Physical Activity Rewires the Human Brain against Neurodegeneration
Abstract
:1. Introduction
2. Results
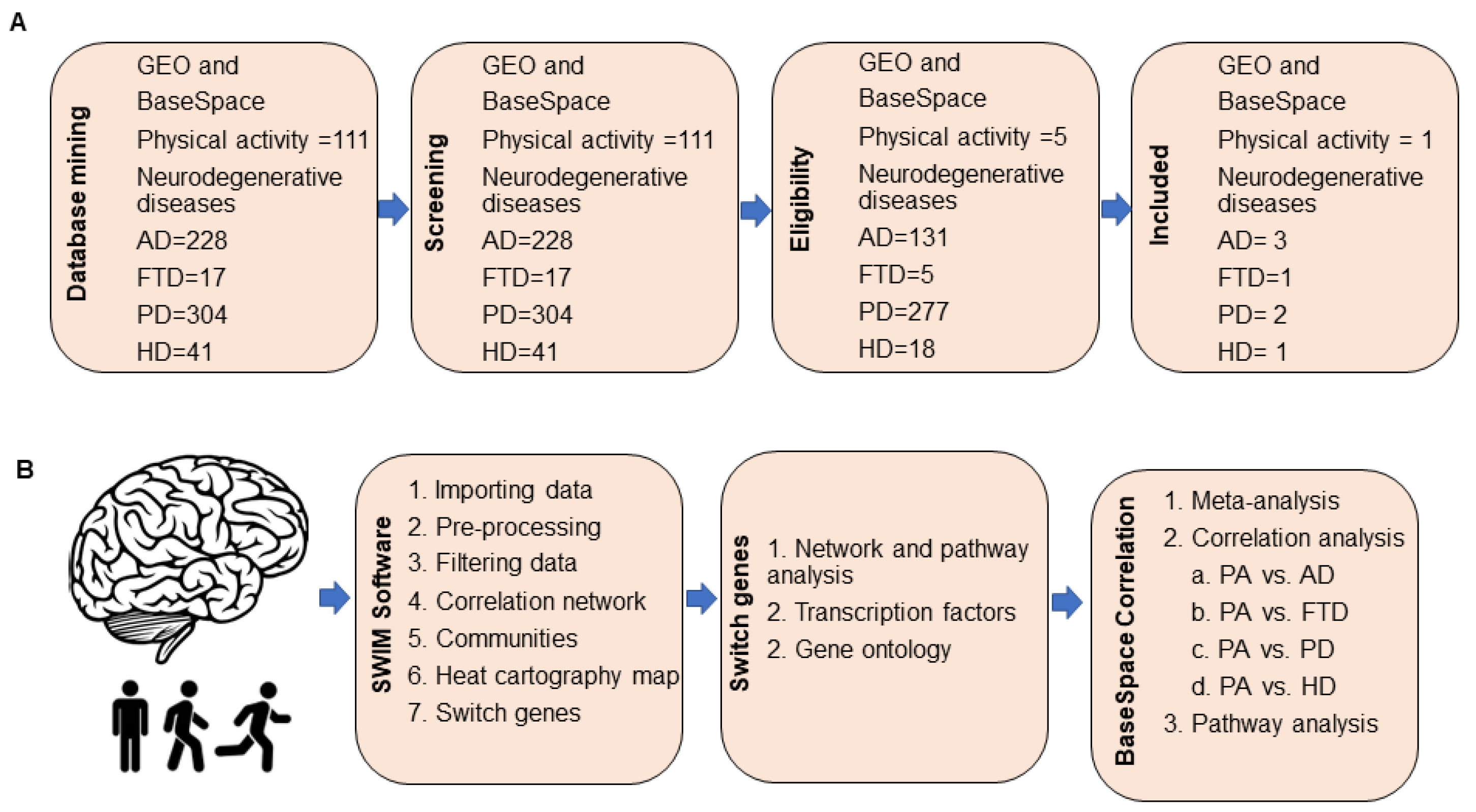
2.1. Database Mining for Transcriptomic Studies
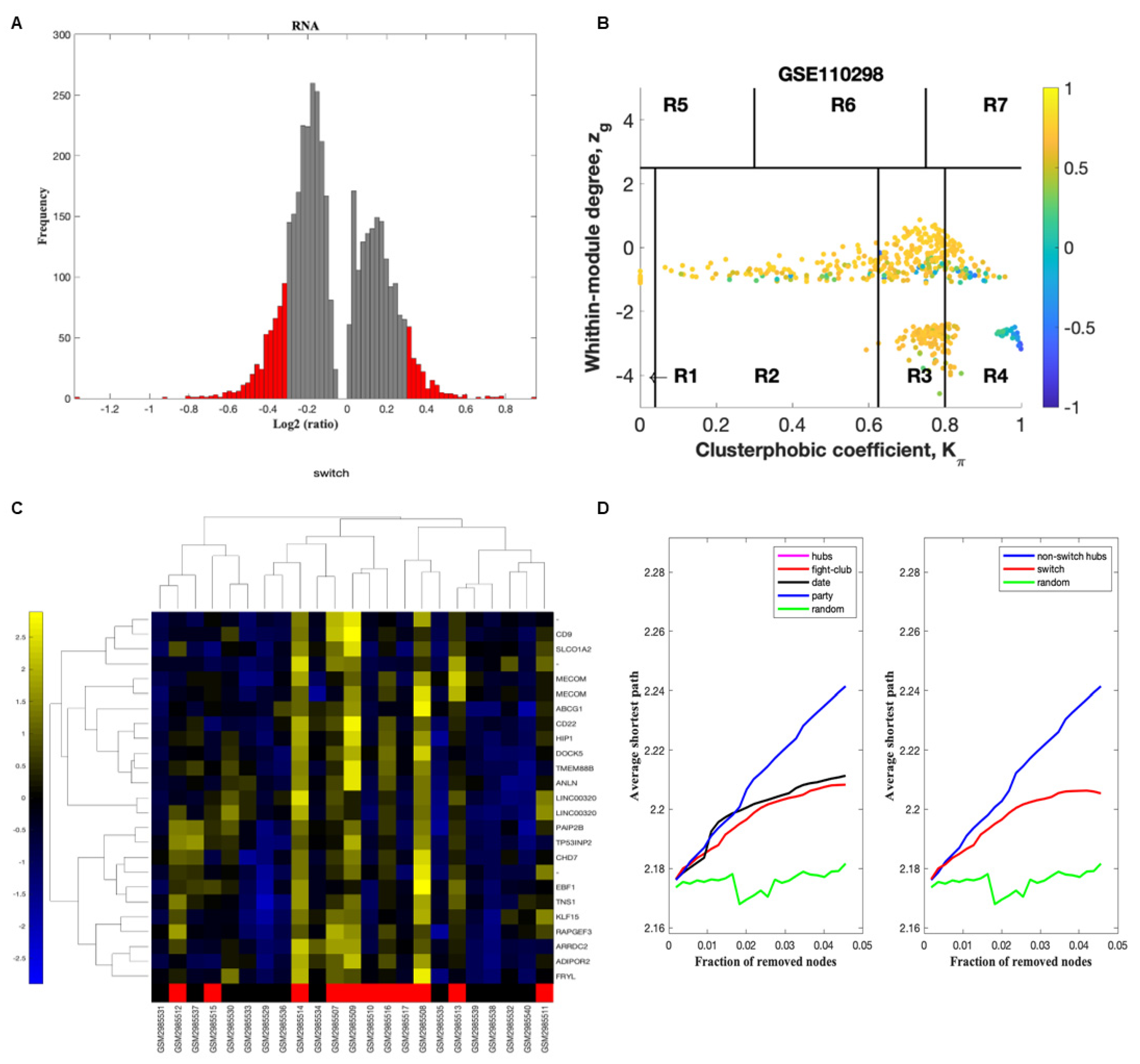
2.2. Identification of Switch Genes in the Hippocampus of Physically Active Subjects
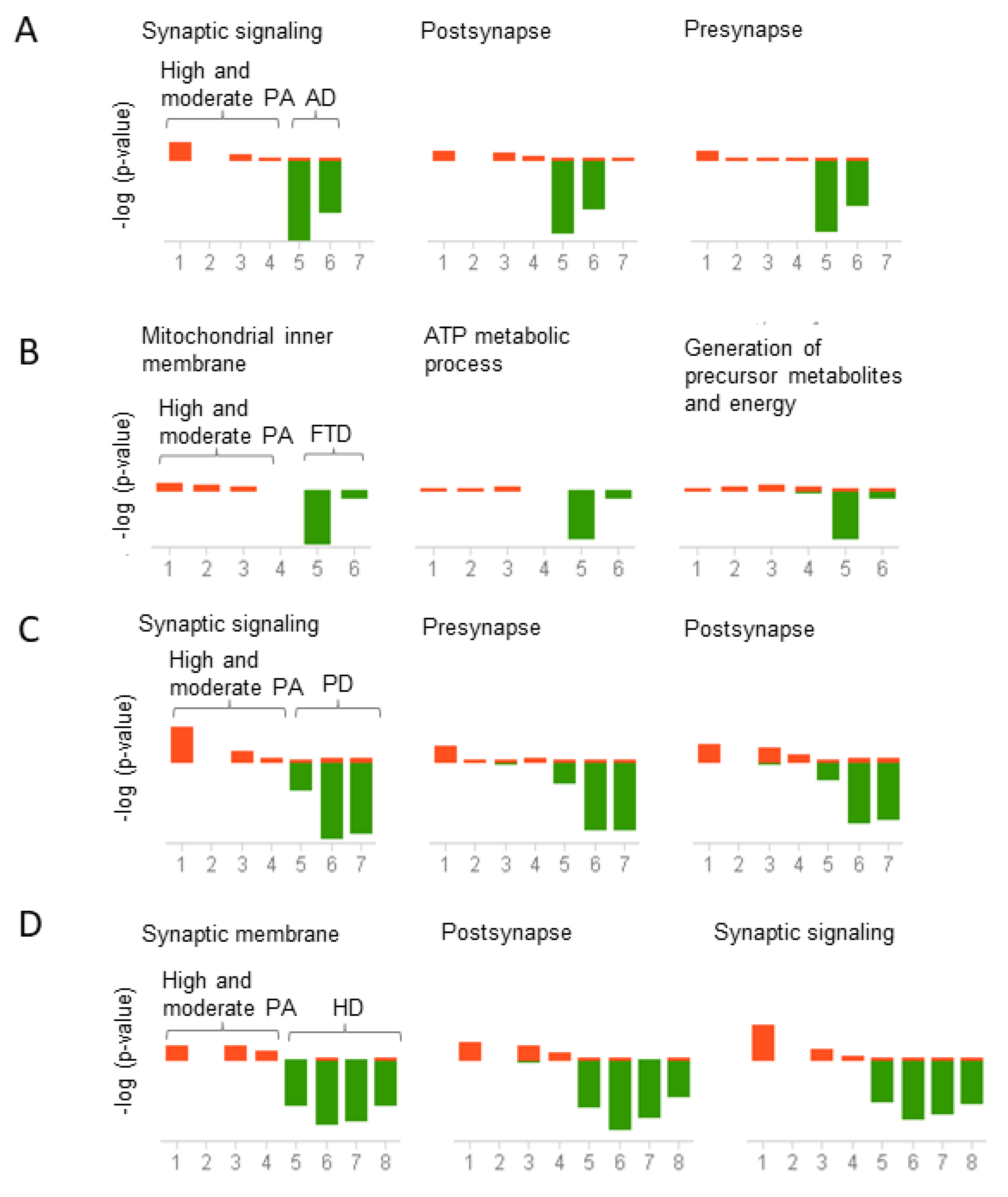
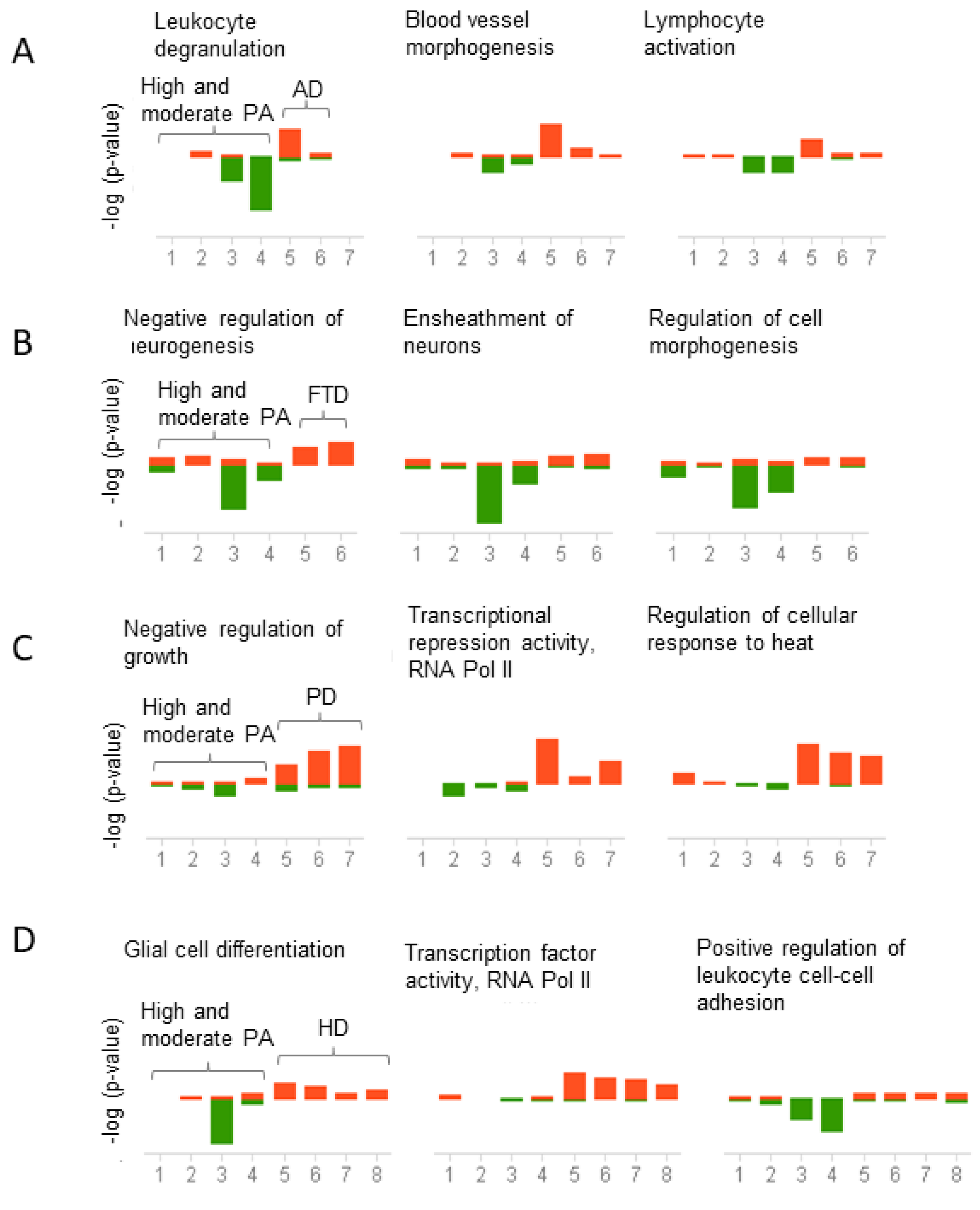
2.3. Biological and Functional Analysis of Switch Genes
2.4. Physical Activity Switch Genes Associated with Neurodegeneration
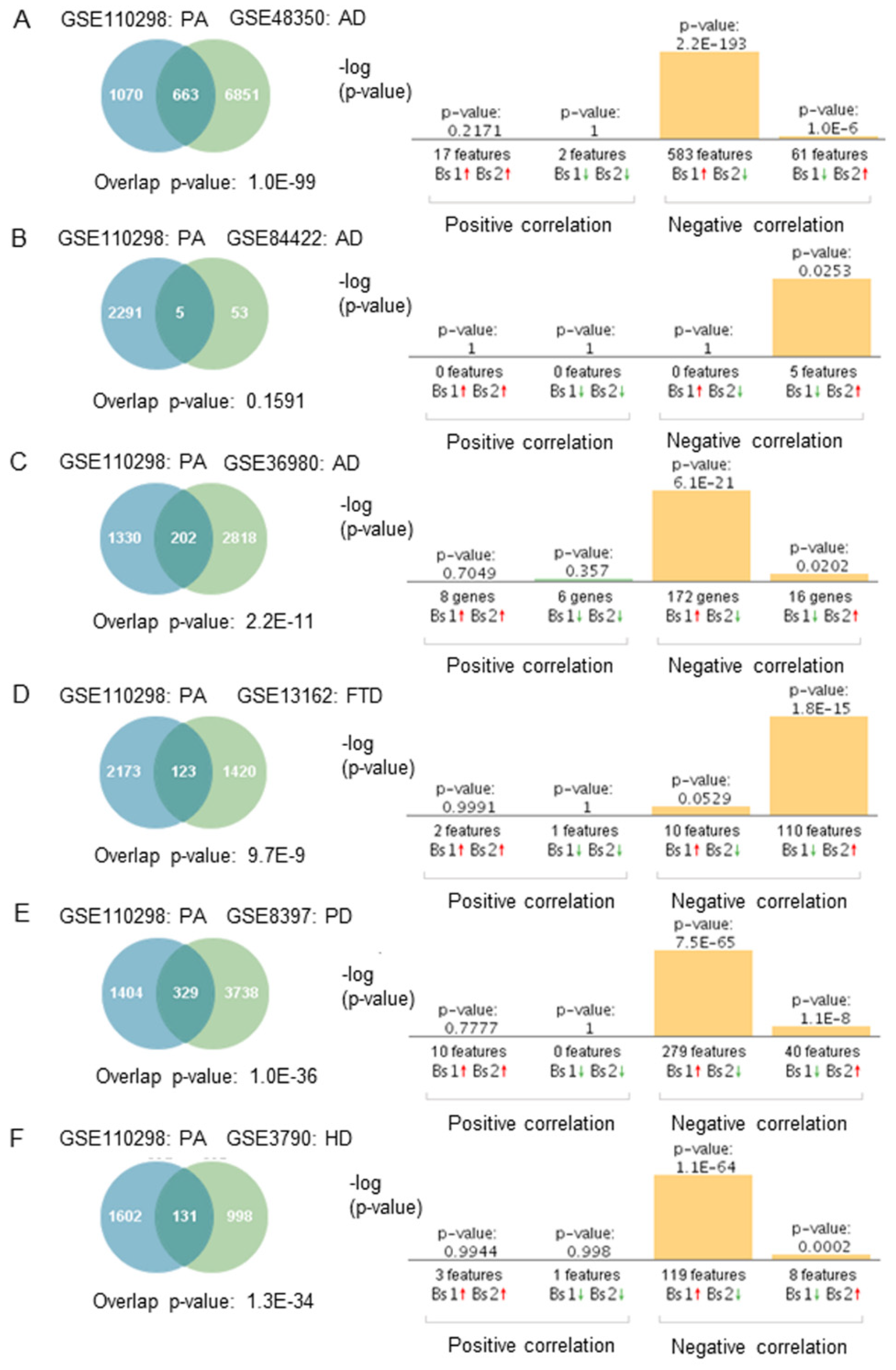
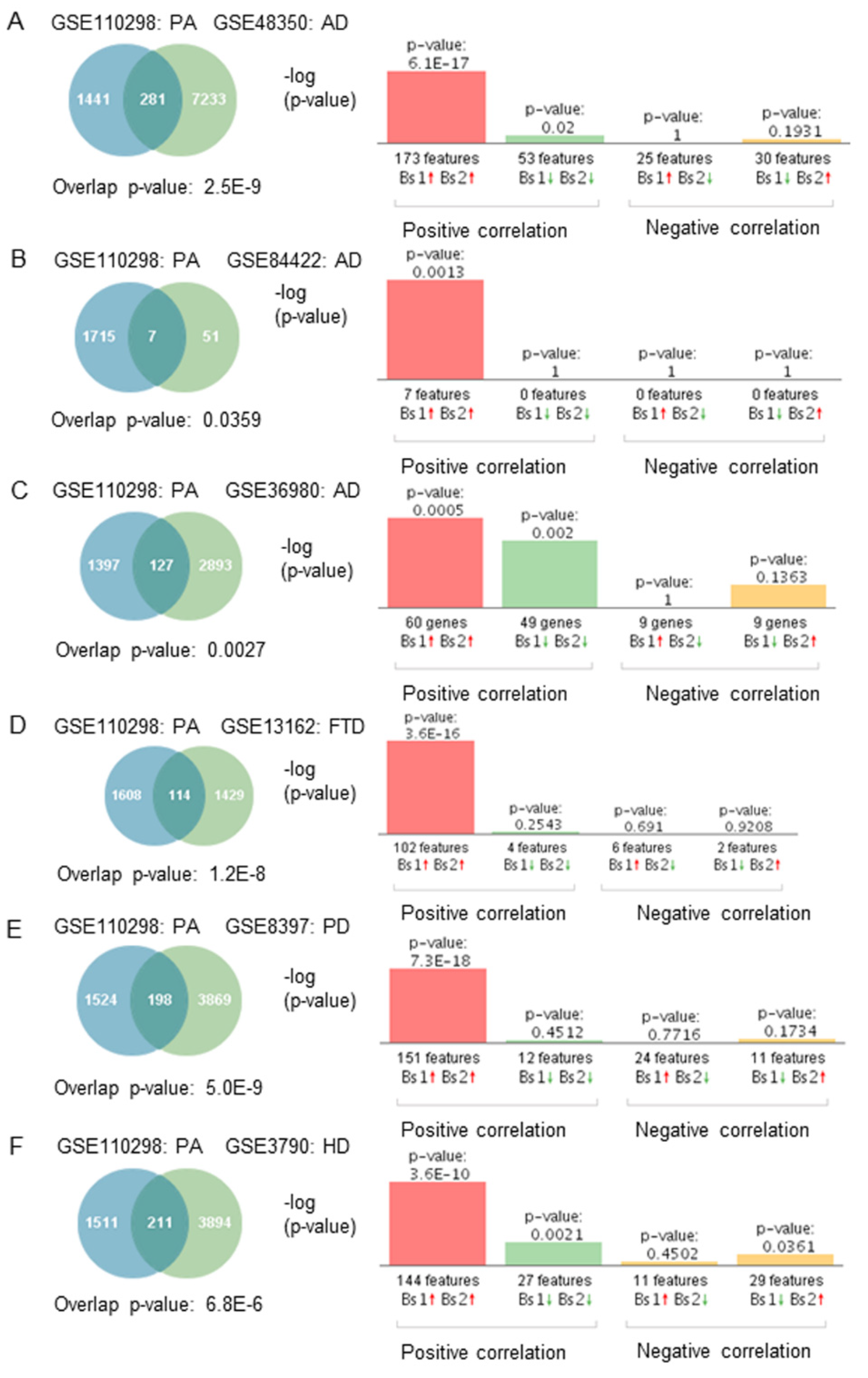
2.5. Gene Expression Correlation between Physical Activity and Neurodegenerative Diseases
3. Discussion
3.1. Switch Genes Associated with Physical Activity
3.2. Transcription Factors Regulation of Switch Genes
3.3. Switch Genes Associated with Neurodegeneration
3.4. Physical Activity Opposes Transcriptional Changes Associated with Neurodegeneration
4. Materials and Methods
4.1. Microarray Dataset Selection
4.2. Description of Microarrays
4.3. Swim Analysis to Identify Switch Genes
4.4. Pathway Analysis
4.5. Gene-Transcription Factors Interaction Analysis
4.6. Gene Expression and Correlation Analyses
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Erickson, K.I.; Weinstein, A.M.; Lopez, O.L. Physical activity, brain plasticity, and Alzheimer’s disease. Arch. Med. Res. 2012, 43, 615–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchman, A.S.; Boyle, P.A.; Yu, L.; Shah, R.C.; Wilson, R.S.; Bennett, D.A. Total daily physical activity and the risk of AD and cognitive decline in older adults. Neurology 2012, 78, 1323–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weuve, J.; Kang, J.H.; Manson, J.E.; Breteler, M.M.; Ware, J.H.; Grodstein, F. Physical activity, including walking, and cognitive function in older women. JAMA 2004, 292, 1454–1461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stubbs, B.; Chen, L.J.; Chang, C.Y.; Sun, W.J.; Ku, P.W. Accelerometer-assessed light physical activity is protective of future cognitive ability: A longitudinal study among community dwelling older adults. Exp. Gerontol. 2017, 91, 104–109. [Google Scholar] [CrossRef]
- Blondell, S.J.; Hammersley-Mather, R.; Veerman, J.L. Does physical activity prevent cognitive decline and dementia?: A systematic review and meta-analysis of longitudinal studies. BMC Public Health 2014, 14, 510. [Google Scholar] [CrossRef] [Green Version]
- Bhatti, G.K.; Reddy, A.P.; Reddy, P.H.; Bhatti, J.S. Lifestyle Modifications and Nutritional Interventions in Aging-Associated Cognitive Decline and Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 369. [Google Scholar] [CrossRef]
- Berchtold, N.C.; Prieto, G.A.; Phelan, M.; Gillen, D.L.; Baldi, P.; Bennett, D.A.; Buchman, A.S.; Cotman, C.W. Hippocampal gene expression patterns linked to late-life physical activity oppose age and AD-related transcriptional decline. Neurobiol. Aging 2019, 78, 142–154. [Google Scholar] [CrossRef]
- Sanfilippo, C.; Musumeci, G.; Castrogiovanni, P.; Fazio, F.; Li Volti, G.; Barbagallo, I.; Maugeri, G.; Ravalli, S.; Imbesi, R.; Di Rosa, M. Hippocampal transcriptome deconvolution reveals differences in cell architecture of not demented elderly subjects underwent late-life physical activity. J. Chem. Neuroanat. 2021, 113, 101934. [Google Scholar] [CrossRef]
- Casaletto, K.B.; Lindbergh, C.A.; VandeBunte, A.; Neuhaus, J.; Schneider, J.A.; Buchman, A.S.; Honer, W.G.; Bennett, D.A. Microglial Correlates of Late Life Physical Activity: Relationship with Synaptic and Cognitive Aging in Older Adults. J. Neurosci. 2022, 42, 288–298. [Google Scholar] [CrossRef]
- Bennett, D.A.; Schneider, J.A.; Buchman, A.S.; Barnes, L.L.; Boyle, P.A.; Wilson, R.S. Overview and findings from the rush Memory and Aging Project. Curr. Alzheimer Res. 2012, 9, 646–663. [Google Scholar] [CrossRef]
- Potashkin, J.A.; Bottero, V.; Santiago, J.A.; Quinn, J.P. Computational identification of key genes that may regulate gene expression reprogramming in Alzheimer’s patients. PLoS ONE 2019, 14, e0222921. [Google Scholar] [CrossRef]
- Potashkin, J.A.; Bottero, V.; Santiago, J.A.; Quinn, J.P. Bioinformatic Analysis Reveals Phosphodiesterase 4D-Interacting Protein as a Key Frontal Cortex Dementia Switch Gene. Int. J. Mol. Sci. 2020, 21, 3787. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.A.; Quinn, J.P.; Potashkin, J.A. Network Analysis Identifies Sex-Specific Gene Expression Changes in Blood of Amyotrophic Lateral Sclerosis Patients. Int. J. Mol. Sci. 2021, 22, 7150. [Google Scholar] [CrossRef] [PubMed]
- Fiscon, G.; Conte, F.; Farina, L.; Paci, P. Network-Based Approaches to Explore Complex Biological Systems towards Network Medicine. Genes 2018, 9, 437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, J.; Gill, E.E.; Hancock, R.E. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015, 10, 823–844. [Google Scholar] [CrossRef] [PubMed]
- Bottero, V.; Santiago, J.A.; Quinn, J.P.; Potashkin, J.A. Key Disease Mechanisms Linked to Amyotrophic Lateral Sclerosis in Spinal Cord Motor Neurons. Front. Mol. Neurosci. 2022, 15, 825031. [Google Scholar] [CrossRef]
- Dunstan, D.W.; Thorp, A.A.; Healy, G.N. Prolonged sitting: Is it a distinct coronary heart disease risk factor? Curr. Opin. Cardiol. 2011, 26, 412–419. [Google Scholar] [CrossRef]
- Bergens, O.; Nilsson, A.; Papaioannou, K.G.; Kadi, F. Sedentary Patterns and Systemic Inflammation: Sex-Specific Links in Older Adults. Front. Physiol. 2021, 12, 625950. [Google Scholar] [CrossRef]
- Guzman-Martinez, L.; Maccioni, R.B.; Andrade, V.; Navarrete, L.P.; Pastor, M.G.; Ramos-Escobar, N. Neuroinflammation as a Common Feature of Neurodegenerative Disorders. Front. Pharmacol. 2019, 10, 1008. [Google Scholar] [CrossRef] [Green Version]
- Whittemore, K.; Vera, E.; Martinez-Nevado, E.; Sanpera, C.; Blasco, M.A. Telomere shortening rate predicts species life span. Proc. Natl. Acad. Sci. USA 2019, 116, 15122–15127. [Google Scholar] [CrossRef] [Green Version]
- Rossiello, F.; Jurk, D.; Passos, J.F.; d’Adda di Fagagna, F. Telomere dysfunction in ageing and age-related diseases. Nat. Cell Biol. 2022, 24, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Thadathil, N.; Delotterie, D.F.; Xiao, J.; Hori, R.; McDonald, M.P.; Khan, M.M. DNA Double-Strand Break Accumulation in Alzheimer’s Disease: Evidence from Experimental Models and Postmortem Human Brains. Mol. Neurobiol. 2021, 58, 118–131. [Google Scholar] [CrossRef]
- Shanbhag, N.M.; Evans, M.D.; Mao, W.; Nana, A.L.; Seeley, W.W.; Adame, A.; Rissman, R.A.; Masliah, E.; Mucke, L. Early neuronal accumulation of DNA double strand breaks in Alzheimer’s disease. Acta. Neuropathol. Commun. 2019, 7, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, Y.S.; You, J.S.; Kim, T.K.; Ahn, W.J.; Kim, M.J.; Son, K.H.; Ricarte, D.; Ortiz, D.; Lee, S.J.; Lee, H.J. Senescence and impaired DNA damage responses in alpha-synucleinopathy models. Exp. Mol. Med. 2022, 54, 115–128. [Google Scholar] [CrossRef] [PubMed]
- SenGupta, T.; Palikaras, K.; Esbensen, Y.Q.; Konstantinidis, G.; Galindo, F.J.N.; Achanta, K.; Kassahun, H.; Stavgiannoudaki, I.; Bohr, V.A.; Akbari, M.; et al. Base excision repair causes age-dependent accumulation of single-stranded DNA breaks that contribute to Parkinson disease pathology. Cell Rep. 2021, 36, 109668. [Google Scholar] [CrossRef] [PubMed]
- Northrop, A.; Byers, H.A.; Radhakrishnan, S.K. Regulation of NRF1, a master transcription factor of proteasome genes: Implications for cancer and neurodegeneration. Mol. Biol. Cell. 2020, 31, 2158–2163. [Google Scholar] [CrossRef]
- Rauen, T.; Hedrich, C.M.; Tenbrock, K.; Tsokos, G.C. cAMP responsive element modulator: A critical regulator of cytokine production. Trends Mol. Med. 2013, 19, 262–269. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Liu, X.; Zhong, Y. Interleukin-17A: The Key Cytokine in Neurodegenerative Diseases. Front. Aging Neurosci. 2020, 12, 566922. [Google Scholar] [CrossRef]
- Liu, Z.; Qiu, A.W.; Huang, Y.; Yang, Y.; Chen, J.N.; Gu, T.T.; Cao, B.B.; Qiu, Y.H.; Peng, Y.P. IL-17A exacerbates neuroinflammation and neurodegeneration by activating microglia in rodent models of Parkinson’s disease. Brain Behav. Immun. 2019, 81, 630–645. [Google Scholar] [CrossRef]
- Badanjak, K.; Fixemer, S.; Smajic, S.; Skupin, A.; Grunewald, A. The Contribution of Microglia to Neuroinflammation in Parkinson’s Disease. Int. J. Mol. Sci. 2021, 22, 4676. [Google Scholar] [CrossRef]
- Kulkarni, B.; Cruz-Martins, N.; Kumar, D. Microglia in Alzheimer’s Disease: An Unprecedented Opportunity as Prospective Drug Target. Mol. Neurobiol. 2022, 59, 2678–2693. [Google Scholar] [CrossRef] [PubMed]
- Pluvinage, J.V.; Haney, M.S.; Smith, B.A.H.; Sun, J.; Iram, T.; Bonanno, L.; Li, L.; Lee, D.P.; Morgens, D.W.; Yang, A.C.; et al. CD22 blockade restores homeostatic microglial phagocytosis in ageing brains. Nature 2019, 568, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Nakano, M.; Mitsuishi, Y.; Hara, N.; Mano, T.; Iwata, A.; Murayama, S.; Suzuki, T.; Ikeuchi, T.; Nishimura, M. Transcriptional downregulation of FAM3C/ILEI in the Alzheimer’s brain. Hum. Mol. Genet. 2021, 31, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, J.; Kim, J.H.; Lee, G.; Park, T.E.; Yoon, K.J.; Kim, Y.K.; Lim, C. LSM12-EPAC1 defines a neuroprotective pathway that sustains the nucleocytoplasmic RAN gradient. PLoS Biol. 2020, 18, e3001002. [Google Scholar] [CrossRef]
- Crerar, H.; Scott-Solomon, E.; Bodkin-Clarke, C.; Andreassi, C.; Hazbon, M.; Logie, E.; Cano-Jaimez, M.; Gaspari, M.; Kuruvilla, R.; Riccio, A. Regulation of NGF Signaling by an Axonal Untranslated mRNA. Neuron 2019, 102, 553–563. [Google Scholar] [CrossRef] [Green Version]
- Amadoro, G.; Latina, V.; Balzamino, B.O.; Squitti, R.; Varano, M.; Calissano, P.; Micera, A. Nerve Growth Factor-Based Therapy in Alzheimer’s Disease and Age-Related Macular Degeneration. Front. Neurosci. 2021, 15, 735928. [Google Scholar] [CrossRef]
- Triaca, V.; Ruberti, F.; Canu, N. NGF and the Amyloid Precursor Protein in Alzheimer’s Disease: From Molecular Players to Neuronal Circuits. Adv. Exp. Med. Biol. 2021, 1331, 145–165. [Google Scholar] [CrossRef]
- Fan, L.; Sweet, D.R.; Prosdocimo, D.A.; Vinayachandran, V.; Chan, E.R.; Zhang, R.; Ilkayeva, O.; Lu, Y.; Keerthy, K.S.; Booth, C.E.; et al. Muscle Kruppel-like factor 15 regulates lipid flux and systemic metabolic homeostasis. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Gray, S.; Wang, B.; Orihuela, Y.; Hong, E.G.; Fisch, S.; Haldar, S.; Cline, G.W.; Kim, J.K.; Peroni, O.D.; Kahn, B.B.; et al. Regulation of gluconeogenesis by Kruppel-like factor 15. Cell Metab. 2007, 5, 305–312. [Google Scholar] [CrossRef] [Green Version]
- Santiago, J.A.; Potashkin, J.A. Shared dysregulated pathways lead to Parkinson’s disease and diabetes. Trends Mol. Med. 2013, 19, 176–186. [Google Scholar] [CrossRef]
- Santiago, J.A.; Potashkin, J.A. System-based approaches to decode the molecular links in Parkinson’s disease and diabetes. Neurobiol. Dis. 2014, 72 Pt A, 84–91. [Google Scholar] [CrossRef]
- Santiago, J.A.; Bottero, V.; Potashkin, J.A. Transcriptomic and Network Analysis Highlight the Association of Diabetes at Different Stages of Alzheimer’s Disease. Front. Neurosci. 2019, 13, 1273. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.A.; Potashkin, J.A. The Impact of Disease Comorbidities in Alzheimer’s Disease. Front. Aging Neurosci. 2021, 13, 631770. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.A.; Bottero, V.; Potashkin, J.A. Biological and Clinical Implications of Comorbidities in Parkinson’s Disease. Front. Aging Neurosci. 2017, 9, 394. [Google Scholar] [CrossRef] [PubMed]
- Mehrazad Saber, Z.; Takeuchi, Y.; Sawada, Y.; Aita, Y.; Ho, M.H.; Karkoutly, S.; Tao, D.; Katabami, K.; Ye, C.; Murayama, Y.; et al. High protein diet-induced metabolic changes are transcriptionally regulated via KLF15-dependent and independent pathways. Biochem. Biophys. Res. Commun. 2021, 582, 35–42. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed Moustafa, J.S.; Eleftherohorinou, H.; de Smith, A.J.; Andersson-Assarsson, J.C.; Alves, A.C.; Hadjigeorgiou, E.; Walters, R.G.; Asher, J.E.; Bottolo, L.; Buxton, J.L.; et al. Novel association approach for variable number tandem repeats (VNTRs) identifies DOCK5 as a susceptibility gene for severe obesity. Hum. Mol. Genet. 2012, 21, 3727–3738. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, D.; Nico, J.; Johnston, A.D.; Tobias, T.A.M.; Jorge, Y.; Macian, F.; Greally, J.M. CDC42-related genes are upregulated in helper T cells from obese asthmatic children. J. Allergy Clin. Immunol. 2018, 141, 539–548. [Google Scholar] [CrossRef] [Green Version]
- Lai, Y.; Zhao, A.; Tan, M.; Yang, M.; Lin, Y.; Li, S.; Song, J.; Zheng, H.; Zhu, Z.; Liu, D.; et al. DOCK5 regulates energy balance and hepatic insulin sensitivity by targeting mTORC1 signaling. EMBO Rep. 2020, 21, e49473. [Google Scholar] [CrossRef]
- Mariani, E.; Frabetti, F.; Tarozzi, A.; Pelleri, M.C.; Pizzetti, F.; Casadei, R. Meta-Analysis of Parkinson’s Disease Transcriptome Data Using TRAM Software: Whole Substantia Nigra Tissue and Single Dopamine Neuron Differential Gene Expression. PLoS ONE 2016, 11, e0161567. [Google Scholar] [CrossRef]
- Shen, L.; He, J.; Zhao, Y.; Niu, L.; Chen, L.; Tang, G.; Jiang, Y.; Hao, X.; Bai, L.; Li, X.; et al. MicroRNA-126b-5p Exacerbates Development of Adipose Tissue and Diet-Induced Obesity. Int. J. Mol. Sci. 2021, 22, 10261. [Google Scholar] [CrossRef]
- Xu, D.; Li, Y.; Yang, F.; Sun, C.R.; Pan, J.; Wang, L.; Chen, Z.P.; Fang, S.C.; Yao, X.; Hou, W.T.; et al. Structure and transport mechanism of the human cholesterol transporter ABCG1. Cell Rep. 2022, 38, 110298. [Google Scholar] [CrossRef] [PubMed]
- Sano, O.; Tsujita, M.; Shimizu, Y.; Kato, R.; Kobayashi, A.; Kioka, N.; Remaley, A.T.; Michikawa, M.; Ueda, K.; Matsuo, M. ABCG1 and ABCG4 Suppress gamma-Secretase Activity and Amyloid beta Production. PLoS ONE 2016, 11, e0155400. [Google Scholar] [CrossRef] [Green Version]
- Matsuo, M. ABCA1 and ABCG1 as potential therapeutic targets for the prevention of atherosclerosis. J. Pharmacol. Sci. 2022, 148, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Su, X.; Gao, H.; Li, X.; Qu, Y. The Roles of Lpar1 in Central Nervous System Disorders and Diseases. Front. Neurosci. 2021, 15, 710473. [Google Scholar] [CrossRef]
- Fink, K.L.; Lopez-Giraldez, F.; Kim, I.J.; Strittmatter, S.M.; Cafferty, W.B.J. Identification of Intrinsic Axon Growth Modulators for Intact CNS Neurons after Injury. Cell Rep. 2017, 18, 2687–2701. [Google Scholar] [CrossRef] [PubMed]
- Moller, T.; Contos, J.J.; Musante, D.B.; Chun, J.; Ransom, B.R. Expression and function of lysophosphatidic acid receptors in cultured rodent microglial cells. J. Biol. Chem. 2001, 276, 25946–25952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plastira, I.; Bernhart, E.; Joshi, L.; Koyani, C.N.; Strohmaier, H.; Reicher, H.; Malle, E.; Sattler, W. MAPK signaling determines lysophosphatidic acid (LPA)-induced inflammation in microglia. J. Neuroinflammation 2020, 17, 127. [Google Scholar] [CrossRef] [PubMed]
- Doh-ura, K.; Mekada, E.; Ogomori, K.; Iwaki, T. Enhanced CD9 expression in the mouse and human brains infected with transmissible spongiform encephalopathies. J. Neuropathol. Exp. Neurol. 2000, 59, 774–785. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Jee, S.C.; Kim, S.; Hwang, K.H.; Sung, J.S. Identification and Characterization of mRNA Biomarkers for Sodium Cyanide Exposure. Toxics 2021, 9, 288. [Google Scholar] [CrossRef]
- Jensen, M.S.; Ahlemeyer, B.; Ravati, A.; Thakur, P.; Mennel, H.D.; Krieglstein, J. Preconditioning-induced protection against cyanide-induced neurotoxicity is mediated by preserving mitochondrial function. Neurochem. Int. 2002, 40, 285–293. [Google Scholar] [CrossRef]
- Picca, A.; Guerra, F.; Calvani, R.; Marini, F.; Biancolillo, A.; Landi, G.; Beli, R.; Landi, F.; Bernabei, R.; Bentivoglio, A.R.; et al. Mitochondrial Signatures in Circulating Extracellular Vesicles of Older Adults with Parkinson’s Disease: Results from the EXosomes in PArkiNson’s Disease (EXPAND) Study. J. Clin. Med. 2020, 9, 504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.A.; Kim, S.J.; Chung, K.C. Huntingtin-interacting protein 1-mediated neuronal cell death occurs through intrinsic apoptotic pathways and mitochondrial alterations. FEBS Lett. 2006, 580, 5275–5282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roostaei, T.; Nazeri, A.; Felsky, D.; De Jager, P.L.; Schneider, J.A.; Pollock, B.G.; Bennett, D.A.; Voineskos, A.N.; Alzheimer’s Disease Neuroimaging, I. Genome-wide interaction study of brain beta-amyloid burden and cognitive impairment in Alzheimer’s disease. Mol. Psychiatry 2017, 22, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.A.; Chen, Z.; Won, H.; Huang, A.Y.; Lowe, J.K.; Wojta, K.; Yokoyama, J.S.; Bensimon, G.; Leigh, P.N.; Payan, C.; et al. Joint genome-wide association study of progressive supranuclear palsy identifies novel susceptibility loci and genetic correlation to neurodegenerative diseases. Mol. Neurodegener. 2018, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Golanska, E.; Sieruta, M.; Gresner, S.M.; Pfeffer, A.; Chodakowska-Zebrowska, M.; Sobow, T.M.; Klich, I.; Mossakowska, M.; Szybinska, A.; Barcikowska, M.; et al. APBB2 genetic polymorphisms are associated with severe cognitive impairment in centenarians. Exp. Gerontol. 2013, 48, 391–394. [Google Scholar] [CrossRef]
- Li, Y.; Hollingworth, P.; Moore, P.; Foy, C.; Archer, N.; Powell, J.; Nowotny, P.; Holmans, P.; O’Donovan, M.; Tacey, K.; et al. Genetic association of the APP binding protein 2 gene (APBB2) with late onset Alzheimer disease. Hum. Mutat. 2005, 25, 270–277. [Google Scholar] [CrossRef]
- Hohman, T.J.; Chibnik, L.; Bush, W.S.; Jefferson, A.L.; De Jaeger, P.L.; Thornton-Wells, T.A.; Bennett, D.A.; Schneider, J.A. GSK3beta Interactions with Amyloid Genes: An Autopsy Verification and Extension. Neurotox. Res. 2015, 28, 232–238. [Google Scholar] [CrossRef] [Green Version]
- Hohman, T.J.; Koran, M.E.; Thornton-Wells, T.A.; Alzheimer’s Neuroimaging, I. Interactions between GSK3beta and amyloid genes explain variance in amyloid burden. Neurobiol. Aging 2014, 35, 460–465. [Google Scholar] [CrossRef] [Green Version]
- Galli, S.; Stancheva, S.H.; Dufor, T.; Gibb, A.J.; Salinas, P.C. Striatal Synapse Degeneration and Dysfunction Are Reversed by Reactivation of Wnt Signaling. Front. Synaptic Neurosci. 2021, 13, 670467. [Google Scholar] [CrossRef]
- Milnerwood, A.J.; Raymond, L.A. Early synaptic pathophysiology in neurodegeneration: Insights from Huntington’s disease. Trends Neurosci. 2010, 33, 513–523. [Google Scholar] [CrossRef]
- Janezic, S.; Threlfell, S.; Dodson, P.D.; Dowie, M.J.; Taylor, T.N.; Potgieter, D.; Parkkinen, L.; Senior, S.L.; Anwar, S.; Ryan, B.; et al. Deficits in dopaminergic transmission precede neuron loss and dysfunction in a new Parkinson model. Proc. Natl. Acad. Sci. USA 2013, 110, E4016–E4025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berchtold, N.C.; Coleman, P.D.; Cribbs, D.H.; Rogers, J.; Gillen, D.L.; Cotman, C.W. Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer’s disease. Neurobiol. Aging 2013, 34, 1653–1661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casaletto, K.; Ramos-Miguel, A.; VandeBunte, A.; Memel, M.; Buchman, A.; Bennett, D.; Honer, W. Late-life physical activity relates to brain tissue synaptic integrity markers in older adults. Alzheimers Dement. 2022. [Google Scholar] [CrossRef]
- Wang, T.; Liu, H.; Itoh, K.; Oh, S.; Zhao, L.; Murata, D.; Sesaki, H.; Hartung, T.; Na, C.H.; Wang, J. C9orf72 regulates energy homeostasis by stabilizing mitochondrial complex I assembly. Cell Metab. 2021, 33, 531–546. [Google Scholar] [CrossRef] [PubMed]
- Desseille, C.; Deforges, S.; Biondi, O.; Houdebine, L.; D’Amico, D.; Lamaziere, A.; Caradeuc, C.; Bertho, G.; Bruneteau, G.; Weill, L.; et al. Specific Physical Exercise Improves Energetic Metabolism in the Skeletal Muscle of Amyotrophic-Lateral- Sclerosis Mice. Front. Mol. Neurosci. 2017, 10, 332. [Google Scholar] [CrossRef]
- Noori, A.; Mezlini, A.M.; Hyman, B.T.; Serrano-Pozo, A.; Das, S. Systematic review and meta-analysis of human transcriptomics reveals neuroinflammation, deficient energy metabolism, and proteostasis failure across neurodegeneration. Neurobiol. Dis. 2021, 149, 105225. [Google Scholar] [CrossRef]
- Wang, M.; Roussos, P.; McKenzie, A.; Zhou, X.; Kajiwara, Y.; Brennand, K.J.; De Luca, G.C.; Crary, J.F.; Casaccia, P.; Buxbaum, J.D.; et al. Integrative network analysis of nineteen brain regions identifies molecular signatures and networks underlying selective regional vulnerability to Alzheimer’s disease. Genome. Med. 2016, 8, 104. [Google Scholar] [CrossRef] [Green Version]
- Chen-Plotkin, A.S.; Geser, F.; Plotkin, J.B.; Clark, C.M.; Kwong, L.K.; Yuan, W.; Grossman, M.; Van Deerlin, V.M.; Trojanowski, J.Q.; Lee, V.M. Variations in the progranulin gene affect global gene expression in frontotemporal lobar degeneration. Hum. Mol. Genet. 2008, 17, 1349–1362. [Google Scholar] [CrossRef] [Green Version]
- Papapetropoulos, S.; Ffrench-Mullen, J.; McCorquodale, D.; Qin, Y.; Pablo, J.; Mash, D.C. Multiregional gene expression profiling identifies MRPS6 as a possible candidate gene for Parkinson’s disease. Gene Expr. J. Liver Res. 2006, 13, 205–215. [Google Scholar] [CrossRef]
- Moran, L.B.; Duke, D.C.; Deprez, M.; Dexter, D.T.; Pearce, R.K.; Graeber, M.B. Whole genome expression profiling of the medial and lateral substantia nigra in Parkinson’s disease. Neurogenetics 2006, 7, 1–11. [Google Scholar] [CrossRef]
- Hodges, A.; Strand, A.D.; Aragaki, A.K.; Kuhn, A.; Sengstag, T.; Hughes, G.; Elliston, L.A.; Hartog, C.; Goldstein, D.R.; Thu, D.; et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet. 2006, 15, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Paci, P.; Colombo, T.; Fiscon, G.; Gurtner, A.; Pavesi, G.; Farina, L. SWIM: A computational tool to unveiling crucial nodes in complex biological networks. Sci. Rep. 2017, 7, 44797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kupershmidt, I.; Su, Q.J.; Grewal, A.; Sundaresh, S.; Halperin, I.; Flynn, J.; Shekar, M.; Wang, H.; Park, J.; Cui, W.; et al. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS ONE 2010, 5, e13066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santiago, J.A.; Quinn, J.P.; Potashkin, J.A. Physical Activity Rewires the Human Brain against Neurodegeneration. Int. J. Mol. Sci. 2022, 23, 6223. https://doi.org/10.3390/ijms23116223
Santiago JA, Quinn JP, Potashkin JA. Physical Activity Rewires the Human Brain against Neurodegeneration. International Journal of Molecular Sciences. 2022; 23(11):6223. https://doi.org/10.3390/ijms23116223
Chicago/Turabian StyleSantiago, Jose A., James P. Quinn, and Judith A. Potashkin. 2022. "Physical Activity Rewires the Human Brain against Neurodegeneration" International Journal of Molecular Sciences 23, no. 11: 6223. https://doi.org/10.3390/ijms23116223
APA StyleSantiago, J. A., Quinn, J. P., & Potashkin, J. A. (2022). Physical Activity Rewires the Human Brain against Neurodegeneration. International Journal of Molecular Sciences, 23(11), 6223. https://doi.org/10.3390/ijms23116223





