Centronuclear Myopathy Caused by Defective Membrane Remodelling of Dynamin 2 and BIN1 Variants
Abstract
:1. Introduction
2. T-Tubules: Sarcolemmal Invaginations Essential for E-C Coupling
3. BIN1: A BAR Domain Protein-Inducing Membrane Curvature
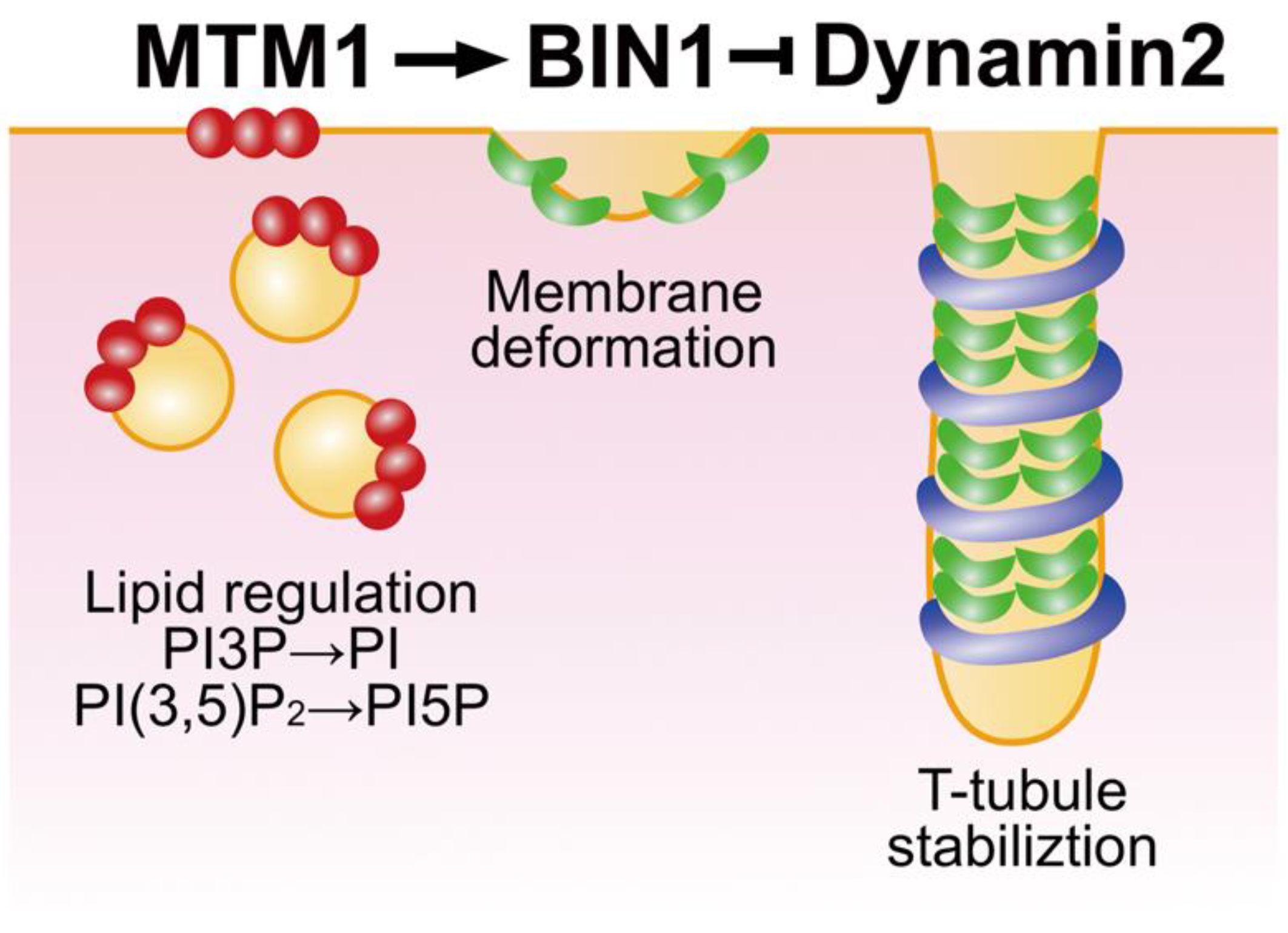
3.1. BIN1 Functions in T-Tubule Biogenesis
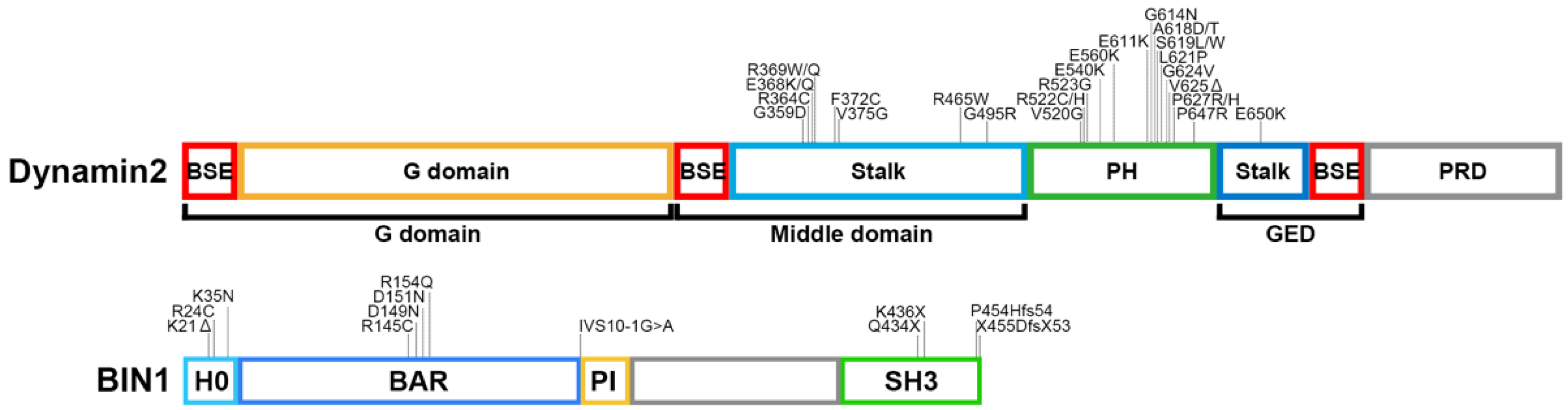
3.2. CNM Pathogenesis Caused by Defective Membrane Remodelling of BIN1 Variants
4. Dynamin: A Membrane Fission Catalyser in Endocytosis
4.1. Structure and Function of Dynamin
4.2. Dynamin 2 Functions in T-Tubule Biogenesis
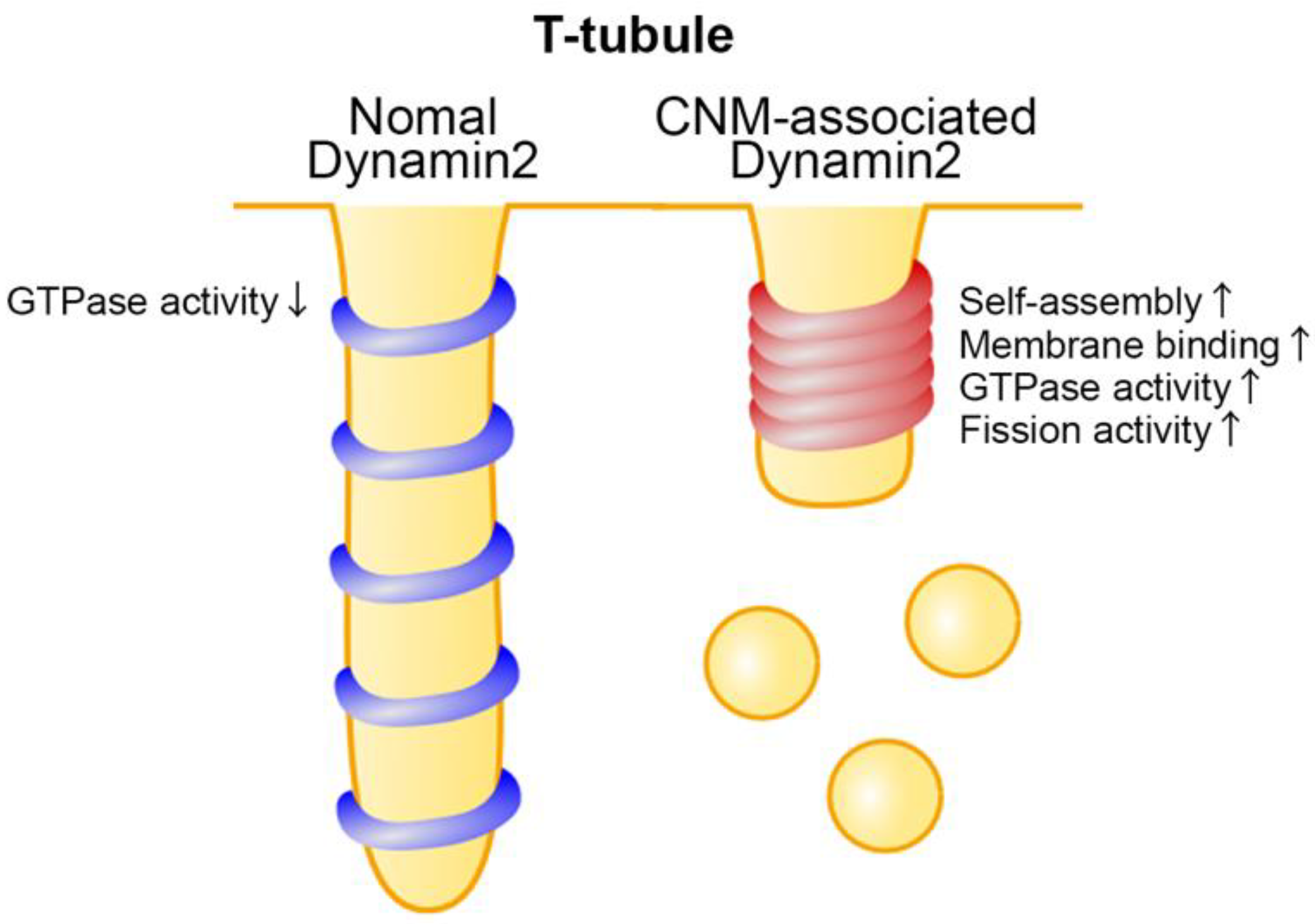
4.3. Dysregulation of T-Tubule Function by CNM-Associated Dynamin 2 Variants
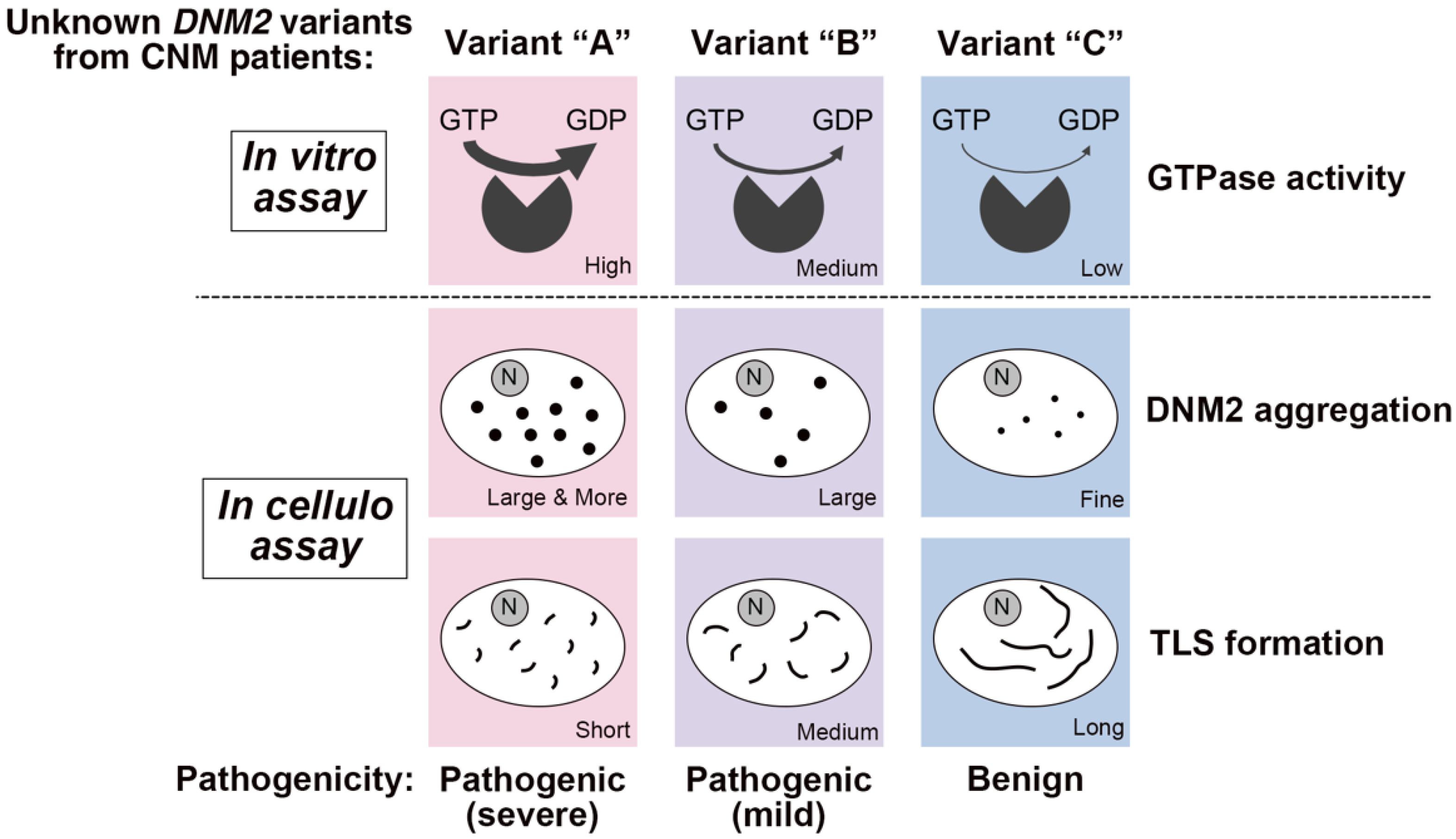
4.4. Correlation between Membrane Fission Activity and Symptom Severities by CNM-Associated Dynamin 2 Variants
4.5. Other Functions of Dynamin 2 in Skeletal Muscle
4.6. Therapeutic Approaches for CNM
5. Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jungbluth, H.; Wallgren-Pettersson, C.; Laporte, J. Centronuclear (myotubular) myopathy. Orphanet. J. Rare Dis. 2008, 3, 26. [Google Scholar] [CrossRef] [PubMed]
- Jungbluth, H.; Gautel, M. Pathogenic mechanisms in centronuclear myopathies. Front. Aging Neurosci. 2014, 6, 339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, N.B. Centronuclear myopathies: A widening concept. Neuromuscul. Disord. 2010, 20, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Majczenko, K.; Davidson, A.E.; Camelo-Piragua, S.; Agrawal, P.B.; Manfready, R.A.; Li, X.; Joshi, S.; Xu, J.; Peng, W.; Beggs, A.H.; et al. Dominant mutation of CCDC78 in a unique congenital myopathy with prominent internal nuclei and atypical cores. Am. J. Hum. Genet. 2012, 91, 365–371. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, P.B.; Pierson, C.R.; Joshi, M.; Liu, X.; Ravenscroft, G.; Moghadaszadeh, B.; Talabere, T.; Viola, M.; Swanson, L.C.; Haliloglu, G.; et al. SPEG interacts with myotubularin, and its deficiency causes centronuclear myopathy with dilated cardiomyopathy. Am. J. Hum. Genet. 2014, 95, 218–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Qusairi, L.; Laporte, J. T-tubule biogenesis and triad formation in skeletal muscle and implication in human diseases. Skelet. Muscle 2011, 1, 26. [Google Scholar] [CrossRef] [PubMed]
- Dowling, J.J.; Gibbs, E.M.; Feldman, E.L. Membrane traffic and muscle: Lessons from human disease. Traffic 2008, 9, 1035–1043. [Google Scholar] [CrossRef] [Green Version]
- Bitoun, M.; Maugenre, S.; Jeannet, P.Y.; Lacene, E.; Ferrer, X.; Laforet, P.; Martin, J.J.; Laporte, J.; Lochmuller, H.; Beggs, A.H.; et al. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat. Genet. 2005, 37, 1207–1209. [Google Scholar] [CrossRef]
- Nicot, A.S.; Toussaint, A.; Tosch, V.; Kretz, C.; Wallgren-Pettersson, C.; Iwarsson, E.; Kingston, H.; Garnier, J.M.; Biancalana, V.; Oldfors, A.; et al. Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat. Genet. 2007, 39, 1134–1139. [Google Scholar] [CrossRef]
- Fischer, D.; Herasse, M.; Bitoun, M.; Barragan-Campos, H.M.; Chiras, J.; Laforet, P.; Fardeau, M.; Eymard, B.; Guicheney, P.; Romero, N.B. Characterization of the muscle involvement in dynamin 2-related centronuclear myopathy. Brain 2006, 129, 1463–1469. [Google Scholar] [CrossRef]
- Bohm, J.; Yis, U.; Ortac, R.; Cakmakci, H.; Kurul, S.H.; Dirik, E.; Laporte, J. Case report of intrafamilial variability in autosomal recessive centronuclear myopathy associated to a novel BIN1 stop mutation. Orphanet. J. Rare Dis. 2010, 5, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Oca, R.; Cowling, B.S.; Laporte, J. Common Pathogenic Mechanisms in Centronuclear and Myotubular Myopathies and Latest Treatment Advances. Int. J. Mol. Sci. 2021, 22, 11377. [Google Scholar] [CrossRef] [PubMed]
- Dowling, J.J.; Weihl, C.C.; Spencer, M.J. Molecular and cellular basis of genetically inherited skeletal muscle disorders. Nat. Rev. Mol. Cell Biol. 2021, 22, 713–732. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Maani, N.; Dowling, J.J. Dynamin 2 (DNM2) as Cause of, and Modifier for, Human Neuromuscular Disease. Neurotherapeutics 2018, 15, 966–975. [Google Scholar] [CrossRef] [Green Version]
- Cowling, B.S.; Toussaint, A.; Muller, J.; Laporte, J. Defective membrane remodeling in neuromuscular diseases: Insights from animal models. PLoS Genet. 2012, 8, e1002595. [Google Scholar] [CrossRef]
- Durieux, A.C.; Prudhon, B.; Guicheney, P.; Bitoun, M. Dynamin 2 and human diseases. J. Mol. Med. 2010, 88, 339–350. [Google Scholar] [CrossRef] [Green Version]
- Franzini-Armstrong, C. The relationship between form and function throughout the history of excitation-contraction coupling. J. Gen. Physiol. 2018, 150, 189–210. [Google Scholar] [CrossRef] [Green Version]
- Rosemblatt, M.; Hidalgo, C.; Vergara, C.; Ikemoto, N. Immunological and biochemical properties of transverse tubule membranes isolated from rabbit skeletal muscle. J. Biol. Chem. 1981, 256, 8140–8148. [Google Scholar] [CrossRef]
- Lee, E.; Marcucci, M.; Daniell, L.; Pypaert, M.; Weisz, O.A.; Ochoa, G.C.; Farsad, K.; Wenk, M.R.; De Camilli, P. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science 2002, 297, 1193–1196. [Google Scholar] [CrossRef] [Green Version]
- Flucher, B.E. How is SR calcium release in muscle modulated by PIP(4,5)2? J. Gen. Physiol 2015, 145, 361–364. [Google Scholar] [CrossRef] [Green Version]
- Barrientos, G.; Llanos, P.; Hidalgo, J.; Bolanos, P.; Caputo, C.; Riquelme, A.; Sanchez, G.; Quest, A.F.; Hidalgo, C. Cholesterol removal from adult skeletal muscle impairs excitation-contraction coupling and aging reduces caveolin-3 and alters the expression of other triadic proteins. Front. Physiol. 2015, 6, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calderon, J.C.; Bolanos, P.; Caputo, C. The excitation-contraction coupling mechanism in skeletal muscle. Biophys. Rev. 2014, 6, 133–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fill, M.; Copello, J.A. Ryanodine receptor calcium release channels. Physiol. Rev. 2002, 82, 893–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Tae, H.S.; Norris, N.C.; Karunasekara, Y.; Pouliquin, P.; Board, P.G.; Dulhunty, A.F.; Casarotto, M.G. A dihydropyridine receptor alpha1s loop region critical for skeletal muscle contraction is intrinsically unstructured and binds to a SPRY domain of the type 1 ryanodine receptor. Int. J. Biochem. Cell Biol. 2009, 41, 677–686. [Google Scholar] [CrossRef]
- Hu, H.; Wang, Z.; Wei, R.; Fan, G.; Wang, Q.; Zhang, K.; Yin, C.C. The molecular architecture of dihydropyrindine receptor/L-type Ca2+ channel complex. Sci. Rep. 2015, 5, 8370. [Google Scholar] [CrossRef] [Green Version]
- Dowling, J.J.; Lawlor, M.W.; Dirksen, R.T. Triadopathies: An emerging class of skeletal muscle diseases. Neurotherapeutics 2014, 11, 773–785. [Google Scholar] [CrossRef] [Green Version]
- Prokic, I.; Cowling, B.S.; Laporte, J. Amphiphysin 2 (BIN1) in physiology and diseases. J. Mol. Med. 2014, 92, 453–463. [Google Scholar] [CrossRef]
- Frost, A.; Unger, V.M.; De Camilli, P. The BAR domain superfamily: Membrane-molding macromolecules. Cell 2009, 137, 191–196. [Google Scholar] [CrossRef] [Green Version]
- Suetsugu, S.; Toyooka, K.; Senju, Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev. Biol. 2010, 21, 340–349. [Google Scholar] [CrossRef]
- Casal, E.; Federici, L.; Zhang, W.; Fernandez-Recio, J.; Priego, E.M.; Miguel, R.N.; DuHadaway, J.B.; Prendergast, G.C.; Luisi, B.F.; Laue, E.D. The crystal structure of the BAR domain from human Bin1/amphiphysin II and its implications for molecular recognition. Biochemistry 2006, 45, 12917–12928. [Google Scholar] [CrossRef] [Green Version]
- Sakamuro, D.; Elliott, K.J.; Wechsler-Reya, R.; Prendergast, G.C. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat. Genet. 1996, 14, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.H.; David, C.; Ochoa, G.C.; Freyberg, Z.; Daniell, L.; Grabs, D.; Cremona, O.; De Camilli, P. Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle. J. Cell Biol. 1997, 137, 1355–1367. [Google Scholar] [CrossRef] [PubMed]
- Prokic, I.; Cowling, B.S.; Kutchukian, C.; Kretz, C.; Tasfaout, H.; Gache, V.; Hergueux, J.; Wendling, O.; Ferry, A.; Toussaint, A.; et al. Differential physiological role of BIN1 isoforms in skeletal muscle development, function and regeneration. Dis. Model. Mech. 2020, 13, dmm044354. [Google Scholar] [CrossRef] [PubMed]
- Cowling, B.S.; Prokic, I.; Tasfaout, H.; Rabai, A.; Humbert, F.; Rinaldi, B.; Nicot, A.S.; Kretz, C.; Friant, S.; Roux, A.; et al. Amphiphysin (BIN1) negatively regulates dynamin 2 for normal muscle maturation. J. Clin. Investig. 2017, 127, 4477–4487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tjondrokoesoemo, A.; Park, K.H.; Ferrante, C.; Komazaki, S.; Lesniak, S.; Brotto, M.; Ko, J.K.; Zhou, J.; Weisleder, N.; Ma, J. Disrupted membrane structure and intracellular Ca2+ signaling in adult skeletal muscle with acute knockdown of Bin1. PLoS ONE 2011, 6, e25740. [Google Scholar] [CrossRef]
- Razzaq, A.; Robinson, I.M.; McMahon, H.T.; Skepper, J.N.; Su, Y.; Zelhof, A.C.; Jackson, A.P.; Gay, N.J.; O’Kane, C.J. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev. 2001, 15, 2967–2979. [Google Scholar] [CrossRef] [Green Version]
- Isas, J.M.; Ambroso, M.R.; Hegde, P.B.; Langen, J.; Langen, R. Tubulation by amphiphysin requires concentration-dependent switching from wedging to scaffolding. Structure 2015, 23, 873–881. [Google Scholar] [CrossRef] [Green Version]
- Adam, J.; Basnet, N.; Mizuno, N. Structural insights into the cooperative remodeling of membranes by amphiphysin/BIN1. Sci. Rep. 2015, 5, 15452. [Google Scholar] [CrossRef] [Green Version]
- Picas, L.; Viaud, J.; Schauer, K.; Vanni, S.; Hnia, K.; Fraisier, V.; Roux, A.; Bassereau, P.; Gaits-Iacovoni, F.; Payrastre, B.; et al. BIN1/M-Amphiphysin2 induces clustering of phosphoinositides to recruit its downstream partner dynamin. Nat. Commun. 2014, 5, 5647. [Google Scholar] [CrossRef] [Green Version]
- Gowrisankaran, S.; Wang, Z.; Morgan, D.G.; Milosevic, I.; Mim, C. Cells Control BIN1-Mediated Membrane Tubulation by Altering the Membrane Charge. J. Mol. Biol. 2020, 432, 1235–1250. [Google Scholar] [CrossRef]
- Wu, T.; Baumgart, T. BIN1 membrane curvature sensing and generation show autoinhibition regulated by downstream ligands and PI(4,5)P2. Biochemistry 2014, 53, 7297–7309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drager, N.M.; Nachman, E.; Winterhoff, M.; Bruhmann, S.; Shah, P.; Katsinelos, T.; Boulant, S.; Teleman, A.A.; Faix, J.; Jahn, T.R. Bin1 directly remodels actin dynamics through its BAR domain. EMBO Rep. 2017, 18, 2051–2066. [Google Scholar] [CrossRef] [PubMed]
- Hong, T.; Yang, H.; Zhang, S.S.; Cho, H.C.; Kalashnikova, M.; Sun, B.; Zhang, H.; Bhargava, A.; Grabe, M.; Olgin, J.; et al. Cardiac BIN1 folds T-tubule membrane, controlling ion flux and limiting arrhythmia. Nat. Med. 2014, 20, 624–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Shi, Z.; Baumgart, T. Mutations in BIN1 associated with centronuclear myopathy disrupt membrane remodeling by affecting protein density and oligomerization. PLoS ONE 2014, 9, e93060. [Google Scholar] [CrossRef]
- Bohm, J.; Vasli, N.; Maurer, M.; Cowling, B.S.; Shelton, G.D.; Kress, W.; Toussaint, A.; Prokic, I.; Schara, U.; Anderson, T.J.; et al. Altered splicing of the BIN1 muscle-specific exon in humans and dogs with highly progressive centronuclear myopathy. PLoS Genet. 2013, 9, e1003430. [Google Scholar] [CrossRef]
- Fugier, C.; Klein, A.F.; Hammer, C.; Vassilopoulos, S.; Ivarsson, Y.; Toussaint, A.; Tosch, V.; Vignaud, A.; Ferry, A.; Messaddeq, N.; et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 2011, 17, 720–725. [Google Scholar] [CrossRef] [Green Version]
- Fujise, K.; Okubo, M.; Abe, T.; Yamada, H.; Nishino, I.; Noguchi, S.; Takei, K.; Takeda, T. Mutant BIN1-Dynamin 2 complexes dysregulate membrane remodeling in the pathogenesis of centronuclear myopathy. J. Biol. Chem. 2021, 296, 100077. [Google Scholar] [CrossRef]
- Bohm, J.; Biancalana, V.; Malfatti, E.; Dondaine, N.; Koch, C.; Vasli, N.; Kress, W.; Strittmatter, M.; Taratuto, A.L.; Gonorazky, H.; et al. Adult-onset autosomal dominant centronuclear myopathy due to BIN1 mutations. Brain 2014, 137, 3160–3170. [Google Scholar] [CrossRef] [Green Version]
- Toussaint, A.; Cowling, B.S.; Hnia, K.; Mohr, M.; Oldfors, A.; Schwab, Y.; Yis, U.; Maisonobe, T.; Stojkovic, T.; Wallgren-Pettersson, C.; et al. Defects in amphiphysin 2 (BIN1) and triads in several forms of centronuclear myopathies. Acta Neuropathol. 2011, 121, 253–266. [Google Scholar] [CrossRef]
- Royer, B.; Hnia, K.; Gavriilidis, C.; Tronchere, H.; Tosch, V.; Laporte, J. The myotubularin-amphiphysin 2 complex in membrane tubulation and centronuclear myopathies. EMBO Rep. 2013, 14, 907–915. [Google Scholar] [CrossRef] [Green Version]
- Falcone, S.; Roman, W.; Hnia, K.; Gache, V.; Didier, N.; Laine, J.; Aurade, F.; Marty, I.; Nishino, I.; Charlet-Berguerand, N.; et al. N-WASP is required for Amphiphysin-2/BIN1-dependent nuclear positioning and triad organization in skeletal muscle and is involved in the pathophysiology of centronuclear myopathy. EMBO Mol. Med. 2014, 6, 1455–1475. [Google Scholar] [CrossRef] [PubMed]
- Antonny, B.; Burd, C.; De Camilli, P.; Chen, E.; Daumke, O.; Faelber, K.; Ford, M.; Frolov, V.A.; Frost, A.; Hinshaw, J.E.; et al. Membrane fission by dynamin: What we know and what we need to know. EMBO J. 2016, 35, 2270–2284. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, S.M.; De Camilli, P. Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 2012, 13, 75–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, S.L.; Frolov, V.A. Dynamin: Functional design of a membrane fission catalyst. Annu. Rev. Cell Dev. Biol. 2011, 27, 79–105. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Garcia, F.; McNiven, M.A. Differential distribution of dynamin isoforms in mammalian cells. Mol. Biol. Cell 1998, 9, 2595–2609. [Google Scholar] [CrossRef] [Green Version]
- Cook, T.; Mesa, K.; Urrutia, R. Three dynamin-encoding genes are differentially expressed in developing rat brain. J. Neurochem. 1996, 67, 927–931. [Google Scholar] [CrossRef] [PubMed]
- Cook, T.A.; Urrutia, R.; McNiven, M.A. Identification of dynamin 2, an isoform ubiquitously expressed in rat tissues. Proc. Natl. Acad. Sci. USA 1994, 91, 644–648. [Google Scholar] [CrossRef] [Green Version]
- Reubold, T.F.; Eschenburg, S.; Becker, A.; Leonard, M.; Schmid, S.L.; Vallee, R.B.; Kull, F.J.; Manstein, D.J. Crystal structure of the GTPase domain of rat dynamin 1. Proc. Natl. Acad. Sci. USA 2005, 102, 13093–13098. [Google Scholar] [CrossRef] [Green Version]
- Faelber, K.; Posor, Y.; Gao, S.; Held, M.; Roske, Y.; Schulze, D.; Haucke, V.; Noe, F.; Daumke, O. Crystal structure of nucleotide-free dynamin. Nature 2011, 477, 556–560. [Google Scholar] [CrossRef]
- Zhenga, J.; Cahill, S.M.; Lemmon, M.A.; Fushmana, D.; Schlessinger, J.; Cowburn, D. Identification of the binding site for acidic phospholipids on the pH domain of dynamin: Implications for stimulation of GTPase activity. J. Mol. Biol. 1996, 255, 14–21. [Google Scholar] [CrossRef]
- Bethoney, K.A.; King, M.C.; Hinshaw, J.E.; Ostap, E.M.; Lemmon, M.A. A possible effector role for the pleckstrin homology (PH) domain of dynamin. Proc. Natl. Acad. Sci. USA 2009, 106, 13359–13364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrotra, N.; Nichols, J.; Ramachandran, R. Alternate pleckstrin homology domain orientations regulate dynamin-catalyzed membrane fission. Mol. Biol. Cell 2014, 25, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Reubold, T.F.; Faelber, K.; Plattner, N.; Posor, Y.; Ketel, K.; Curth, U.; Schlegel, J.; Anand, R.; Manstein, D.J.; Noe, F.; et al. Crystal structure of the dynamin tetramer. Nature 2015, 525, 404–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Lee, D.M.; Jimah, J.R.; Gerassimov, N.; Yang, C.; Kim, S.; Luvsanjav, D.; Winkelman, J.; Mettlen, M.; Abrams, M.E.; et al. Dynamin regulates the dynamics and mechanical strength of the actin cytoskeleton as a multifilament actin-bundling protein. Nat. Cell Biol. 2020, 22, 674–688. [Google Scholar] [CrossRef]
- Takei, K.; McPherson, P.S.; Schmid, S.L.; De Camilli, P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature 1995, 374, 186–190. [Google Scholar] [CrossRef]
- Chuang, M.C.; Lin, S.S.; Ohniwa, R.L.; Lee, G.H.; Su, Y.A.; Chang, Y.C.; Tang, M.J.; Liu, Y.W. Tks5 and Dynamin-2 enhance actin bundle rigidity in invadosomes to promote myoblast fusion. J. Cell Biol. 2019, 218, 1670–1685. [Google Scholar] [CrossRef] [Green Version]
- Takei, K.; Haucke, V.; Slepnev, V.; Farsad, K.; Salazar, M.; Chen, H.; De Camilli, P. Generation of coated intermediates of clathrin-mediated endocytosis on protein-free liposomes. Cell 1998, 94, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Sweitzer, S.M.; Hinshaw, J.E. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell 1998, 93, 1021–1029. [Google Scholar] [CrossRef] [Green Version]
- Chappie, J.S.; Acharya, S.; Leonard, M.; Schmid, S.L.; Dyda, F. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature 2010, 465, 435–440. [Google Scholar] [CrossRef] [Green Version]
- Ganichkin, O.M.; Vancraenenbroeck, R.; Rosenblum, G.; Hofmann, H.; Mikhailov, A.S.; Daumke, O.; Noel, J.K. Quantification and demonstration of the collective constriction-by-ratchet mechanism in the dynamin molecular motor. Proc. Natl. Acad. Sci. USA 2021, 118, e2101144118. [Google Scholar] [CrossRef]
- Colom, A.; Redondo-Morata, L.; Chiaruttini, N.; Roux, A.; Scheuring, S. Dynamic remodeling of the dynamin helix during membrane constriction. Proc. Natl. Acad. Sci. USA 2017, 114, 5449–5454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, T.; Kozai, T.; Yang, H.; Ishikuro, D.; Seyama, K.; Kumagai, Y.; Abe, T.; Yamada, H.; Uchihashi, T.; Ando, T.; et al. Dynamic clustering of dynamin-amphiphysin helices regulates membrane constriction and fission coupled with GTP hydrolysis. eLife 2018, 7, e30246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roux, A.; Uyhazi, K.; Frost, A.; De Camilli, P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature 2006, 441, 528–531. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Chen, K.; Dong, B.; Yang, M.; Filbrun, S.L.; Myoung, Y.; Huang, T.X.; Gu, Y.; Wang, G.; Fang, N. Dynamin-dependent vesicle twist at the final stage of clathrin-mediated endocytosis. Nat. Cell Biol. 2021, 23, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Diatloff-Zito, C.; Gordon, A.J.; Duchaud, E.; Merlin, G. Isolation of an ubiquitously expressed cDNA encoding human dynamin II, a member of the large GTP-binding protein family. Gene 1995, 163, 301–306. [Google Scholar] [CrossRef]
- Cowling, B.S.; Toussaint, A.; Amoasii, L.; Koebel, P.; Ferry, A.; Davignon, L.; Nishino, I.; Mandel, J.L.; Laporte, J. Increased expression of wild-type or a centronuclear myopathy mutant of dynamin 2 in skeletal muscle of adult mice leads to structural defects and muscle weakness. Am. J. Pathol. 2011, 178, 2224–2235. [Google Scholar] [CrossRef] [Green Version]
- Biancalana, V.; Romero, N.B.; Thuestad, I.J.; Ignatius, J.; Kataja, J.; Gardberg, M.; Heron, D.; Malfatti, E.; Oldfors, A.; Laporte, J. Some DNM2 mutations cause extremely severe congenital myopathy and phenocopy myotubular myopathy. Acta Neuropathol. Commun. 2018, 6, 93. [Google Scholar] [CrossRef]
- Bohm, J.; Biancalana, V.; Dechene, E.T.; Bitoun, M.; Pierson, C.R.; Schaefer, E.; Karasoy, H.; Dempsey, M.A.; Klein, F.; Dondaine, N.; et al. Mutation spectrum in the large GTPase dynamin 2, and genotype-phenotype correlation in autosomal dominant centronuclear myopathy. Hum. Mutat. 2012, 33, 949–959. [Google Scholar] [CrossRef] [Green Version]
- Casar-Borota, O.; Jacobsson, J.; Libelius, R.; Oldfors, C.H.; Malfatti, E.; Romero, N.B.; Oldfors, A. A novel dynamin-2 gene mutation associated with a late-onset centronuclear myopathy with necklace fibres. Neuromuscul. Disord. 2015, 25, 345–348. [Google Scholar] [CrossRef]
- Hohendahl, A.; Roux, A.; Galli, V. Structural insights into the centronuclear myopathy-associated functions of BIN1 and dynamin 2. J. Struct. Biol. 2016, 196, 37–47. [Google Scholar] [CrossRef] [Green Version]
- Marks, B.; Stowell, M.H.; Vallis, Y.; Mills, I.G.; Gibson, A.; Hopkins, C.R.; McMahon, H.T. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature 2001, 410, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Warnock, D.E.; Hinshaw, J.E.; Schmid, S.L. Dynamin self-assembly stimulates its GTPase activity. J. Biol. Chem. 1996, 271, 22310–22314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallis, Y.; Wigge, P.; Marks, B.; Evans, P.R.; McMahon, H.T. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr. Biol. 1999, 9, 257–260. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, E.M.; Davidson, A.E.; Telfer, W.R.; Feldman, E.L.; Dowling, J.J. The myopathy-causing mutation DNM2-S619L leads to defective tubulation in vitro and in developing zebrafish. Dis. Model. Mech. 2014, 7, 157–161. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Smith, L.; Volpatti, J.; Fabian, L.; Dowling, J.J. Insights into wild-type dynamin 2 and the consequences of DNM2 mutations from transgenic zebrafish. Hum. Mol. Genet. 2019, 28, 4186–4196. [Google Scholar] [CrossRef]
- Wang, L.; Barylko, B.; Byers, C.; Ross, J.A.; Jameson, D.M.; Albanesi, J.P. Dynamin 2 mutants linked to centronuclear myopathies form abnormally stable polymers. J. Biol. Chem. 2010, 285, 22753–22757. [Google Scholar] [CrossRef] [Green Version]
- Kenniston, J.A.; Lemmon, M.A. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010, 29, 3054–3067. [Google Scholar] [CrossRef] [Green Version]
- Chin, Y.H.; Lee, A.; Kan, H.W.; Laiman, J.; Chuang, M.C.; Hsieh, S.T.; Liu, Y.W. Dynamin-2 mutations associated with centronuclear myopathy are hypermorphic and lead to T-tubule fragmentation. Hum. Mol. Genet. 2015, 24, 5542–5554. [Google Scholar] [CrossRef] [Green Version]
- Kutchukian, C.; Szentesi, P.; Allard, B.; Trochet, D.; Beuvin, M.; Berthier, C.; Tourneur, Y.; Guicheney, P.; Csernoch, L.; Bitoun, M.; et al. Impaired excitation-contraction coupling in muscle fibres from the dynamin2(R465W) mouse model of centronuclear myopathy. J. Physiol. 2017, 595, 7369–7382. [Google Scholar] [CrossRef] [Green Version]
- Hinostroza, F.; Neely, A.; Araya-Duran, I.; Maraboli, V.; Canan, J.; Rojas, M.; Aguayo, D.; Latorre, R.; Gonzalez-Nilo, F.D.; Cardenas, A.M. Dynamin-2 R465W mutation induces long range perturbation in highly ordered oligomeric structures. Sci. Rep. 2020, 10, 18151. [Google Scholar] [CrossRef]
- Fujise, K.; Okubo, M.; Abe, T.; Yamada, H.; Takei, K.; Nishino, I.; Takeda, T.; Noguchi, S. Imaging-based evaluation of pathogenicity by novel DNM2 variants associated with centronuclear myopathy. Hum. Mutat. 2021, 43, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Antonescu, C.N.; Diaz, M.; Femia, G.; Planas, J.V.; Klip, A. Clathrin-dependent and independent endocytosis of glucose transporter 4 (GLUT4) in myoblasts: Regulation by mitochondrial uncoupling. Traffic 2008, 9, 1173–1190. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Jamett, A.M.; Baez-Matus, X.; Olivares, M.J.; Hinostroza, F.; Guerra-Fernandez, M.J.; Vasquez-Navarrete, J.; Bui, M.T.; Guicheney, P.; Romero, N.B.; Bevilacqua, J.A.; et al. Dynamin-2 mutations linked to Centronuclear Myopathy impair actin-dependent trafficking in muscle cells. Sci. Rep. 2017, 7, 4580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Hasani, H.; Kunamneni, R.K.; Dawson, K.; Hinck, C.S.; Muller-Wieland, D.; Cushman, S.W. Roles of the N- and C-termini of GLUT4 in endocytosis. J. Cell Sci. 2002, 115, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Puri, C.; Manni, M.M.; Vicinanza, M.; Hilcenko, C.; Zhu, Y.; Runwal, G.; Stamatakou, E.; Menzies, F.M.; Mamchaoui, K.; Bitoun, M.; et al. A DNM2 Centronuclear Myopathy Mutation Reveals a Link between Recycling Endosome Scission and Autophagy. Dev. Cell 2020, 53, 154–168.e6. [Google Scholar] [CrossRef] [PubMed]
- Schulze, R.J.; Weller, S.G.; Schroeder, B.; Krueger, E.W.; Chi, S.; Casey, C.A.; McNiven, M.A. Lipid droplet breakdown requires dynamin 2 for vesiculation of autolysosomal tubules in hepatocytes. J. Cell Biol. 2013, 203, 315–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mesaki, K.; Tanabe, K.; Obayashi, M.; Oe, N.; Takei, K. Fission of tubular endosomes triggers endosomal acidification and movement. PLoS ONE 2011, 6, e19764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birgisdottir, A.B.; Johansen, T. Autophagy and endocytosis—Interconnections and interdependencies. J. Cell Sci. 2020, 133, jcs228114. [Google Scholar] [CrossRef]
- Durieux, A.C.; Vassilopoulos, S.; Laine, J.; Fraysse, B.; Brinas, L.; Prudhon, B.; Castells, J.; Freyssenet, D.; Bonne, G.; Guicheney, P.; et al. A centronuclear myopathy—Dynamin 2 mutation impairs autophagy in mice. Traffic 2012, 13, 869–879. [Google Scholar] [CrossRef] [Green Version]
- Kounakis, K.; Chaniotakis, M.; Markaki, M.; Tavernarakis, N. Emerging Roles of Lipophagy in Health and Disease. Front. Cell Dev. Biol. 2019, 7, 185. [Google Scholar] [CrossRef]
- Tinelli, E.; Pereira, J.A.; Suter, U. Muscle-specific function of the centronuclear myopathy and Charcot-Marie-Tooth neuropathy-associated dynamin 2 is required for proper lipid metabolism, mitochondria, muscle fibers, neuromuscular junctions and peripheral nerves. Hum. Mol. Genet. 2013, 22, 4417–4429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.E.; Westrate, L.M.; Wu, H.; Page, C.; Voeltz, G.K. Multiple dynamin family members collaborate to drive mitochondrial division. Nature 2016, 540, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Marette, A.; Burdett, E.; Douen, A.; Vranic, M.; Klip, A. Insulin induces the translocation of GLUT4 from a unique intracellular organelle to transverse tubules in rat skeletal muscle. Diabetes 1992, 41, 1562–1569. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hansen, P.A.; Marshall, B.A.; Holloszy, J.O.; Mueckler, M. Insulin unmasks a COOH-terminal Glut4 epitope and increases glucose transport across T-tubules in skeletal muscle. J. Cell Biol. 1996, 135, 415–430. [Google Scholar] [CrossRef] [Green Version]
- Ploug, T.; van Deurs, B.; Ai, H.; Cushman, S.W.; Ralston, E. Analysis of GLUT4 distribution in whole skeletal muscle fibers: Identification of distinct storage compartments that are recruited by insulin and muscle contractions. J. Cell Biol. 1998, 142, 1429–1446. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.S.; Hsieh, T.L.; Liou, G.G.; Li, T.N.; Lin, H.C.; Chang, C.W.; Wu, H.Y.; Yao, C.K.; Liu, Y.W. Dynamin-2 Regulates Postsynaptic Cytoskeleton Organization and Neuromuscular Junction Development. Cell Rep. 2020, 33, 108310. [Google Scholar] [CrossRef]
- Paterson, E.K.; Courtneidge, S.A. Invadosomes are coming: New insights into function and disease relevance. FEBS J. 2018, 285, 8–27. [Google Scholar] [CrossRef]
- Cao, F.; Zhou, Y.; Liu, X.; Yu, C.H. Podosome formation promotes plasma membrane invagination and integrin-beta3 endocytosis on a viscous RGD-membrane. Commun. Biol. 2020, 3, 117. [Google Scholar] [CrossRef] [Green Version]
- Ochoa, G.C.; Slepnev, V.I.; Neff, L.; Ringstad, N.; Takei, K.; Daniell, L.; Kim, W.; Cao, H.; McNiven, M.; Baron, R.; et al. A functional link between dynamin and the actin cytoskeleton at podosomes. J. Cell Biol. 2000, 150, 377–390. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Fujise, K.; Wint, H.; Senju, Y.; Suetsugu, S.; Yamada, H.; Takei, K.; Takeda, T. Dynamin 2 and BAR domain protein pacsin 2 cooperatively regulate formation and maturation of podosomes. Biochem. Biophys. Res. Commun. 2021, 571, 145–151. [Google Scholar] [CrossRef]
- Ervasti, J.M. Costameres: The Achilles’ heel of Herculean muscle. J. Biol. Chem. 2003, 278, 13591–13594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peter, A.K.; Cheng, H.; Ross, R.S.; Knowlton, K.U.; Chen, J. The costamere bridges sarcomeres to the sarcolemma in striated muscle. Prog. Pediatr. Cardiol. 2011, 31, 83–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rybakova, I.N.; Patel, J.R.; Ervasti, J.M. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J. Cell Biol. 2000, 150, 1209–1214. [Google Scholar] [CrossRef] [PubMed]
- Franck, A.; Laine, J.; Moulay, G.; Lemerle, E.; Trichet, M.; Gentil, C.; Benkhelifa-Ziyyat, S.; Lacene, E.; Bui, M.T.; Brochier, G.; et al. Clathrin plaques and associated actin anchor intermediate filaments in skeletal muscle. Mol. Biol. Cell 2019, 30, 579–590. [Google Scholar] [CrossRef]
- Vassilopoulos, S.; Gentil, C.; Laine, J.; Buclez, P.O.; Franck, A.; Ferry, A.; Precigout, G.; Roth, R.; Heuser, J.E.; Brodsky, F.M.; et al. Actin scaffolding by clathrin heavy chain is required for skeletal muscle sarcomere organization. J. Cell Biol. 2014, 205, 377–393. [Google Scholar] [CrossRef] [Green Version]
- Massana Munoz, X.; Buono, S.; Koebel, P.; Laporte, J.; Cowling, B.S. Different in vivo impacts of dynamin 2 mutations implicated in Charcot-Marie-Tooth neuropathy or centronuclear myopathy. Hum. Mol. Genet. 2019, 28, 4067–4077. [Google Scholar] [CrossRef]
- Roman, W.; Martins, J.P.; Carvalho, F.A.; Voituriez, R.; Abella, J.V.G.; Santos, N.C.; Cadot, B.; Way, M.; Gomes, E.R. Myofibril contraction and crosslinking drive nuclear movement to the periphery of skeletal muscle. Nat. Cell Biol. 2017, 19, 1189–1201. [Google Scholar] [CrossRef]
- Yarar, D.; To, W.; Abo, A.; Welch, M.D. The Wiskott-Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr. Biol. 1999, 9, 555–558. [Google Scholar] [CrossRef] [Green Version]
- Machesky, L.M.; Mullins, R.D.; Higgs, H.N.; Kaiser, D.A.; Blanchoin, L.; May, R.C.; Hall, M.E.; Pollard, T.D. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA 1999, 96, 3739–3744. [Google Scholar] [CrossRef] [Green Version]
- Egile, C.; Loisel, T.P.; Laurent, V.; Li, R.; Pantaloni, D.; Sansonetti, P.J.; Carlier, M.F. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J. Cell Biol. 1999, 146, 1319–1332. [Google Scholar] [CrossRef] [Green Version]
- Kierdaszuk, B.; Berdynski, M.; Karolczak, J.; Redowicz, M.J.; Zekanowski, C.; Kaminska, A.M. A novel mutation in the DNM2 gene impairs dynamin 2 localization in skeletal muscle of a patient with late onset centronuclear myopathy. Neuromuscul. Disord. 2013, 23, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Fongy, A.; Falcone, S.; Laine, J.; Prudhon, B.; Martins-Bach, A.; Bitoun, M. Nuclear defects in skeletal muscle from a Dynamin 2-linked centronuclear myopathy mouse model. Sci. Rep. 2019, 9, 1580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, C.F.; Bitoun, M.; Vainzof, M. Satellite cells deficiency and defective regeneration in dynamin 2-related centronuclear myopathy. FASEB J. 2021, 35, e21346. [Google Scholar] [CrossRef] [PubMed]
- Buono, S.; Ross, J.A.; Tasfaout, H.; Levy, Y.; Kretz, C.; Tayefeh, L.; Matson, J.; Guo, S.; Kessler, P.; Monia, B.P.; et al. Reducing dynamin 2 (DNM2) rescues DNM2-related dominant centronuclear myopathy. Proc. Natl. Acad. Sci. USA 2018, 115, 11066–11071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trochet, D.; Prudhon, B.; Beuvin, M.; Peccate, C.; Lorain, S.; Julien, L.; Benkhelifa-Ziyyat, S.; Rabai, A.; Mamchaoui, K.; Ferry, A.; et al. Allele-specific silencing therapy for Dynamin 2-related dominant centronuclear myopathy. EMBO Mol. Med. 2018, 10, 239–253. [Google Scholar] [CrossRef]
- Munoz, X.M.; Kretz, C.; Silva-Rojas, R.; Ochala, J.; Menuet, A.; Romero, N.B.; Cowling, B.S.; Laporte, J. Physiological impact and disease reversion for the severe form of centronuclear myopathy linked to dynamin. JCI Insight 2020, 5, e137899. [Google Scholar] [CrossRef]
- Cowling, B.S.; Chevremont, T.; Prokic, I.; Kretz, C.; Ferry, A.; Coirault, C.; Koutsopoulos, O.; Laugel, V.; Romero, N.B.; Laporte, J. Reducing dynamin 2 expression rescues X-linked centronuclear myopathy. J. Clin. Investig. 2014, 124, 1350–1363. [Google Scholar] [CrossRef] [Green Version]
- Tasfaout, H.; Lionello, V.M.; Kretz, C.; Koebel, P.; Messaddeq, N.; Bitz, D.; Laporte, J.; Cowling, B.S. Single Intramuscular Injection of AAV-shRNA Reduces DNM2 and Prevents Myotubular Myopathy in Mice. Mol. Ther. 2018, 26, 1082–1092. [Google Scholar] [CrossRef] [Green Version]
- Silva-Rojas, R.; Nattarayan, V.; Jaque-Fernandez, F.; Gomez-Oca, R.; Menuet, A.; Reiss, D.; Goret, M.; Messaddeq, N.; Lionello, V.M.; Kretz, C.; et al. Mice with muscle-specific deletion of Bin1 recapitulate centronuclear myopathy and acute downregulation of dynamin 2 improves their phenotypes. Mol. Ther. 2022, 30, 868–880. [Google Scholar] [CrossRef]
- Espinosa, K.G.; Geissah, S.; Groom, L.; Volpatti, J.; Scott, I.C.; Dirksen, R.T.; Zhao, M.; Dowling, J.J. Characterization of a novel zebrafish model of SPEG-related centronuclear myopathy. Dis. Model. Mech. 2022, 15, dmm049437. [Google Scholar] [CrossRef]
- Hall, T.E.; Martel, N.; Ariotti, N.; Xiong, Z.; Lo, H.P.; Ferguson, C.; Rae, J.; Lim, Y.W.; Parton, R.G. In vivo cell biological screening identifies an endocytic capture mechanism for T-tubule formation. Nat. Commun. 2020, 11, 3711. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujise, K.; Noguchi, S.; Takeda, T. Centronuclear Myopathy Caused by Defective Membrane Remodelling of Dynamin 2 and BIN1 Variants. Int. J. Mol. Sci. 2022, 23, 6274. https://doi.org/10.3390/ijms23116274
Fujise K, Noguchi S, Takeda T. Centronuclear Myopathy Caused by Defective Membrane Remodelling of Dynamin 2 and BIN1 Variants. International Journal of Molecular Sciences. 2022; 23(11):6274. https://doi.org/10.3390/ijms23116274
Chicago/Turabian StyleFujise, Kenshiro, Satoru Noguchi, and Tetsuya Takeda. 2022. "Centronuclear Myopathy Caused by Defective Membrane Remodelling of Dynamin 2 and BIN1 Variants" International Journal of Molecular Sciences 23, no. 11: 6274. https://doi.org/10.3390/ijms23116274
APA StyleFujise, K., Noguchi, S., & Takeda, T. (2022). Centronuclear Myopathy Caused by Defective Membrane Remodelling of Dynamin 2 and BIN1 Variants. International Journal of Molecular Sciences, 23(11), 6274. https://doi.org/10.3390/ijms23116274





