Targeting PIK3CA Actionable Mutations in the Circulome: A Proof of Concept in Metastatic Breast Cancer
Abstract
:1. Introduction
2. Results
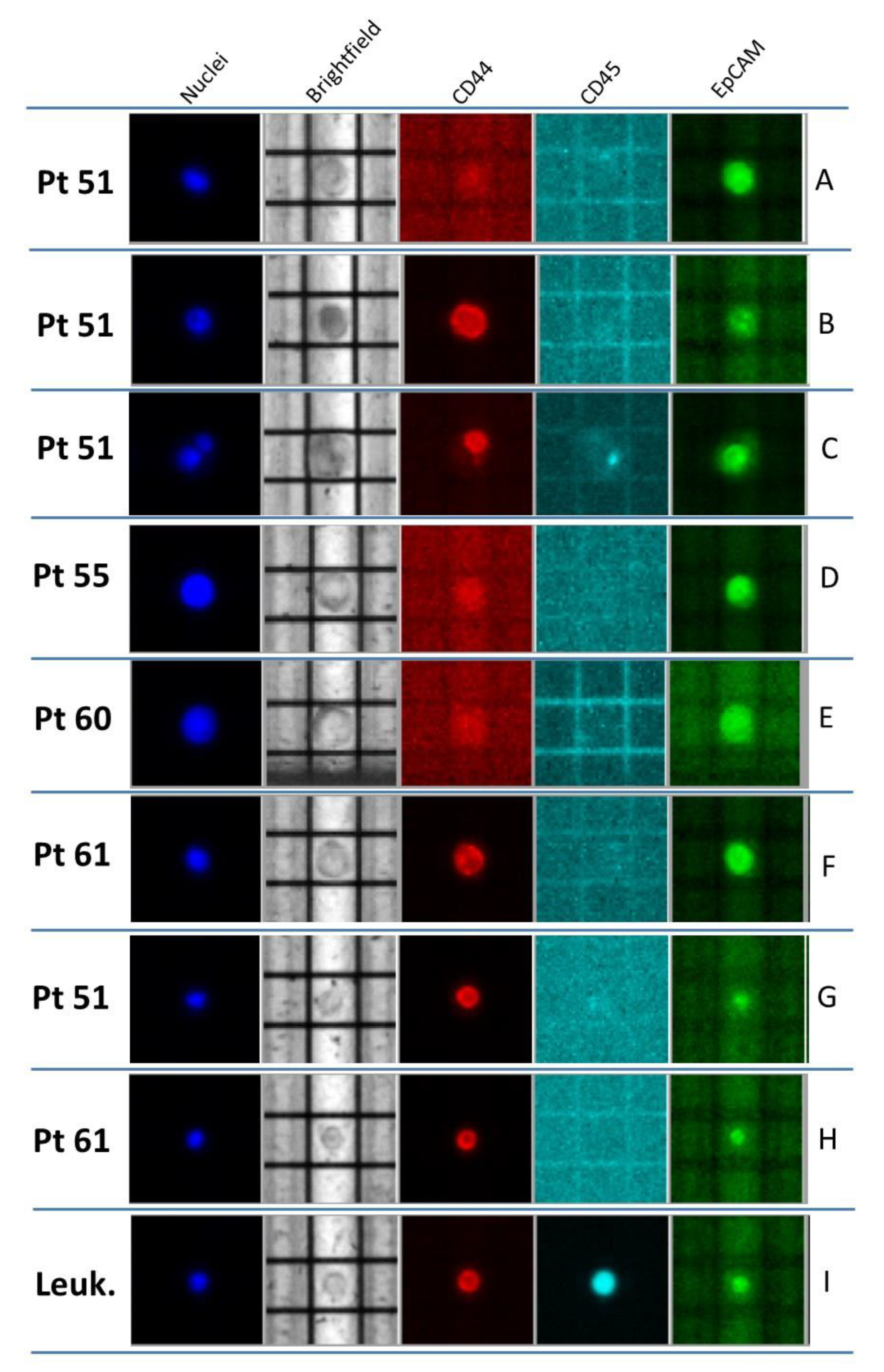
2.1. Case 51
2.2. Case 55
2.3. Case 60
2.4. Case 61
3. Discussion
4. Materials and Methods
4.1. Patient Selection
4.2. Tissue Mutational Analysis: Custom-Designed NGS Panel Testing
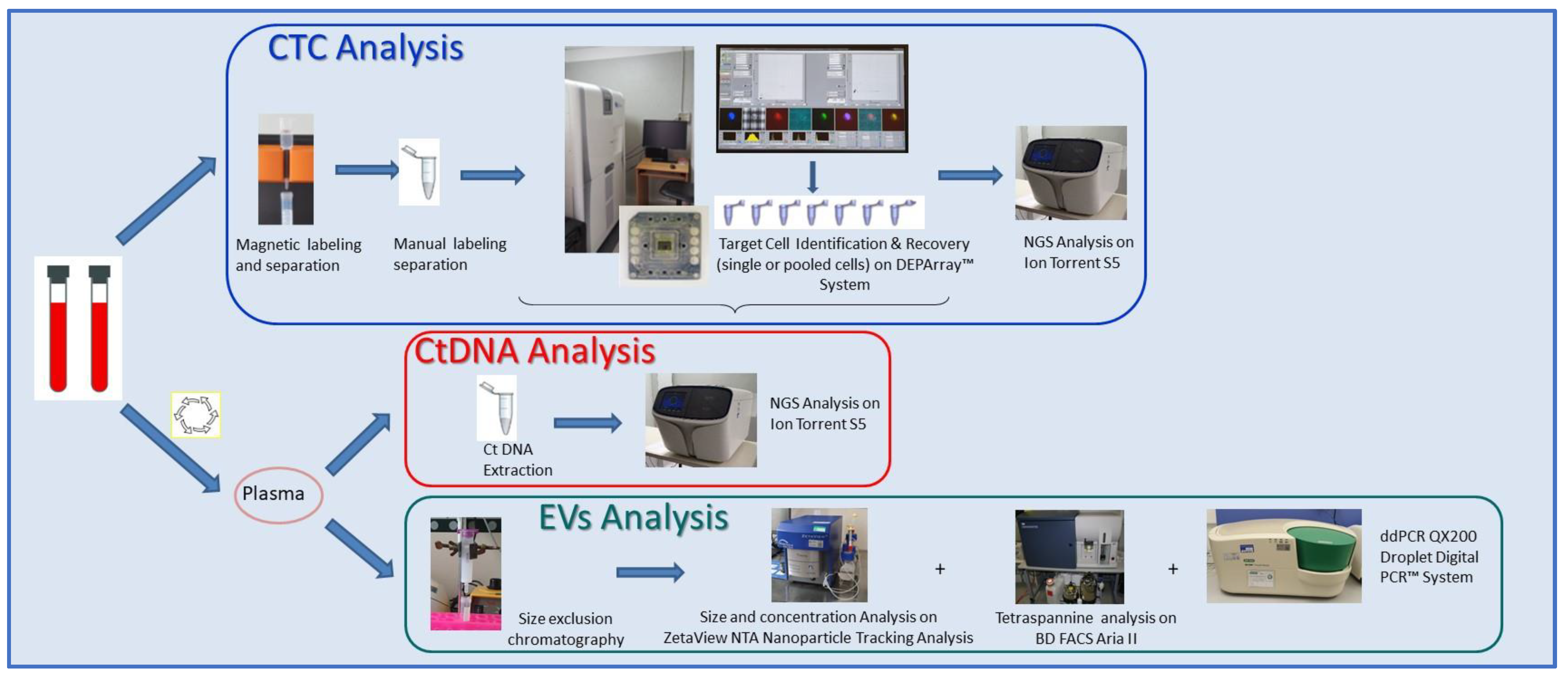
4.3. Circulome Processing Workflow
4.4. Isolation, Extraction, and Quantification of ctDNA
4.5. CTC Selection, Recovery, and Characterization
4.6. EV Isolation and Characterization
4.7. ctDNA Mutational Analysis
4.8. CTC Whole-Genome Amplification-Free Library Preparation and Sequencing
4.9. PIK3CA Status by Droplet Digital PCR (ddPCR)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinzani, P.; D’Argenio, V.; Del Re, M.; Pellegrini, C.; Cucchiara, F.; Salvianti, F.; Galbiati, S. Updates on liquid biopsy: Current trends and future perspectives for clinical application in solid tumors. Clin. Chem. Lab. Med. 2021, 59, 1181–1200. [Google Scholar] [CrossRef]
- Eslami-S, Z.; Cortés-Hernández, L.E.; Cayrefourcq, L.; Alix-Panabières, C. The Different Facets of Liquid Biopsy: A Kaleidoscopic View. Cold Spring Harb. Perspect. Med. 2020, 10, a037333. [Google Scholar] [CrossRef]
- De Rubis, G.; Rajeev Krishnan, S.; Bebawy, M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef] [Green Version]
- Tellez-Gabriel, M.; Knutsen, E.; Perander, M. Current Status of Circulating Tumor Cells, Circulating Tumor DNA, and Exosomes in Breast Cancer Liquid Biopsies. Int. J. Mol. Sci. 2020, 21, 9457. [Google Scholar] [CrossRef]
- Cayrefourcq, L.; Alix-Panabières, C. Clinical relevance of liquid biopsy in breast cancer: Update in 2020. Expert Rev. Mol. Diagn. 2020, 20, 913–919. [Google Scholar] [CrossRef]
- Buono, G.; Gerratana, L.; Bulfoni, M.; Provinciali, N.; Basile, D.; Giuliano, M.; Corvaja, C.; Arpino, G.; Del Mastro, L.; De Placido, S.; et al. Circulating tumor DNA analysis in breast cancer: Is it ready for prime-time? Cancer Treat. Rev. 2019, 73, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Heitzer, E.; van den Broek, D.; Denis, M.G.; Hofman, P.; Hubank, M.; Mouliere, F.; Paz-Ares, L.; Schuuring, E.; Sültmann, H.; Vainer, G.; et al. Recommendations for a practical implementation of circulating tumor DNA mutation testing in metastatic non-small-cell lung cancer. ESMO Open 2022, 7, 100399. [Google Scholar] [CrossRef]
- Alix-Panabierès, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef]
- Bidard, F.C.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- Huang, X.; Gao, P.; Song, Y.; Sun, J.; Chen, X.; Zhao, J.; Xu, H.; Wang, Z. Meta-analysis of the prognostic value of circulating tumor cells detected with the CellSearch System in colorectal cancer. BMC Cancer 2015, 15, 202. [Google Scholar] [CrossRef] [Green Version]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.-M.; Greystoke, A.; Ward, T.H.; Ferraldeschi, R.; Hughes, A.; Clack, G.; et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 2011, 29, 1556–1563. [Google Scholar] [CrossRef]
- Sparano, J.; O’Neill, A.; Alpaugh, K.; Wolff, A.C.; Northfelt, D.W.; Dang, C.T.; Sledge, G.W.; Miller, K.D. Association of Circulating Tumor Cells With Late Recurrence of Estrogen Receptor-Positive Breast Cancer: A Secondary Analysis of a Randomized Clinical Trial. JAMA Oncol. 2018, 4, 1700–1706. [Google Scholar] [CrossRef]
- Habli, Z.; Alchamaa, W.; Saab, R.; Kadara, H.; Khraiche, M.L. Circulating Tumor Cell Detection Technologies and Clinical Utility: Challenges and Opportunities. Cancers 2020, 12, 1930. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [Green Version]
- Reduzzi, C.; Motta, R.; Bertolini, G.; Miodini, P.; Martinetti, A.; Sottotetti, E.; Daidone, M.G.; Cappelletti, V. Development of a Protocol for Single-Cell Analysis of Circulating Tumor Cells in Patients with Solid Tumors. Adv. Exp. Med. Biol. 2017, 994, 83–103. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Doyle, M.L.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef] [Green Version]
- Landman, A. 16th World Conference on Lung Cancer. Lancet Oncol. 2015, 16, 1290. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef] [Green Version]
- LeBleu, V.S.; Kalluri, R. Exosomes as a Multicomponent Biomarker Platform in Cancer. Trends Cancer 2020, 6, 767–774. [Google Scholar] [CrossRef]
- Thery, L.; Meddis, A.; Cabel, L.; Proudhon, C.; Latouche, A.; Pierga, J.-Y.; Bidard, F.-C. Circulating Tumor Cells in Early Breast Cancer. JNCI Cancer Spectr. 2019, 3, pkz026. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef]
- De Luca, F.; Rotunno, G.; Salvianti, F.; Galardi, F.; Pestrin, M.; Gabellini, S.; Simi, L.; Mancini, I.; Vannucchi, A.M.; Pazzagli, M.; et al. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget 2016, 7, 26107–26119. [Google Scholar] [CrossRef]
- Ma, F.; Zhu, W.; Guan, Y.; Yang, L.; Xia, X.; Chen, S.; Li, Q.; Guan, X.; Yi, Z.; Qian, H.; et al. ctDNA dynamics: A novel indicator to track resistance in metastatic breast cancer treated with anti-HER2 therapy. Oncotarget 2016, 7, 66020–66031. [Google Scholar] [CrossRef]
- Fribbens, C.; Garcia Murillas, I.; Beaney, M.; Hrebien, S.; O’Leary, B.; Kilburn, L.; Howarth, K.; Epstein, M.; Green, E.; Rosenfeld, N.; et al. Tracking evolution of aromatase inhibitor resistance with circulating tumour DNA analysis in metastatic breast cancer. Ann. Oncol. 2018, 29, 145–153. [Google Scholar] [CrossRef]
- Lucci, A.; Hall, C.S.; Lodhi, A.K.; Bhattacharyya, A.; Anderson, A.E.; Xiao, L.; Bedrosian, I.; Kuerer, H.M.; Krishnamurthy, S. Circulating tumour cells in non-metastatic breast cancer: A prospective study. Lancet Oncol. 2012, 13, 688–695. [Google Scholar] [CrossRef]
- Rack, B.; Schindlbeck, C.; Jückstock, J.; Andergassen, U.; Hepp, P.; Zwingers, T.; Friedl, T.W.P.; Lorenz, R.; Tesch, H.; Fasching, P.A.; et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J. Natl. Cancer Inst. 2014, 106, dju066. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [Green Version]
- Bidard, F.-C.; Jacot, W.; Dureau, S.; Brain, E.; Bachelot, T.; Bourgeois, H.; Goncalves, A.; Ladoire, S.; Naman, H.; Dalenc, F.; et al. Clinical utility of circulating tumor cell count as a tool to chose between first line hormone therapy and chemotherapy for ER+ HER2- metastatic breast cancer: Results of the phase III STIC CTC trial. Cancer Res. 2019, 79 (Suppl. S4), GS3-07. [Google Scholar]
- Cristofanilli, M.; Pierga, J.Y.; Reuben, J.; Rademaker, A.; Davis, A.A.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): International expert consensus paper. Crit. Rev. Oncol. Hematol. 2019, 134, 39–45. [Google Scholar] [CrossRef]
- Budd, G.T.; Cristofanilli, M.; Ellis, M.J.; Stopeck, A.; Borden, E.; Miller, M.C.; Matera, J.; Repollet, M.; Doyle, G.V.; Terstappen, L.W.M.M.; et al. Circulating tumor cells versus imaging--predicting overall survival in metastatic breast cancer. Clin. Cancer Res. 2006, 12, 6403–6409. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.C.; Shields, P.G.; Warren, R.D.; Cohen, P.; Wilkinson, M.; Ottaviano, Y.L.; Rao, S.B.; Eng-Wong, J.; Seillier-Moiseiwitsch, F.; Noone, A.M.; et al. Circulating tumor cells: A useful predictor of treatment efficacy in metastatic breast cancer. J. Clin. Oncol. 2009, 27, 5153–5159. [Google Scholar] [CrossRef] [Green Version]
- Stevic, I.; Müller, V.; Weber, K.; Fasching, P.A.; Karn, T.; Marmé, F.; Schem, C.; Stickeler, E.; Denkert, C.; van Mackelenbergh, M.; et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 2018, 16, 179. [Google Scholar] [CrossRef] [Green Version]
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun. Biol. 2019, 2, 325. [Google Scholar] [CrossRef] [Green Version]
- Del Re, M.; Crucitta, S.; Lorenzini, G.; De Angelis, C.; Diodati, L.; Cavallero, D.; Bargagna, I.; Cinacchi, P.; Fratini, B.; Salvadori, B.; et al. PI3K mutations detected in liquid biopsy are associated to reduced sensitivity to CDK4/6 inhibitors in metastatic breast cancer patients. Pharmacol. Res. 2021, 163, 105241. [Google Scholar] [CrossRef]
- Fritsch, C.; Huang, A.; Chatenay-Rivauday, C.; Schnell, C.; Reddy, A.; Liu, M.; Kauffmann, A.; Guthy, D.; Erdmann, D.; De Pover, A.; et al. Characterization of the novel and specific PI3Kα inhibitor NVP-BYL719 and development of the patient stratification strategy for clinical trials. Mol. Cancer Ther. 2014, 13, 1117–1129. [Google Scholar] [CrossRef] [Green Version]
- Juric, D.; Janku, F.; Rodón, J.; Burris, H.A.; Mayer, I.A.; Schuler, M.; Seggewiss-Bernhardt, R.; Gil-Martin, M.; Middleton, M.R.; Baselga, J. Alpelisib Plus Fulvestrant in PIK3CA-Altered and PIK3CA-Wild-Type Estrogen Receptor-Positive Advanced Breast Cancer: A Phase 1b Clinical Trial. JAMA Oncol. 2019, 5, e184475. [Google Scholar] [CrossRef] [Green Version]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef]
- Piñeiro, R.; Martínez-Pena, I.; López-López, R. Relevance of CTC Clusters in Breast Cancer Metastasis. Adv. Exp. Med. Biol. 2020, 1220, 93–115. [Google Scholar] [CrossRef] [Green Version]
- Shaw, J.A.; Guttery, D.S.; Hills, A.; Fernandez-Garcia, D.; Page, K.; Rosales, B.M.; Goddard, K.S.; Hastings, R.K.; Luo, J.; Ogle, O.; et al. Mutation Analysis of Cell-Free DNA and Single Circulating Tumor Cells in Metastatic Breast Cancer Patients with High Circulating Tumor Cell Counts. Clin. Cancer Res. 2017, 23, 88–96. [Google Scholar] [CrossRef] [Green Version]
- Nakai, M.; Yamada, T.; Sekiya, K.; Sato, A.; Hankyo, M.; Kuriyama, S.; Takahashi, G.; Kurita, T.; Yanagihara, K.; Yoshida, H.; et al. Use of Liquid Biopsy to Detect PIK3CA Mutation in Metastatic Breast Cancer. J. Nippon Med. Sch. 2022, 89, 66–71. [Google Scholar] [CrossRef]
- Bredemeier, M.; Kasimir-Bauer, S.; Kolberg, H.-C.; Herold, T.; Synoracki, S.; Hauch, S.; Edimiris, P.; Bankfalvi, A.; Tewes, M.; Kimmig, R.; et al. Comparison of the PI3KCA pathway in circulating tumor cells and corresponding tumor tissue of patients with metastatic breast cancer. Mol. Med. Rep. 2017, 15, 2957–2968. [Google Scholar] [CrossRef] [Green Version]
- Higgins, M.J.; Jelovac, D.; Barnathan, E.; Blair, B.; Slater, S.; Powers, P.; Zorzi, J.; Jeter, S.C.; Oliver, G.R.; Fetting, J.; et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin. Cancer Res. 2012, 18, 3462–3469. [Google Scholar] [CrossRef] [Green Version]
- Reduzzi, C.; Vismara, M.; Silvestri, M.; Celio, L.; Niger, M.; Peverelli, G.; De Braud, F.; Daidone, M.G.; Cappelletti, V. A novel circulating tumor cell subpopulation for treatment monitoring and molecular characterization in biliary tract cancer. Int. J. Cancer 2020, 146, 3495–3503. [Google Scholar] [CrossRef]
- De Luca, G.; Cardinali, B.; Del Mastro, L.; Lastraioli, S.; Carli, F.; Ferrarini, M.; Calin, G.A.; Garuti, A.; Mazzitelli, C.; Zupo, S.; et al. Optimization of a WGA-Free Molecular Tagging-Based NGS Protocol for CTCs Mutational Profiling. Int. J. Mol. Sci. 2020, 21, 4364. [Google Scholar] [CrossRef]
- Bulfoni, M.; Gerratana, L.; Del Ben, F.; Marzinotto, S.; Sorrentino, M.; Turetta, M.; Scoles, G.; Toffoletto, B.; Isola, M.; Beltrami, C.A.; et al. In patients with metastatic breast cancer the identification of circulating tumor cells in epithelial-to-mesenchymal transition is associated with a poor prognosis. Breast Cancer Res. 2016, 18, 30. [Google Scholar] [CrossRef] [Green Version]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular vesicle heterogeneity: Subpopulations, isolation techniques, and diverse functions in cancer progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef] [Green Version]
- Keup, C.; Suryaprakash, V.; Storbeck, M.; Hoffmann, O.; Kimming, R.; Kasimir-Bauer, S. Longitudinal Multi-Parametric Liquid Biopsy Approach Identifies Unique Features of Circulating Tumor Cell, Extracellular Vesicle, and Cell-Free DNA Characterization for Disease Monitoring in Metastatic Breast Cancer Patients. Cells 2021, 10, 212. [Google Scholar] [CrossRef]
- Kalluri, R. The Biology and Function of Exosomes in Cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Gorgun, C.; Reverberi, D.; Rotta, G.; Villa, F.; Quarto, R.; Tasso, R. Isolation and Flow Cytometry Characterization of Extracellular- Vesicle Subpopulations Derived from Human Mesenchymal Stromal Cells. Curr. Protoc. Stem Cell Biol. 2019, 48, e76. [Google Scholar] [CrossRef] [Green Version]
- Dono, M.; De Luca, G.; Lastraioli, S.; Anselmi, G.; Dal Bello, M.G.; Coco, S.; Vanni, I.; Grossi, F.; Vigani, A.; Genova, C.; et al. Tag-based next generation sequencing: A feasible and reliable assay for EGFR T790M mutation detection in circulating tumor DNA of non small cell lung cancer patients. Mol. Med. 2019, 25, 15. [Google Scholar] [CrossRef]


| Pt | Age at First Diagnosis | Biopsy and staging | NAC | Surgery and Restaging | AC, RT, HT | DFS | Site of 1st Recurrence | 1st-Line Therapy | PFS I | 2nd-Line Therapy | PFS II |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PT#51 | 2013 40 years | / | No | Ductal BC pT2, G2 pN3a ER: 40% PgR: 40% Ki67:8% | EC (4 cycles) + T (12 cycles) + RT + LHRHa and Tam | 4 ys | Bones | CDK4/6 Inhibitors + AI | 22 ms | Alpelisib + fulvestrant | NA |
| Pt#55 | 2004 43 years | Ductal BC cT4b cN1 ER: 75% PgR: 5% Ki67: 24% | E + C + D (6 cycles) | Ductal BC ypT1c, G2 pN1a ER: 15% PgR: 0% Ki67: 25% | C + M + F (3 cycles) + RT + Tam for 2 ys, then AI for 3 ys | 8 ys | Bones | CDK4/6 Inhibitors + AI | 12 ms | Alpelisib + fulvestrant | 7 ms |
| Pt#60 | 2018 65 years | Ductal BC M1 ER: 90% PgR: 70%Ki67: 20% | No | / | / | / | Bones | CDK4/6 Inhibitors + AI | 21 ms | Alpelisib + fulvestrant | 7 ms |
| Pt#61 | 2015 41 years | Ductal BC ER: 96% PgR: 86% Ki67: 18% | EC (4 cycles) + T (12 cycles) | Ductal BC ypT1c, G2 pN1a ER: 93% PgR: 16% KI67: 6% | RT + LHRHa and AI | 4 ys | Bones, nodes, liver | CDK4/6 Inhibitors + fulvestrant | 5 ms | Alpelisib + fulvestrant | 3 ms |
| Pt ID | Tissue | CTCs | ctDNA | EVs | ||
|---|---|---|---|---|---|---|
| Mutational Status by NGS | Conventional CTCs | Non-Conventional CTCs | Mutational Status by NGS | PIK3CA Status by ddPCR | PIK3CA Status by ddPCR | |
| #51 | PIK3CA, Hys1047Arg | PIK3CA, Hys1047Arg | not tested | PIK3CA, Hys1047Arg (2.3%) | PIK3CA, Hys1047Arg (2.2%) | PIK3CA, Hys1047Arg (1.4%) |
| #55 | PIK3CA, Glu542Lys | WT | PIK3CA, Glu545Lys | PIK3CA, Glu542Lys (7.5%) Glu545Lys (4.0%) | PIK3CA, Glu542Lys (8.1 %) Glu545Lys (4.0%) | PIK3CA, Glu545Lys * |
| #60 | PIK3CA, Hys1047Leu | WT | WT | PIK3CA, Hys1047Leu (0.7%); ESR1, Asp538Gly (0.39%) | PIK3CA, Hys1047Leu (0.7%) | WT |
| #61 | PIK3CA, Glu542Lys; TP53, Gly245Ser | PIK3CA, Glu542Lys; TP53, Gly245Ser | PIK3CA, Glu542Lys; TP53, Gly245Ser | PIK3CA, Glu542Lys (51.6%); TP53, Gly245Ser (33.5%) | PIK3CA, Glu542Lys (48.8%) | PIK3CA Glu542Lys (54.6%) |
| Pt ID | Conventional CTCs | EVs | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Number | EpCAM− | EpCAM+ | CD44 | Number | Size | CD9 | CD81 | CD63 | |
| #51 | 41 | 21 | 17 | 3 | 5.90 × 1010 | 157.8 | Neg | 82.4% | 13.2% |
| #55 | 18 | 13 | 5 | 0 | 9.90 × 1011 | 159.6 | Neg | 82.2% | 17.0% |
| #60 | 19 | 3 | 16 | 0 | 5.70 × 1010 | 152.1 | Neg | 34.3% | 20.4% |
| #61 | 8 | 5 | 2 | 1 | 2.00 × 1010 | 173.7 | Neg | 89.5% | 47.90% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardinali, B.; De Luca, G.; Tasso, R.; Coco, S.; Garuti, A.; Buzzatti, G.; Sciutto, A.; Arecco, L.; Villa, F.; Carli, F.; et al. Targeting PIK3CA Actionable Mutations in the Circulome: A Proof of Concept in Metastatic Breast Cancer. Int. J. Mol. Sci. 2022, 23, 6320. https://doi.org/10.3390/ijms23116320
Cardinali B, De Luca G, Tasso R, Coco S, Garuti A, Buzzatti G, Sciutto A, Arecco L, Villa F, Carli F, et al. Targeting PIK3CA Actionable Mutations in the Circulome: A Proof of Concept in Metastatic Breast Cancer. International Journal of Molecular Sciences. 2022; 23(11):6320. https://doi.org/10.3390/ijms23116320
Chicago/Turabian StyleCardinali, Barbara, Giuseppa De Luca, Roberta Tasso, Simona Coco, Anna Garuti, Giulia Buzzatti, Andrea Sciutto, Luca Arecco, Federico Villa, Franca Carli, and et al. 2022. "Targeting PIK3CA Actionable Mutations in the Circulome: A Proof of Concept in Metastatic Breast Cancer" International Journal of Molecular Sciences 23, no. 11: 6320. https://doi.org/10.3390/ijms23116320
APA StyleCardinali, B., De Luca, G., Tasso, R., Coco, S., Garuti, A., Buzzatti, G., Sciutto, A., Arecco, L., Villa, F., Carli, F., Reverberi, D., Quarto, R., Dono, M., & Del Mastro, L. (2022). Targeting PIK3CA Actionable Mutations in the Circulome: A Proof of Concept in Metastatic Breast Cancer. International Journal of Molecular Sciences, 23(11), 6320. https://doi.org/10.3390/ijms23116320








