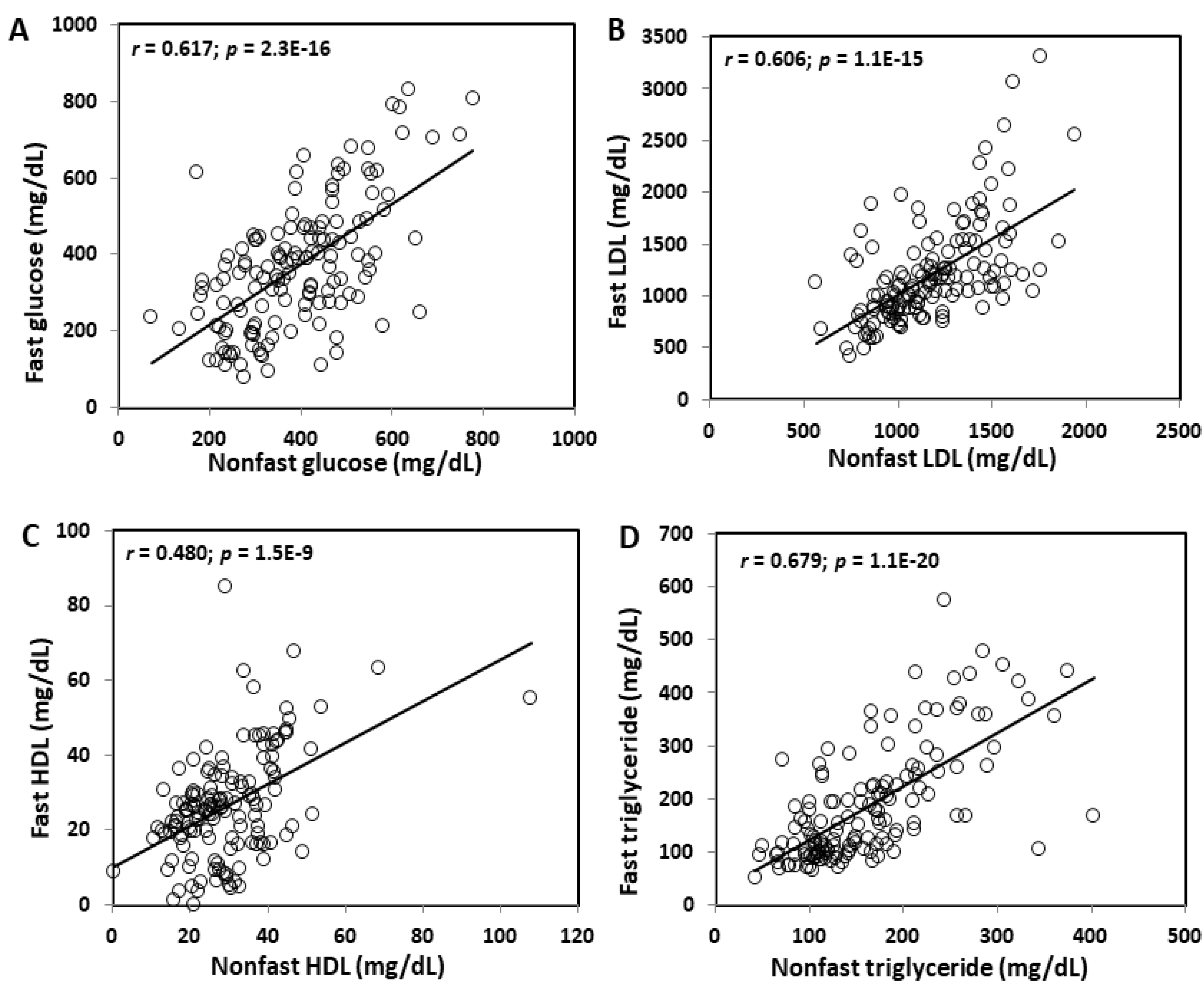
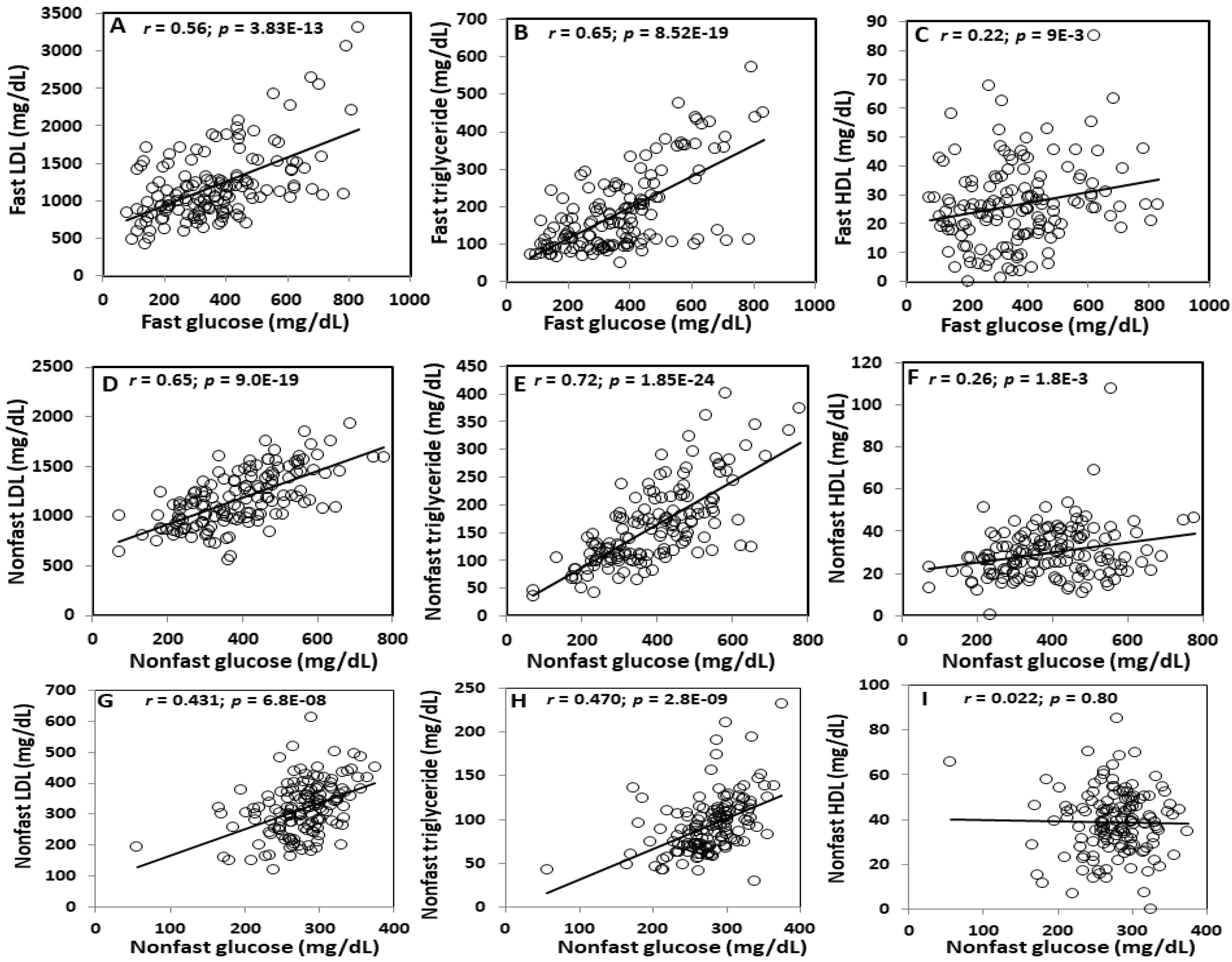
2.4. Correlations between Plasma Glucose and Lipid Levels
2.5. Causal Relationship between Plasma Glucose and Lipids
2.6. Prioritization of Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Glucose Assay
4.3. Lipid Assay
4.4. Genotyping
4.5. QTL Analysis
4.6. Evaluating Causal Relationships between Traits Based on Overlapping QTL
4.7. Prioritization of Candidate Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stumvoll, M.; Goldstein, B.J.; Haeften, T.W. van Type 2 Diabetes: Principles of Pathogenesis and Therapy. Lancet 2005, 365, 1333–1346. [Google Scholar] [CrossRef]
- Lin, X.; Xu, Y.; Pan, X.; Xu, J.; Ding, Y.; Sun, X.; Song, X.; Ren, Y.; Shan, P.-F. Global, Regional, and National Burden and Trend of Diabetes in 195 Countries and Territories: An Analysis from 1990 to 2025. Sci. Rep. 2020, 10, 14790. [Google Scholar] [CrossRef] [PubMed]
- Ali, O. Genetics of Type 2 Diabetes. World J. Diabetes 2013, 4, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Meigs, J.B.; Cupples, L.A.; Wilson, P.W. Parental Transmission of Type 2 Diabetes: The Framingham Offspring Study. Diabetes 2000, 49, 2201–2207. [Google Scholar] [CrossRef] [PubMed]
- Medici, F.; Hawa, M.; Ianari, A.; Pyke, D.A.; Leslie, R.D. Concordance Rate for Type II Diabetes Mellitus in Monozygotic Twins: Actuarial Analysis. Diabetologia 1999, 42, 146–150. [Google Scholar] [CrossRef]
- Moller, D.E.; Flier, J.S. Detection of an Alteration in the Insulin-Receptor Gene in a Patient with Insulin Resistance, Acanthosis Nigricans, and the Polycystic Ovary Syndrome (Type A Insulin Resistance). N. Engl. J. Med. 1988, 319, 1526–1529. [Google Scholar] [CrossRef]
- Thomas, H.; Jaschkowitz, K.; Bulman, M.; Frayling, T.M.; Mitchell, S.M.; Roosen, S.; Lingott-Frieg, A.; Tack, C.J.; Ellard, S.; Ryffel, G.U.; et al. A Distant Upstream Promoter of the HNF-4alpha Gene Connects the Transcription Factors Involved in Maturity-Onset Diabetes of the Young. Hum. Mol. Genet. 2001, 10, 2089–2097. [Google Scholar] [CrossRef]
- Salopuro, T.; Pulkkinen, L.; Lindstrom, J.; Eriksson, J.G.; Valle, T.T.; Hamalainen, H.; Ilanne-Parikka, P.; Keinanen-Kiukaanniemi, S.; Tuomilehto, J.; Laakso, M.; et al. Genetic Variation in Leptin Receptor Gene Is Associated with Type 2 Diabetes and Body Weight: The Finnish Diabetes Prevention Study. Int. J. Obes. 2005, 29, 1245–1251. [Google Scholar] [CrossRef]
- Naggert, J.K.; Fricker, L.D.; Varlamov, O.; Nishina, P.M.; Rouille, Y.; Steiner, D.F.; Carroll, R.J.; Paigen, B.J.; Leiter, E.H. Hyperproinsulinaemia in Obese Fat/Fat Mice Associated with a Carboxypeptidase E Mutation Which Reduces Enzyme Activity. Nat. Genet. 1995, 10, 135–142. [Google Scholar] [CrossRef]
- Agarwal, A.K.; Arioglu, E.; Almeida, S.D.; Akkoc, N.; Taylor, S.I.; Bowcock, A.M.; Barnes, R.I.; Garg, A. AGPAT2 Is Mutated in Congenital Generalized Lipodystrophy Linked to Chromosome 9q34. Nat. Genet. 2002, 31, 21–23. [Google Scholar] [CrossRef]
- Barroso, I.; Gurnell, M.; Crowley, V.E.; Agostini, M.; Schwabe, J.W.; Soos, M.A.; Maslen, G.L.; Williams, T.D.; Lewis, H.; Schafer, A.J.; et al. Dominant Negative Mutations in Human PPARgamma Associated with Severe Insulin Resistance, Diabetes Mellitus and Hypertension. Nature 1999, 402, 880–883. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Rochford, J.J.; Wolfrum, C.; Gray, S.L.; Schinner, S.; Wilson, J.C.; Soos, M.A.; Murgatroyd, P.R.; Williams, R.M.; Acerini, C.L.; et al. A Family with Severe Insulin Resistance and Diabetes Due to a Mutation in AKT2. Science 2004, 304, 1325–1328. [Google Scholar] [CrossRef] [PubMed]
- Xue, A.; Wu, Y.; Zhu, Z.; Zhang, F.; Kemper, K.E.; Zheng, Z.; Yengo, L.; Lloyd-Jones, L.R.; Sidorenko, J.; Wu, Y.; et al. Genome-Wide Association Analyses Identify 143 Risk Variants and Putative Regulatory Mechanisms for Type 2 Diabetes. Nat. Commun. 2018, 9, 2941. [Google Scholar] [CrossRef]
- Peters, L.L.; Robledo, R.F.; Bult, C.J.; Churchill, G.A.; Paigen, B.J.; Svenson, K.L. The Mouse as a Model for Human Biology: A Resource Guide for Complex Trait Analysis. Nat. Rev. 2007, 8, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Pennacchio, L.A. Insights from Human/Mouse Genome Comparisons. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2003, 14, 429–436. [Google Scholar] [CrossRef]
- Attie, A.D.; Churchill, G.A.; Nadeau, J.H. How Mice Are Indispensable for Understanding Obesity and Diabetes Genetics. Curr. Opin. Endocrinol. Diabetes Obes. 2017, 24, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, Y.; Plump, A.S.; Raines, E.W.; Breslow, J.L.; Ross, R. ApoE-Deficient Mice Develop Lesions of All Phases of Atherosclerosis throughout the Arterial Tree. Arterioscler. Thromb. J. Vasc. Biol. Am. Heart Assoc. 1994, 14, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Q.; Chai, W.; Chen, M.H.; Liu, Z.; Shi, W. Hyperglycemia in Apolipoprotein E-Deficient Mouse Strains with Different Atherosclerosis Susceptibility. Cardiovasc. Diabetol. 2011, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, J.; Chen, M.H.; Liu, Z.; Shi, W. Variation in Type 2 Diabetes-Related Phenotypes among Apolipoprotein E-Deficient Mouse Strains. PLoS ONE 2015, 10, e0120935. [Google Scholar] [CrossRef]
- Wilson, P.W.; Meigs, J.B.; Sullivan, L.; Fox, C.S.; Nathan, D.M.; RB, S.D. Prediction of Incident Diabetes Mellitus in Middle-Aged Adults: The Framingham Offspring Study. Arch. Intern. Med. 2007, 167, 1068–1074. [Google Scholar] [CrossRef]
- Peng, J.; Zhao, F.; Yang, X.; Pan, X.; Xin, J.; Wu, M.; Peng, Y.G. Association between Dyslipidemia and Risk of Type 2 Diabetes Mellitus in Middle-Aged and Older Chinese Adults: A Secondary Analysis of a Nationwide Cohort. BMJ Open 2021, 11, e042821. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.H.; Bae, J.C.; Park, S.E.; Rhee, E.J.; Park, C.Y.; Oh, K.W.; Park, S.W.; Kim, S.W.; Lee, W.-Y. Association of Lipid and Lipoprotein Profiles with Future Development of Type 2 Diabetes in Nondiabetic Korean Subjects: A 4-Year Retrospective, Longitudinal Study. J. Clin. Endocrinol. Metab. 2011, 96, E2050–E2054. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Wang, Q.; Choi, W.; Li, J. Mapping and Congenic Dissection of Genetic Loci Contributing to Hyperglycemia and Dyslipidemia in Mice. PLoS ONE 2016, 11, e0148462. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Grainger, A.T.; Manichaikul, A.; Farber, E.; Onengut-Gumuscu, S.; Shi, W. Genetic Linkage of Hyperglycemia and Dyslipidemia in an Intercross between BALB/CJ and SM/J Apoe-Deficient Mouse Strains. BMC Genet. 2015, 16, 133. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.J.; Tang, X.; He, J.; Shi, W. Hyperlipidemia Influences the Accuracy of Glucometer-Measured Blood Glucose Concentrations in Genetically Diverse Mice. Am. J. Med. Sci. 2021, 362, 297–302. [Google Scholar] [CrossRef]
- Sattar, N.; Preiss, D.; Murray, H.M.; Welsh, P.; Buckley, B.M.; de Craen, A.J.M.; Seshasai, S.R.K.; McMurray, J.J.; Freeman, D.J.; Jukema, J.W.; et al. Statins and Risk of Incident Diabetes: A Collaborative Meta-Analysis of Randomised Statin Trials. Lancet Lond. Engl. 2010, 375, 735–742. [Google Scholar] [CrossRef]
- Swerdlow, D.I.; Preiss, D.; Kuchenbaecker, K.B.; Holmes, M.V.; Engmann, J.E.L.; Shah, T.; Sofat, R.; Stender, S.; Johnson, P.C.D.; Scott, R.A.; et al. HMG-Coenzyme A Reductase Inhibition, Type 2 Diabetes, and Bodyweight: Evidence from Genetic Analysis and Randomised Trials. Lancet Lond. Engl. 2015, 385, 351–361. [Google Scholar] [CrossRef]
- Andersson, C.; Lyass, A.; Larson, M.G.; Robins, S.J.; Vasan, R.S. Low-Density-Lipoprotein Cholesterol Concentrations and Risk of Incident Diabetes: Epidemiological and Genetic Insights from the Framingham Heart Study. Diabetologia 2015, 58, 2774–2780. [Google Scholar] [CrossRef]
- Climent, E.; Pérez-Calahorra, S.; Marco-Benedí, V.; Plana, N.; Sánchez, R.; Ros, E.; Ascaso, J.F.; Puzo, J.; Almagro, F.; Lahoz, C.; et al. Effect of LDL Cholesterol, Statins and Presence of Mutations on the Prevalence of Type 2 Diabetes in Heterozygous Familial Hypercholesterolemia. Sci. Rep. 2017, 7, 5596. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Zhang, Z.; Gilbert, T.R.; Matsumoto, A.H.; Dobrin, S.E.; Shi, W. Quantitative Trait Locus Analysis of Carotid Atherosclerosis in an Intercross between C57BL/6 and C3H Apolipoprotein E-Deficient Mice. Stroke J. Cereb. Circ. 2008, 39, 166–173. [Google Scholar] [CrossRef]
- Lotta, L.A.; Sharp, S.J.; Burgess, S.; Perry, J.R.B.; Stewart, I.D.; Willems, S.M.; Luan, J.; Ardanaz, E.; Arriola, L.; Balkau, B.; et al. Association Between Low-Density Lipoprotein Cholesterol-Lowering Genetic Variants and Risk of Type 2 Diabetes: A Meta-Analysis. JAMA 2016, 316, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.J.; Peloso, G.M.; Yu, H.; Butterworth, A.S.; Wang, X.; Mahajan, A.; Saleheen, D.; Emdin, C.; Alam, D.; Alves, A.C.; et al. Exome-Wide Association Study of Plasma Lipids in >300,000 Individuals. Nat. Genet. 2017, 49, 1758–1766. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Johnson, M.W.; Lusis, A.J. Quantitative Trait Locus Analysis of Plasma Lipoprotein Levels in an Autoimmune Mouse Model: Interactions between Lipoprotein Metabolism, Autoimmune Disease, and Atherogenesis. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gargalovic, P.; Wong, J.; Gu, J.L.; Wu, X.; Qi, H.; Wen, P.; Xi, L.; Tan, B.; Gogliotti, R.; et al. Hyplip2, a New Gene for Combined Hyperlipidemia and Increased Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 1928–1934. [Google Scholar] [CrossRef]
- Wergedal, J.E.; Ackert-Bicknell, C.L.; Beamer, W.G.; Mohan, S.; Baylink, D.J.; Srivastava, A.K. Mapping Genetic Loci That Regulate Lipid Levels in a NZB/B1NJxRF/J Intercross and a Combined Intercross Involving NZB/B1NJ, RF/J, MRL/MpJ, and SJL/J Mouse Strains. J. Lipid Res. 2007, 48, 1724–1734. [Google Scholar] [CrossRef]
- Suto, J.; Matsuura, S.; Yamanaka, H.; Sekikawa, K. Quantitative Trait Loci That Regulate Plasma Lipid Concentration in Hereditary Obese KK and KK-Ay Mice. Biochim. Biophys. Acta 1999, 1453, 385–395. [Google Scholar] [CrossRef][Green Version]
- Characterization of Cq3, a Quantitative Trait Locus That Controls Plasma Cholesterol and Phospholipid Levels in Mice. Available online: https://www.jstage.jst.go.jp/article/jvms/68/4/68_4_303/_article (accessed on 26 March 2022).
- Burkhardt, R.; Sundermann, S.; Ludwig, D.; Ceglarek, U.; Holdt, L.M.; Thiery, J.; Teupser, D. Cosegregation of Aortic Root Atherosclerosis and Intermediate Lipid Phenotypes on Chromosomes 2 and 8 in an Intercross of C57BL/6 and BALBc/ByJ Low-Density Lipoprotein Receptor-/- Mice. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 775–784. [Google Scholar] [CrossRef]
- Colinayo, V.V.; Qiao, J.H.; Wang, X.; Krass, K.L.; Schadt, E.; Lusis, A.J.; Drake, T.A. Genetic Loci for Diet-Induced Atherosclerotic Lesions and Plasma Lipids in Mice. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2003, 14, 464–471. [Google Scholar] [CrossRef]
- Lyons, M.A.; Wittenburg, H.; Li, R.; Walsh, K.A.; Korstanje, R.; Churchill, G.A.; Carey, M.C.; Paigen, B. Quantitative Trait Loci That Determine Lipoprotein Cholesterol Levels in an Intercross of 129S1/SvImJ and CAST/Ei Inbred Mice. Physiol. Genom. 2004, 17, 60–68. [Google Scholar] [CrossRef]
- Purcell-Huynh, D.A.; Weinreb, A.; Castellani, L.W.; Mehrabian, M.; Doolittle, M.H.; Lusis, A.J. Genetic Factors in Lipoprotein Metabolism. Analysis of a Genetic Cross between Inbred Mouse Strains NZB/BINJ and SM/J Using a Complete Linkage Map Approach. J. Clin. Investig. 1995, 96, 1845–1858. [Google Scholar] [CrossRef][Green Version]
- Khan, S.U.; Rahman, H.; Okunrintemi, V.; Riaz, H.; Khan, M.S.; Sattur, S.; Kaluski, E.; Lincoff, A.M.; Martin, S.S.; Blaha, M.J. Association of Lowering Low-Density Lipoprotein Cholesterol With Contemporary Lipid-Lowering Therapies and Risk of Diabetes Mellitus: A Systematic Review and Meta-Analysis. J. Am. Heart Assoc. 2019, 8, e011581. [Google Scholar] [CrossRef]
- Hwang, Y.C.; Ahn, H.Y.; Park, S.W.; Park, C.Y. Apolipoprotein B and Non-HDL Cholesterol Are More Powerful Predictors for Incident Type 2 Diabetes than Fasting Glucose or Glycated Hemoglobin in Subjects with Normal Glucose Tolerance: A 3.3-Year Retrospective Longitudinal Study. Acta Diabetol. 2014, 51, 941–946. [Google Scholar] [CrossRef]
- Ley, S.H.; Harris, S.B.; Connelly, P.W.; Mamakeesick, M.; Gittelsohn, J.; Wolever, T.M.; Hegele, R.A.; Zinman, B.; Hanley, A.J. Association of Apolipoprotein B with Incident Type 2 Diabetes in an Aboriginal Canadian Population. Clin. Chem. 2010, 56, 666–670. [Google Scholar] [CrossRef]
- Daboul, M.W. A Study Measuring the Effect of High Serum Triglyceride and Cholesterol on Glucose Elevation in Human Serum. Oman Med. J. 2011, 26, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Drew, B.G.; Rye, K.A.; Duffy, S.J.; Barter, P.; Kingwell, B.A. The Emerging Role of HDL in Glucose Metabolism. Nat. Rev. 2012, 8, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Farbstein, D.; Levy, A.P. HDL Dysfunction in Diabetes: Causes and Possible Treatments. Expert Rev. Cardiovasc. Ther. 2012, 10, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Fuller, D.T.; Grainger, A.T.; Manichaikul, A.; Shi, W. Genetic Linkage of Oxidative Stress with Cardiometabolic Traits in an Intercross Derived from Hyperlipidemic Mouse Strains. Atherosclerosis 2019, 293, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Pei, H.; James, J.C.; Li, Y.; Matsumoto, A.H.; Helm, G.A.; Shi, W. Circulating Adhesion Molecules in ApoE-Deficient Mouse Strains with Different Atherosclerosis Susceptibility. Biochem. Biophys. Res. Commun. 2005, 329, 1102–1107. [Google Scholar] [CrossRef] [PubMed]
- Bonizzi, A.; Piuri, G.; Corsi, F.; Cazzola, R.; Mazzucchelli, S. HDL Dysfunctionality: Clinical Relevance of Quality Rather Than Quantity. Biomedicines 2021, 9, 729. [Google Scholar] [CrossRef]
- Nordestgaard, B.G.; Langsted, A.; Mora, S.; Kolovou, G.; Baum, H.; Bruckert, E.; Watts, G.F.; Sypniewska, G.; Wiklund, O.; Borén, J.; et al. Fasting Is Not Routinely Required for Determination of a Lipid Profile: Clinical and Laboratory Implications Including Flagging at Desirable Concentration Cut-Points—A Joint Consensus Statement from the European Atherosclerosis Society and European Federation of Clinical Chemistry and Laboratory Medicine. Eur. Heart J. 2016, 37, 1944–1958. [Google Scholar] [CrossRef]
- Sidhu, D.; Naugler, C. Fasting Time and Lipid Levels in a Community-Based Population: A Cross-Sectional Study. Arch. Intern. Med. 2012, 172, 1707–1710. [Google Scholar] [CrossRef]
- Keirns, B.H.; Sciarrillo, C.M.; Koemel, N.A.; Emerson, S.R. Fasting, Non-Fasting and Postprandial Triglycerides for Screening Cardiometabolic Risk. J. Nutr. Sci. 2021, 10, e75. [Google Scholar] [CrossRef]
- LeBoeuf, R.C.; Caldwell, M.; Kirk, E. Regulation by Nutritional Status of Lipids and Apolipoproteins A-I, A-II, and A-IV in Inbred Mice. J. Lipid Res. 1994, 35, 121–133. [Google Scholar] [CrossRef]
- Bansal, S.; Buring, J.E.; Rifai, N.; Mora, S.; Sacks, F.M.; Ridker, P.M. Fasting Compared with Nonfasting Triglycerides and Risk of Cardiovascular Events in Women. JAMA 2007, 298, 309–316. [Google Scholar] [CrossRef]
- Freiberg, J.J.; Tybjaerg-Hansen, A.; Jensen, J.S.; Nordestgaard, B.G. Nonfasting Triglycerides and Risk of Ischemic Stroke in the General Population. JAMA 2008, 300, 2142–2152. [Google Scholar] [CrossRef]
- Shi, L.J.; Chagari, B.; An, A.; Chen, M.-H.; Bao, Y.; Shi, W. Genetic Connection between Hyperglycemia and Carotid Atherosclerosis in Hyperlipidemic Mice. Genes 2022, 13, 510. [Google Scholar] [CrossRef]
- Wiltshire, S.; Hattersley, A.T.; Hitman, G.A.; Walker, M.; Levy, J.C.; Sampson, M.; O’Rahilly, S.; Frayling, T.M.; Bell, J.I.; Lathrop, G.M.; et al. A Genomewide Scan for Loci Predisposing to Type 2 Diabetes in a U.K. Population (the Diabetes UK Warren 2 Repository): Analysis of 573 Pedigrees Provides Independent Replication of a Susceptibility Locus on Chromosome 1q. Am. J. Hum. Genet. 2001, 69, 553–569. [Google Scholar] [CrossRef]
- Grainger, A.T.; Jones, M.B.; Li, J.; Chen, M.-H.; Manichaikul, A.; Shi, W. Genetic Analysis of Atherosclerosis Identifies a Major Susceptibility Locus in the Major Histocompatibility Complex of Mice. Atherosclerosis 2016, 254, 124–132. [Google Scholar] [CrossRef]
- Joner, M.; Morimoto, K.; Kasukawa, H.; Steigerwald, K.; Merl, S.; Nakazawa, G.; John, M.C.; Finn, A.V.; Acampado, E.; Kolodgie, F.D.; et al. Site-Specific Targeting of Nanoparticle Prednisolone Reduces in-Stent Restenosis in a Rabbit Model of Established Atheroma. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1960–1966. [Google Scholar] [CrossRef]
- Broman, K.W.; Sen, S.; Owens, S.E.; Manichaikul, A.; Southard-Smith, E.M.; Churchill, G.A. The X Chromosome in Quantitative Trait Locus Mapping. Genetics 2006, 174, 2151–2158. [Google Scholar] [CrossRef]
- Leduc, M.S.; Hageman, R.S.; Verdugo, R.A.; Tsaih, S.W.; Walsh, K.; Churchill, G.A.; Paigen, B. Integration of QTL and Bioinformatic Tools to Identify Candidate Genes for Triglycerides in Mice. J. Lipid Res. 2011, 52, 1672–1682. [Google Scholar] [CrossRef] [PubMed]
- Delpero, M.; Arends, D.; Sprechert, M.; Krause, F.; Kluth, O.; Schürmann, A.; Brockmann, G.A.; Hesse, D. Identification of Four Novel QTL Linked to the Metabolic Syndrome in the Berlin Fat Mouse. Int. J. Obes. 2021, 46, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.B.; An, A.; Shi, L.J.; Shi, W. Regional Variation in Genetic Control of Atherosclerosis in Hyperlipidemic Mice. G3 Bethesda Md 2020, 10, 4679–4689. [Google Scholar] [CrossRef] [PubMed]
- Grainger, A.T.; Pilar, N.; Li, J.; Chen, M.-H.; Abramson, A.M.; Becker-Pauly, C.; Shi, W. Identification of Mep1a as a Susceptibility Gene for Atherosclerosis in Mice. Genetics 2021, 219, iyab160. [Google Scholar] [CrossRef]
- Garrett, N.E., 3rd; Grainger, A.T.; Li, J.; Chen, M.H.; Shi, W. Genetic Analysis of a Mouse Cross Implicates an Anti-Inflammatory Gene in Control of Atherosclerosis Susceptibility. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2017, 28, 90–99. [Google Scholar] [CrossRef]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT Missense Predictions for Genomes. Nat. Protoc. 2016, 11, 1–9. [Google Scholar] [CrossRef]



| Locus Name | Chr | LOD a | Peak (Mb) | Closest Marker | 95%CI (Mb) b | High Allele | Mode of Inheritance | Allelic Effect c | ||
|---|---|---|---|---|---|---|---|---|---|---|
| BB | H | LL | ||||||||
| Non-HDL (non-fast) | ||||||||||
| Nhdlq18, Hyplip2 | 15 | 6.78 | 59.7 | gUNC25683390 | 23.4–62.4 | LL | Additive | 981 ± 188 | 1217 ± 269 | 1284 ± 273 |
| Non-HDL (fast) | ||||||||||
| Nhdlq18, Hyplip2 | 15 | 3.60 | 64.7 | mJAX00062941 | 3.7–69.7 | LL | Additive | 953 ± 364 | 1211 ± 443 | 1383 ± 537 |
| Cq3 | 3 | 2.61 | 129.5 | UNC6226186 | 46.8–152.8 | LL | Recessive | 1298 ± 476 | 1066 ± 333 | 1388 ± 629 |
| HDL (fast) | ||||||||||
| 2 | 2.62 | 26.4 | gUNC2767893 | 4.4–76.4 | - | Heterosis | 33.2 ± 11.3 | 26.5 ± 9.4 | 35.7 ± 16.5 | |
| HDL (non-fast) | ||||||||||
| 6 | 3.18 | 138.6 | S6R065543032 | 126.7–145.7 | LL | Recessive | 30.8 ± 11.3 | 26.5 ± 9.4 | 35.7 ± 16.5 | |
| Triglyceride (non-fast) | ||||||||||
| Tglq3 | 15 | 5.33 | 53.2 | gUNC25604126 | 16.1–66.3 | LL | Additive | 112 ± 38 | 165 ± 68 | 192 ± 80 |
| Triglyceride (fast) | ||||||||||
| Tglq3 | 15 | 4.18 | 59.0 | gUNC25683390 | 5.7–64.9 | LL | Additive | 122 ± 55 | 202 ± 109 | 216 ± 118 |
| Chr | Position | Gene | dbSNP | Ref | LP_J | RF_J | CAST_EiJ | BALB_cJ | 129P2_OlaHsd | NZB_B1NJ | Csq | AA | AA Coord | SIF T |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 3979418 | Fbxo4 | rs263501543 | C | T * | - | - | - | T * | T * | missense_variant | G/R | 3 | 0.13 |
| 15 | 3980692 | Fbxo4 | rs31620120 | G | A * | - | - | - | A * | A * | upstream_gene_variant | |||
| 15 | 4370841 | Plcxd3 | rs31896942 | G | A | - | - | - | A | A | upstream_gene_variant | |||
| 15 | 4371744 | Plcxd3 | rs32133183 | A | G | - | - | - | G | G | upstream_gene_variant | |||
| 15 | 4371814 | Plcxd3 | rs32535204 | G | A | - | - | - | A | A | upstream_gene_variant | |||
| 15 | 4373929 | Plcxd3 | rs31966655 | A | G | - | - | - | G | G | upstream_gene_variant | |||
| 15 | 25413871 | Basp1 | rs262340099 | C | T | - | - | - | t | T | upstream_gene_variant | |||
| 15 | 32173458 | Tas2r119 | rs51514737 | A | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 32173469 | Tas2r119 | rs46216365 | A | G | - | - | - | G | - | upstream_gene_variant | |||
| 15 | 41819897 | Oxr1 | rs50179186 | T | G | - | G | - | G | G | missense_variant | S/A | 239 | 0.02 |
| 15 | 41820278 | Oxr1 | rs31574788 | A | C | - | C | - | C | C | missense_variant | T/P | 428 | 0.2 |
| 15 | 41820327 | Oxr1 | rs31850612 | C | T | - | - | - | T | T | missense_variant | S/L | 363 | 0.15 |
| 15 | 43283531 | Eif3e | rs49218373 | A | G | - | - | - | G | - | upstream_gene_variant | |||
| 15 | 43285477 | Eif3e | rs46145200 | G | A | - | - | - | A | - | upstream_gene_variant | |||
| 15 | 43285631 | Eif3e | rs52016538 | G | A | - | - | - | A | - | upstream_gene_variant | |||
| 15 | 43286989 | Eif3e | rs52120197 | T | C | - | - | - | C | - | upstream_gene_variant | |||
| 15 | 43287113 | Eif3e | rs578378356 | T | C | - | - | - | C | - | upstream_gene_variant | |||
| 15 | 43287574 | Eif3e | rs585572417 | A | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 43870636 | Tmem74 | rs32033816 | G | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 43870825 | Tmem74 | rs32094059 | A | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 43872938 | Tmem74 | rs52337392 | A | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 43873253 | Tmem74 | rs31706856 | T | C | - | - | - | C | - | upstream_gene_variant | |||
| 15 | 44191397 | Trhr | rs52202529 | T | C | - | - | - | C | - | upstream_gene_variant | |||
| 15 | 44194654 | Trhr | rs32144218 | A | C | - | - | - | C | C | upstream_gene_variant | |||
| 15 | 44194999 | Trhr | rs32516067 | G | A | - | - | - | A | - | upstream_gene_variant | |||
| 15 | 44195817 | Trhr | rs31913481 | A | G | - | - | - | G | - | upstream_gene_variant | |||
| 15 | 44196286 | Trhr | rs33306588 | C | T | - | - | - | T | - | 5_prime_utr_variant | |||
| 15 | 44400055 | Nudcd1 | rs51311008 | G | A | - | - | - | A | - | synonymous_variant | |||
| 15 | 44405503 | Nudcd1 | rs32045629 | G | A | - | - | - | A | - | synonymous_variant | |||
| 15 | 44452953 | Pkhd1l1 | rs3716450 | A | G | - | - | - | G | - | upstream_gene_variant | |||
| 15 | 44453339 | Pkhd1l1 | rs32007056 | G | A | - | - | - | A | - | upstream_gene_variant | |||
| 15 | 44453354 | Pkhd1l1 | rs31937456 | G | A | - | - | - | A | - | upstream_gene_variant | |||
| 15 | 44453478 | Pkhd1l1 | rs31864580 | A | G | - | - | - | G | - | upstream_gene_variant | |||
| 15 | 44453784 | Pkhd1l1 | rs31916062 | A | G | - | - | - | G | - | upstream_gene_variant | |||
| 15 | 44454507 | Pkhd1l1 | rs31571652 | G | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 44454738 | Pkhd1l1 | rs31772927 | G | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 44456205 | Pkhd1l1 | rs50758342 | A | G | - | - | - | G | - | upstream_gene_variant | |||
| 15 | 44457111 | Pkhd1l1 | rs31671052 | C | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 44457216 | Pkhd1l1 | rs31628733 | T | C | - | - | - | C | - | upstream_gene_variant | |||
| 15 | 44457499 | Pkhd1l1 | rs33288852 | G | A | - | - | - | A | - | upstream_gene_variant | |||
| 15 | 44457534 | Pkhd1l1 | rs239874708 | C | G | - | - | - | G | - | upstream_gene_variant | |||
| 15 | 44457535 | Pkhd1l1 | rs259180475 | C | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 44457539 | Pkhd1l1 | rs217867183 | G | A | - | - | - | A | - | upstream_gene_variant | |||
| 15 | 44493144 | Pkhd1l1 | rs33290607 | C | T | - | - | - | T | - | missense_variant | T/I | 335 | 0.22 |
| 15 | 44586472 | Pkhd1l1 | rs32502839 | A | C | - | C | - | C | - | missense_variant | N/T | 3877 | 0.01 |
| 15 | 44746395 | Sybu | rs31952285 | C | G * | - | - | - | G * | - | missense_variant | R/P | 159 | 0 |
| 15 | 44787720 | Sybu | rs32209513 | A | G * | - | G * | - | G * | - | missense_variant | F/L | 63 | 0.9 |
| 15 | 45109081 | Kcnv1 | rs13482546 | C | T | - | - | - | T | - | missense_variant | V/I | 469 | 0.95 |
| 15 | 45114557 | Kcnv1 | rs51296409 | G | A | - | - | - | A | - | synonymous_variant | |||
| 15 | 45115527 | Kcnv1 | rs32212679 | G | T | - | - | - | T | T | upstream_gene_variant | |||
| 15 | 45115974 | Kcnv1 | rs32021420 | C | A | - | - | - | A | - | upstream_gene_variant | |||
| 15 | 45116156 | Kcnv1 | rs31825726 | G | A | - | - | - | A | - | upstream_gene_variant | |||
| 15 | 45116355 | Kcnv1 | rs32046135 | C | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 45117030 | Kcnv1 | rs32376626 | G | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 45118047 | Kcnv1 | rs48752875 | T | A | - | - | A | - | upstream_gene_variant | ||||
| 15 | 45118200 | Kcnv1 | rs45851350 | G | A | - | - | - | A | - | upstream_gene_variant | |||
| 15 | 45118590 | Kcnv1 | rs50814374 | T | C | - | - | - | C | - | upstream_gene_variant | |||
| 15 | 45118760 | Kcnv1 | rs47234445 | T | C | - | - | - | C | - | upstream_gene_variant | |||
| 15 | 54252267 | Tnfrsf11b | rs31629761 | C | T | - | - | - | T | - | synonymous_variant | |||
| 15 | 54252313 | Tnfrsf11b | rs31799791 | A | C | - | - | - | C | - | missense_variant | L/R | 296 | 0.61 |
| 15 | 54256164 | Tnfrsf11b | rs33484516 | C | T | - | - | - | T | - | missense_variant | R/Q | 138 | 0.23 |
| 15 | 54278337 | Tnfrsf11b | rs31923434 | G | A | - | - | - | A | - | 5_prime_utr_variant | |||
| 15 | 54278530 | Tnfrsf11b | rs47057076 | G | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 54278620 | Tnfrsf11b | rs49814729 | A | G | - | - | - | G | - | upstream_gene_variant | |||
| 15 | 54278628 | Tnfrsf11b | rs33490015 | A | G | - | - | - | G | - | upstream_gene_variant | |||
| 15 | 54278937 | Tnfrsf11b | rs51583114 | T | C | - | - | - | C | - | upstream_gene_variant | |||
| 15 | 54278947 | Tnfrsf11b | rs33489239 | A | G | - | - | - | G | - | upstream_gene_variant | |||
| 15 | 54278995 | Tnfrsf11b | rs51682935 | G | A | - | - | - | A | - | upstream_gene_variant | |||
| 15 | 54279003 | Tnfrsf11b | rs46278323 | C | T | - | - | - | T | - | upstream_gene_variant | |||
| 15 | 54279030 | Tnfrsf11b | rs33489235 | G | C | - | - | - | C | - | upstream_gene_variant | |||
| 15 | 54279073 | Tnfrsf11b | rs33488583 | T | C | - | - | - | C | - | upstream_gene_variant | |||
| 15 | 55180943 | Deptor | rs48756035 | C | T | - | - | - | T | T | synonymous_variant | |||
| 15 | 55220217 | Deptor | rs32271813 | G | A * | - | - | - | A * | A * | missense_variant |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, L.J.; Tang, X.; He, J.; Shi, W. Genetic Evidence for a Causal Relationship between Hyperlipidemia and Type 2 Diabetes in Mice. Int. J. Mol. Sci. 2022, 23, 6184. https://doi.org/10.3390/ijms23116184
Shi LJ, Tang X, He J, Shi W. Genetic Evidence for a Causal Relationship between Hyperlipidemia and Type 2 Diabetes in Mice. International Journal of Molecular Sciences. 2022; 23(11):6184. https://doi.org/10.3390/ijms23116184
Chicago/Turabian StyleShi, Lisa J., Xiwei Tang, Jiang He, and Weibin Shi. 2022. "Genetic Evidence for a Causal Relationship between Hyperlipidemia and Type 2 Diabetes in Mice" International Journal of Molecular Sciences 23, no. 11: 6184. https://doi.org/10.3390/ijms23116184
APA StyleShi, L. J., Tang, X., He, J., & Shi, W. (2022). Genetic Evidence for a Causal Relationship between Hyperlipidemia and Type 2 Diabetes in Mice. International Journal of Molecular Sciences, 23(11), 6184. https://doi.org/10.3390/ijms23116184







