Repurposing Histaminergic Drugs in Multiple Sclerosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Retrival
2.2. SAveRUNNER Algorithm
2.3. Gene Set Enrichment Analysis
2.4. Human Brain Tissue
2.5. Immunohistochemistry
2.6. Immunofluorescence
2.7. Confocal Microscopy
2.8. Protein Extraction and Western Blotting
2.9. Statistical Analysis
3. Results
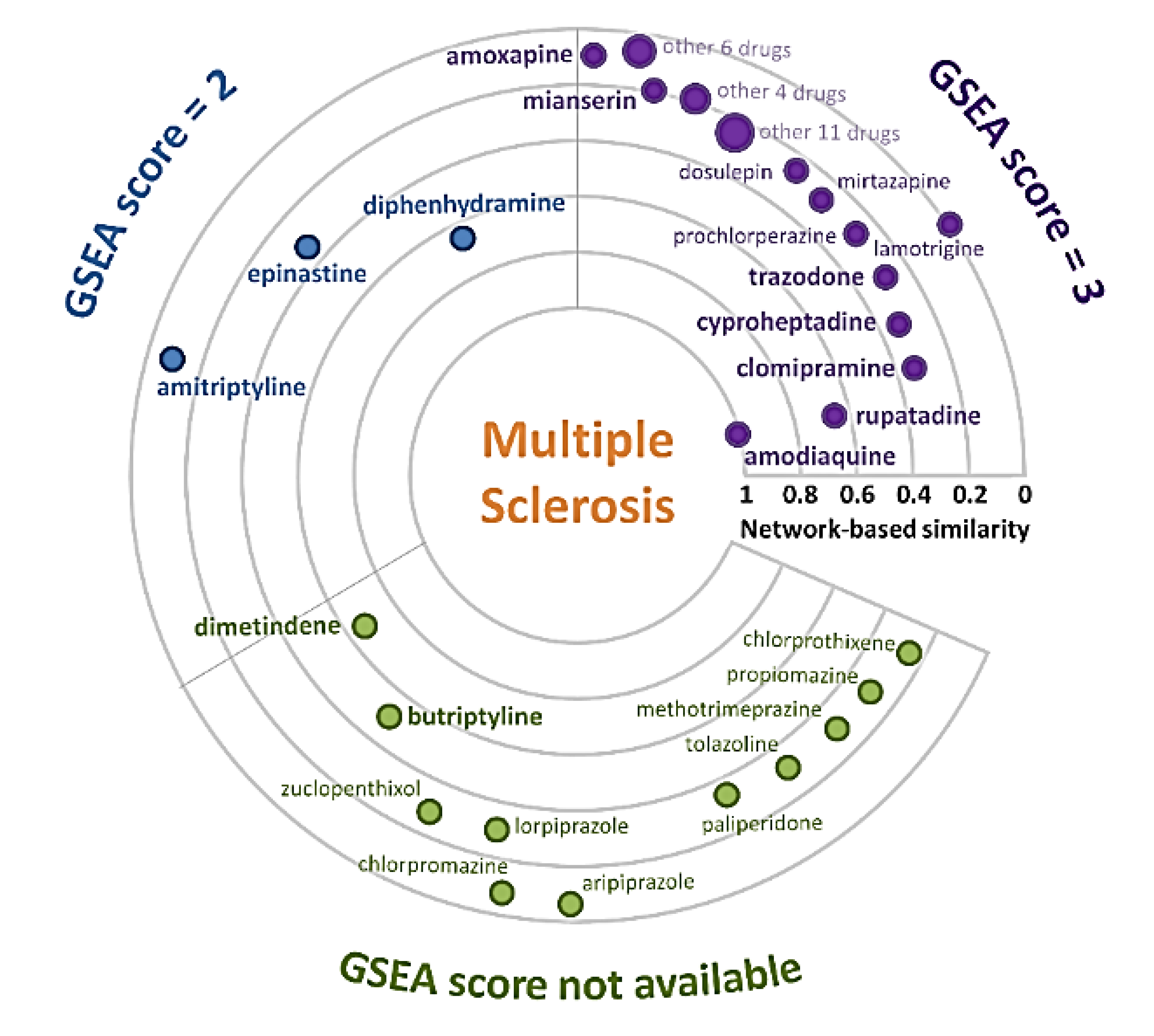
3.1. MS-Drug Network
3.2. Gene Set Enrichment Analysis of Anti-MS Repurposable Drugs
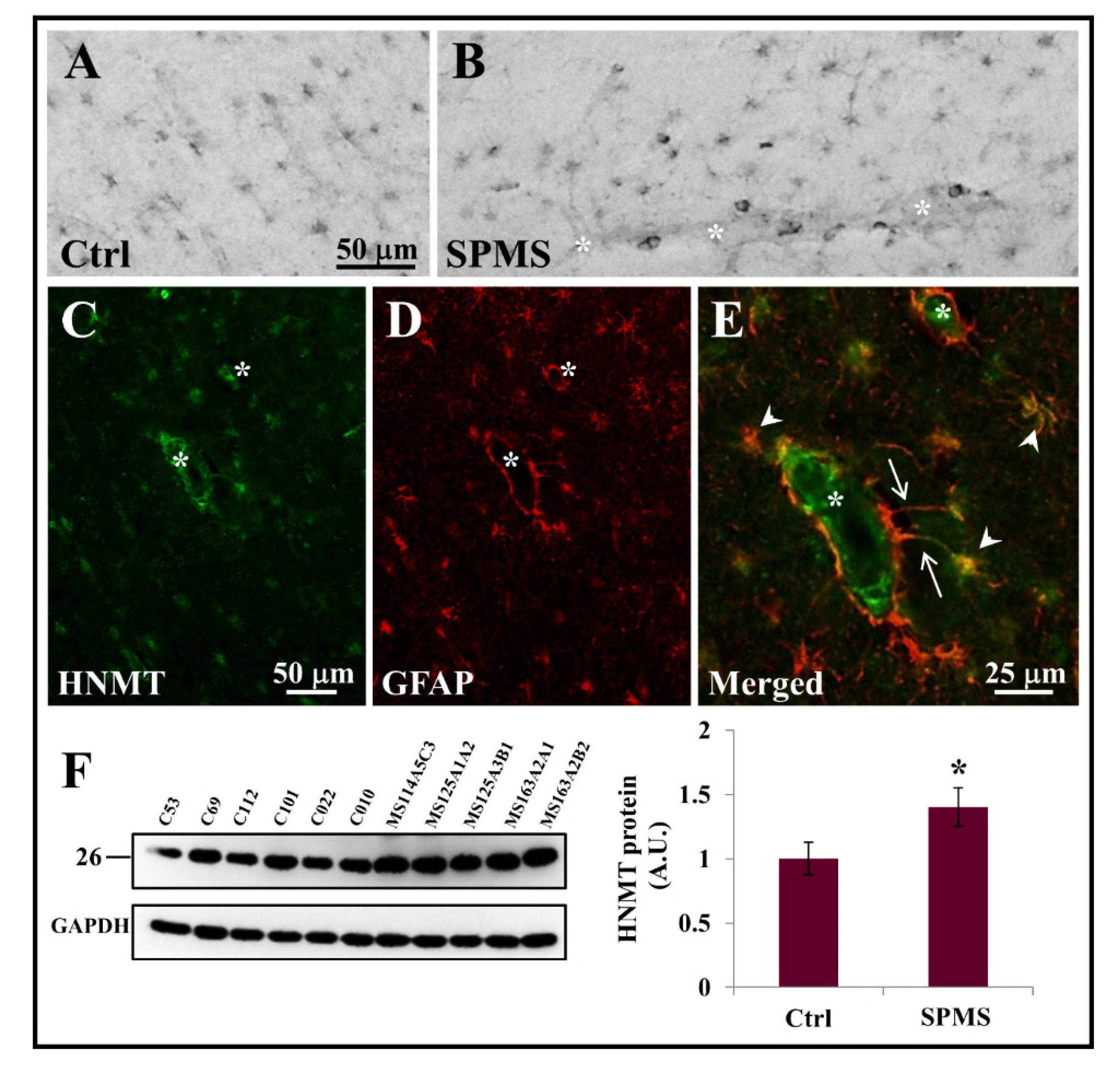
3.3. HNMT Is Present on Astrocytes in the Parenchyma of Secondary Progressive MS Frontal Cortex
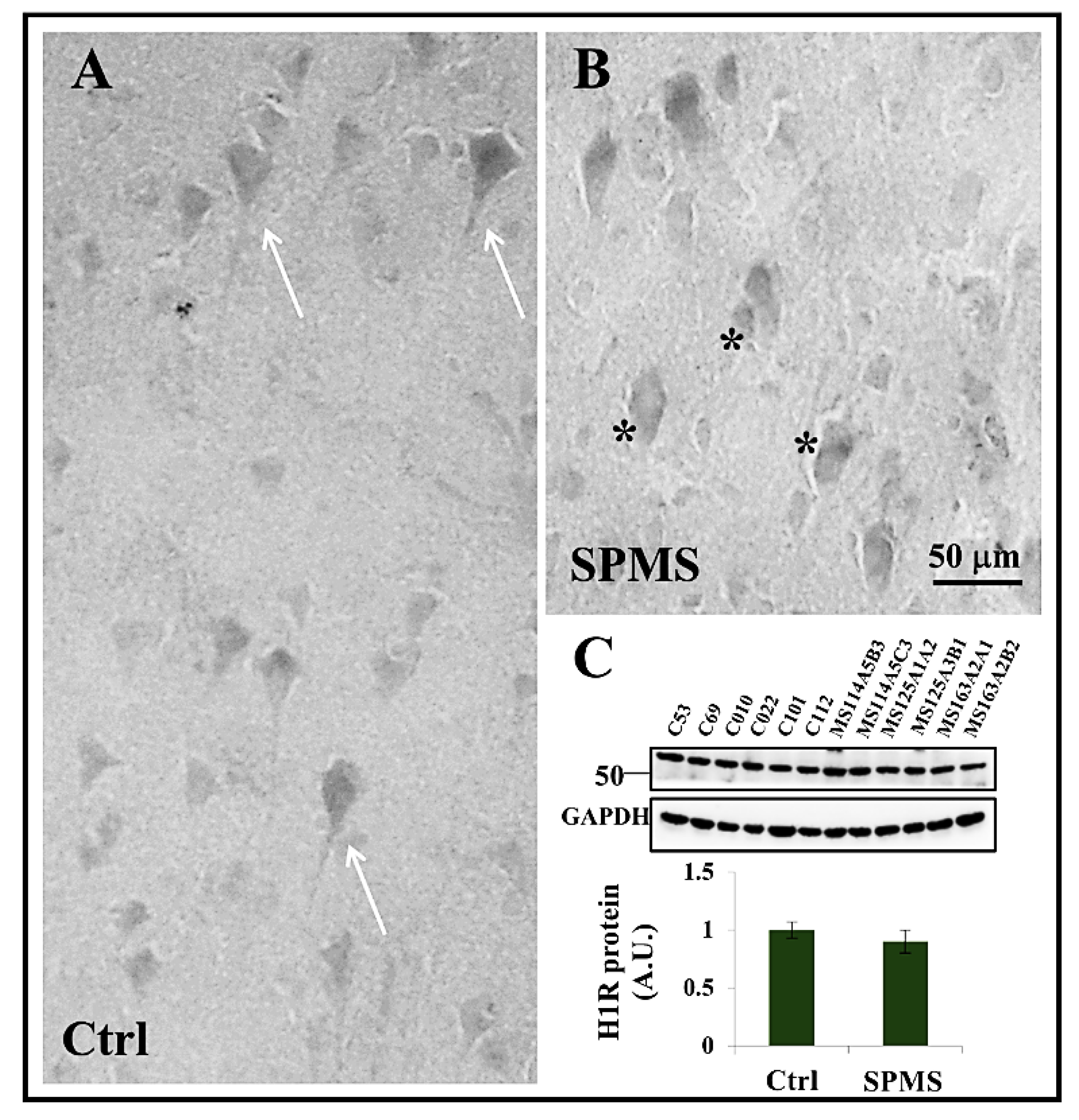
3.4. Expression of Histamine H1 Receptor in Frontal Cortex
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Radandish, M.; Khalilian, P.; Esmaeil, N. The Role of Distinct Subsets of Macrophages in the Pathogenesis of MS and the Impact of Different Therapeutic Agents on These Populations. Front. Immunol. 2021, 12, 3307. [Google Scholar] [CrossRef] [PubMed]
- Pardini, M.; Brown, J.W.L.; Magliozzi, R.; Reynolds, R.; Chard, D.T. Surface-in Pathology in Multiple Sclerosis: A New View on Pathogenesis? Brain J. Neurol. 2021, 144, 1646–1654. [Google Scholar] [CrossRef]
- Absinta, M.; Lassmann, H.; Trapp, B. Mechanisms Underlying Progression in Multiple Sclerosis. Curr. Opin. Neurol. 2020, 33, 277–285. [Google Scholar] [CrossRef]
- Brown, J.; Quattrochi, B.; Everett, C.; Hong, B.-Y.; Cervantes, J. Gut Commensals, Dysbiosis, and Immune Response Imbalance in the Pathogenesis of Multiple Sclerosis. Mult. Scler. Houndmills Basingstoke Engl. 2021, 27, 807–811. [Google Scholar] [CrossRef]
- Cree, B.A.C.; Arnold, D.L.; Chataway, J.; Chitnis, T.; Fox, R.J.; Ramajo, A.P.; Murphy, N.; Lassmann, H. Secondary Progressive Multiple Sclerosis: New Insights. Neurology 2021, 97, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Muresanu, D.F.; Patnaik, R.; Menon, P.K.; Tian, Z.R.; Sahib, S.; Castellani, R.J.; Nozari, A.; Lafuente, J.V.; Buzoianu, A.D.; et al. Histamine H3 and H4 Receptors Modulate Parkinson’s Disease Induced Brain Pathology. Neuroprotective Effects of Nanowired BF-2649 and Clobenpropit with Anti-Histamine-Antibody Therapy. Prog. Brain Res. 2021, 266, 1–73. [Google Scholar] [CrossRef]
- Hu, W.; Chen, Z. The Roles of Histamine and Its Receptor Ligands in Central Nervous System Disorders: An Update. Pharmacol. Ther. 2017, 175, 116–132. [Google Scholar] [CrossRef] [PubMed]
- Volonté, C.; Apolloni, S.; Sabatelli, M. Histamine beyond Its Effects on Allergy: Potential Therapeutic Benefits for the Treatment of Amyotrophic Lateral Sclerosis (ALS). Pharmacol. Ther. 2019, 202, 120–131. [Google Scholar] [CrossRef]
- Ghamari, N.; Zarei, O.; Arias-Montaño, J.-A.; Reiner, D.; Dastmalchi, S.; Stark, H.; Hamzeh-Mivehroud, M. Histamine H3 Receptor Antagonists/Inverse Agonists: Where Do They Go? Pharmacol. Ther. 2019, 200, 69–84. [Google Scholar] [CrossRef]
- Provensi, G.; Costa, A.; Izquierdo, I.; Blandina, P.; Passani, M.B. Brain Histamine Modulates Recognition Memory: Possible Implications in Major Cognitive Disorders. Br. J. Pharmacol. 2020, 177, 539–556. [Google Scholar] [CrossRef]
- Provensi, G.; Passani, M.B.; Costa, A.; Izquierdo, I.; Blandina, P. Neuronal Histamine and the Memory of Emotionally Salient Events. Br. J. Pharmacol. 2020, 177, 557–569. [Google Scholar] [CrossRef]
- Carthy, E.; Ellender, T. Histamine, Neuroinflammation and Neurodevelopment: A Review. Front. Neurosci. 2021, 15, 680214. [Google Scholar] [CrossRef] [PubMed]
- Jadidi-Niaragh, F.; Mirshafiey, A. Histamine and Histamine Receptors in Pathogenesis and Treatment of Multiple Sclerosis. Neuropharmacology 2010, 59, 180–189. [Google Scholar] [CrossRef]
- Passani, M.B.; Ballerini, C. Histamine and Neuroinflammation: Insights from Murine Experimental Autoimmune Encephalomyelitis. Front. Syst. Neurosci. 2012, 6, 32. [Google Scholar] [CrossRef] [Green Version]
- Panula, P.; Nuutinen, S. The Histaminergic Network in the Brain: Basic Organization and Role in Disease. Nat. Rev. Neurosci. 2013, 14, 472–487. [Google Scholar] [CrossRef]
- Volonté, C.; Apolloni, S.; Amadio, S. The Histamine and Multiple Sclerosis Alliance: Pleiotropic Actions and Functional Validation. In Current Topics in Behavioral Neurosciences; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar] [CrossRef]
- Chen, Y.; Zhen, W.; Guo, T.; Zhao, Y.; Liu, A.; Rubio, J.P.; Krull, D.; Richardson, J.C.; Lu, H.; Wang, R. Histamine Receptor 3 Negatively Regulates Oligodendrocyte Differentiation and Remyelination. PLoS ONE 2017, 12, e0189380. [Google Scholar] [CrossRef] [Green Version]
- Passani, M.B.; Blandina, P. Histamine Receptors in the CNS as Targets for Therapeutic Intervention. Trends Pharmacol. Sci. 2011, 32, 242–249. [Google Scholar] [CrossRef] [Green Version]
- Zadeh, A.R.; Falahatian, M.; Alsahebfosoul, F. Serum Levels of Histamine and Diamine Oxidase in Multiple Sclerosis. Am. J. Clin. Exp. Immunol. 2018, 7, 100–105. [Google Scholar]
- Loy, B.D.; Fling, B.W.; Sage, K.M.; Spain, R.I.; Horak, F.B. Serum Histidine Is Lower in Fatigued Women with Multiple Sclerosis. Fatigue Biomed. Health Behav. 2019, 7, 69–80. [Google Scholar] [CrossRef]
- Cacabelos, R.; Torrellas, C.; Fernández-Novoa, L.; Aliev, G. Neuroimmune Crosstalk in CNS Disorders: The Histamine Connection. Curr. Pharm. Des. 2016, 22, 819–848. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R.; Torrellas, C.; Fernández-Novoa, L.; López-Muñoz, F. Histamine and Immune Biomarkers in CNS Disorders. Mediat. Inflamm. 2016, 2016, e1924603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.W.; Leonard, B.E.; Helmeste, D.M. Long COVID, Neuropsychiatric Disorders, Psychotropics, Present and Future. Acta Neuropsychiatr. 2022, 34, 109–126. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Gulbahce, N.; Loscalzo, J. Network Medicine: A Network-Based Approach to Human Disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldera, M.; Buphamalai, P.; Müller, F.; Menche, J. Interactome-Based Approaches to Human Disease. Curr. Opin. Syst. Biol. 2017, 3, 88–94. [Google Scholar] [CrossRef]
- Paci, P.; Fiscon, G.; Conte, F.; Wang, R.-S.; Farina, L.; Loscalzo, J. Gene Co-Expression in the Interactome: Moving from Correlation toward Causation via an Integrated Approach to Disease Module Discovery. NPJ Syst. Biol. Appl. 2021, 7, 3. [Google Scholar] [CrossRef]
- Cheng, F.; Desai, R.J.; Handy, D.E.; Wang, R.; Schneeweiss, S.; Barabási, A.-L.; Loscalzo, J. Network-Based Approach to Prediction and Population-Based Validation of in Silico Drug Repurposing. Nat. Commun. 2018, 9, 2691. [Google Scholar] [CrossRef] [Green Version]
- Paci, P.; Fiscon, G.; Conte, F.; Wang, R.-S.; Handy, D.E.; Farina, L.; Loscalzo, J. Comprehensive Network Medicine-Based Drug Repositioning via Integration of Therapeutic Efficacy and Side Effects. NPJ Syst. Biol. Appl. 2022, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Gysi, D.M.; Valle, Í.; Zitnik, M.; Ameli, A.; Gan, X.; Varol, O.; Ghiassian, S.D.; Patten, J.J.; Davey, R.A.; Loscalzo, J.; et al. Network Medicine Framework for Identifying Drug-Repurposing Opportunities for COVID-19. Proc. Natl. Acad. Sci. USA 2021, 118, e2025581118. [Google Scholar] [CrossRef] [PubMed]
- Fiscon, G.; Paci, P. SAveRUNNER: An R-Based Tool for Drug Repurposing. BMC Bioinform. 2021, 22, 150. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Farina, L.; Paci, P. SAveRUNNER: A Network-Based Algorithm for Drug Repurposing and Its Application to COVID-19. PLoS Comput. Biol. 2021, 17, e1008686. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Amadio, S.; Volonté, C.; Paci, P. Drug Repurposing: A Network-Based Approach to Amyotrophic Lateral Sclerosis. Neurotherapeutics 2021, 18, 1678–1691. [Google Scholar] [CrossRef] [PubMed]
- Chatr-Aryamontri, A.; Breitkreutz, B.-J.; Oughtred, R.; Boucher, L.; Heinicke, S.; Chen, D.; Stark, C.; Breitkreutz, A.; Kolas, N.; O’Donnell, L.; et al. The BioGRID Interaction Database: 2015 Update. Nucleic Acids Res. 2015, 43, D470–D478. [Google Scholar] [CrossRef] [PubMed]
- Peri, S.; Navarro, J.D.; Kristiansen, T.Z.; Amanchy, R.; Surendranath, V.; Muthusamy, B.; Gandhi, T.K.B.; Chandrika, K.N.; Deshpande, N.; Suresh, S.; et al. Human Protein Reference Database as a Discovery Resource for Proteomics. Nucleic Acids Res. 2004, 32, D497–D501. [Google Scholar] [CrossRef]
- Licata, L.; Briganti, L.; Peluso, D.; Perfetto, L.; Iannuccelli, M.; Galeota, E.; Sacco, F.; Palma, A.; Nardozza, A.P.; Santonico, E.; et al. MINT, the Molecular Interaction Database: 2012 Update. Nucleic Acids Res. 2012, 40, D857–D861. [Google Scholar] [CrossRef]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The MIntAct Project--IntAct as a Common Curation Platform for 11 Molecular Interaction Databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breuer, K.; Foroushani, A.K.; Laird, M.R.; Chen, C.; Sribnaia, A.; Lo, R.; Winsor, G.L.; Hancock, R.E.W.; Brinkman, F.S.L.; Lynn, D.J. InnateDB: Systems Biology of Innate Immunity and beyond-Recent Updates and Continuing Curation. Nucleic Acids Res. 2013, 41, D1228–D1233. [Google Scholar] [CrossRef]
- Yu, W.; Clyne, M.; Khoury, M.J.; Gwinn, M. Phenopedia and Genopedia: Disease-Centered and Gene-Centered Views of the Evolving Knowledge of Human Genetic Associations. Bioinformatics 2010, 26, 145–146. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Li, F.; Zhou, Y.; Zhang, Y.; Wang, Z.; Zhang, R.; Zhu, J.; Ren, Y.; Tan, Y.; et al. Therapeutic Target Database 2020: Enriched Resource for Facilitating Research and Early Development of Targeted Therapeutics. Nucleic Acids Res. 2020, 48, D1031–D1041. [Google Scholar] [CrossRef] [Green Version]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-Based Drug Repurposing for Novel Coronavirus 2019-NCoV/SARS-CoV-2. Cell Discov. 2020, 6, e2025581118. [Google Scholar] [CrossRef] [Green Version]
- Amadio, S.; Parisi, C.; Piras, E.; Fabbrizio, P.; Apolloni, S.; Montilli, C.; Luchetti, S.; Ruggieri, S.; Gasperini, C.; Laghi-Pasini, F.; et al. Modulation of P2X7 Receptor during Inflammation in Multiple Sclerosis. Front. Immunol. 2017, 8, 1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amadio, S.; Montilli, C.; Magliozzi, R.; Bernardi, G.; Reynolds, R.; Volonté, C. P2Y12 Receptor Protein in Cortical Gray Matter Lesions in Multiple Sclerosis. Cereb. Cortex 2010, 20, 1263–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, J.Z.; Zandarowska, E. Effect of Amodiaquine on Histamine Level and Histamine-Methyltransferase Activity in the Rat Brain. Arch. Immunol. Ther. Exp. 1980, 28, 927–930. [Google Scholar]
- Yoshikawa, T.; Nakamura, T.; Yanai, K. Histamine N-Methyltransferase in the Brain. Int. J. Mol. Sci. 2019, 20, 737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirota, M.; Dudley, J.T.; Kim, J.; Chiang, A.P.; Morgan, A.A.; Sweet-Cordero, A.; Sage, J.; Butte, A.J. Discovery and Preclinical Validation of Drug Indications Using Compendia of Public Gene Expression Data. Sci. Transl. Med. 2011, 3, 96ra77. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, J.C.; Arrang, J.M.; Garbarg, M.; Pollard, H.; Ruat, M. Histaminergic Transmission in the Mammalian Brain. Physiol. Rev. 1991, 71, 1–51. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, R.; Naganuma, F.; Nakamura, T.; Miwa, H.; Nakayama-Naono, R.; Matsuzawa, T.; Komatsu, Y.; Sato, Y.; Takahashi, Y.; Tatsuoka-Kitano, H.; et al. Contribution of Astrocytic Histamine N-Methyltransferase to Histamine Clearance and Brain Function in Mice. Neuropharmacology 2022, 212, 109065. [Google Scholar] [CrossRef] [PubMed]
- Elmore, B.O.; Bollinger, J.A.; Dooley, D.M. Human Kidney Diamine Oxidase: Heterologous Expression, Purification, and Characterization. JBIC J. Biol. Inorg. Chem. 2002, 7, 565–579. [Google Scholar] [CrossRef]
- Oberheim, N.A.; Wang, X.; Goldman, S.; Nedergaard, M. Astrocytic Complexity Distinguishes the Human Brain. Trends Neurosci. 2006, 29, 547–553. [Google Scholar] [CrossRef]
- Zeinstra, E.; Riele, P.; Langlois, X.; Wilczak, N.; Leysen, J.; De Keyser, J. Aminergic Receptors in Astrogliotic Plaques from Patients with Multiple Sclerosis. Neurosci. Lett. 2002, 331, 87–90. [Google Scholar] [CrossRef]
- Tuomisto, L.; Kilpeläinen, H.; Riekkinen, P. Histamine and Histamine-N-Methyltransferase in the CSF of Patients with Multiple Sclerosis. Agents Actions 1983, 13, 255–257. [Google Scholar] [CrossRef]
- Kallweit, U.; Aritake, K.; Bassetti, C.L.; Blumenthal, S.; Hayaishi, O.; Linnebank, M.; Baumann, C.R.; Urade, Y. Elevated CSF Histamine Levels in Multiple Sclerosis Patients. Fluids Barriers CNS 2013, 10, 19. [Google Scholar] [CrossRef] [Green Version]
- Rozniecki, J.J.; Hauser, S.L.; Stein, M.; Lincoln, R.; Theoharides, T.C. Elevated Mast Cell Tryptase in Cerebrospinal Fluid of Multiple Sclerosis Patients. Ann. Neurol. 1995, 37, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, R.; Eilam, R.; Arnon, R. Astrocytes in Multiple Sclerosis-Essential Constituents with Diverse Multifaceted Functions. Int. J. Mol. Sci. 2021, 22, 5904. [Google Scholar] [CrossRef] [PubMed]
- Gillson, G.; Wright, J.V.; DeLack, E.; Ballasiotes, G. Transdermal Histamine in Multiple Sclerosis: Part One-Clinical Experience. Altern. Med. Rev. J. Clin. Ther. 1999, 4, 424–428. [Google Scholar]
- Gillson, G.; Wright, J.V.; DeLack, E.; Ballasiotes, G. Transdermal Histamine in Multiple Sclerosis, Part Two: A Proposed Theoretical Basis for Its Use. Altern. Med. Rev. J. Clin. Ther. 2000, 5, 224–248. [Google Scholar]
- García-Martín, E.; Martínez, C.; Benito-León, J.; Calleja, P.; Díaz-Sánchez, M.; Pisa, D.; Alonso-Navarro, H.; Ayuso-Peralta, L.; Torrecilla, D.; Agúndez, J.A.G.; et al. Histamine-N-Methyl Transferase Polymorphism and Risk for Multiple Sclerosis. Eur. J. Neurol. 2010, 17, 335–338. [Google Scholar] [CrossRef]
- Chen, Y.; Cao, B.; Ou, R.; Wei, Q.; Chen, X.; Zhao, B.; Wu, Y.; Song, W.; Shang, H.-F. Determining the Effect of the HNMT, STK39, and NMD3 Polymorphisms on the Incidence of Parkinson’s Disease, Amyotrophic Lateral Sclerosis, and Multiple System Atrophy in Chinese Populations. J. Mol. Neurosci. 2018, 64, 574–580. [Google Scholar] [CrossRef]
- Horton, J.R.; Sawada, K.; Nishibori, M.; Cheng, X. Structural Basis for Inhibition of Histamine N-Methyltransferase by Diverse Drugs. J. Mol. Biol. 2005, 353, 334–344. [Google Scholar] [CrossRef] [Green Version]
- Nhama, A.; Nhamússua, L.; Macete, E.; Bassat, Q.; Salvador, C.; Enosse, S.; Candrinho, B.; Carvalho, E.; Nhacolo, A.; Chidimatembue, A.; et al. In Vivo Efficacy and Safety of Artemether–Lumefantrine and Amodiaquine–Artesunate for Uncomplicated Plasmodium Falciparum Malaria in Mozambique, 2018. Malar. J. 2021, 20, 390. [Google Scholar] [CrossRef]
- Oh, S.; Shin, J.H.; Jang, E.J.; Won, H.Y.; Kim, H.K.; Jeong, M.-G.; Kim, K.S.; Hwang, E.S. Anti-Inflammatory Activity of Chloroquine and Amodiaquine through P21-Mediated Suppression of T Cell Proliferation and Th1 Cell Differentiation. Biochem. Biophys. Res. Commun. 2016, 474, 345–350. [Google Scholar] [CrossRef] [Green Version]
- Persoons, L.; Vanderlinden, E.; Vangeel, L.; Wang, X.; Do, N.D.T.; Foo, S.-Y.C.; Leyssen, P.; Neyts, J.; Jochmans, D.; Schols, D.; et al. Broad Spectrum Anti-Coronavirus Activity of a Series of Anti-Malaria Quinoline Analogues. Antivir. Res. 2021, 193, 105127. [Google Scholar] [CrossRef]
- Moon, M.; Jung, E.S.; Jeon, S.G.; Cha, M.-Y.; Jang, Y.; Kim, W.; Lopes, C.; Mook-Jung, I.; Kim, K.-S. Nurr1 (NR4A2) Regulates Alzheimer’s Disease-Related Pathogenesis and Cognitive Function in the 5XFAD Mouse Model. Aging Cell 2019, 18, e12866. [Google Scholar] [CrossRef]
- Kinoshita, K.; Matsumoto, K.; Kurauchi, Y.; Hisatsune, A.; Seki, T.; Katsuki, H. A Nurr1 Agonist Amodiaquine Attenuates Inflammatory Events and Neurological Deficits in a Mouse Model of Intracerebral Hemorrhage. J. Neuroimmunol. 2019, 330, 48–54. [Google Scholar] [CrossRef]
- Musio, S.; Gallo, B.; Scabeni, S.; Lapilla, M.; Poliani, P.L.; Matarese, G.; Ohtsu, H.; Galli, S.J.; Mantegazza, R.; Steinman, L.; et al. A Key Regulatory Role for Histamine in Experimental Autoimmune Encephalomyelitis: Disease Exacerbation in Histidine Decarboxylase-Deficient Mice. J. Immunol. Baltim. 2006, 176, 17–26. [Google Scholar] [CrossRef]
- Saligrama, N.; Case, L.K.; Krementsov, D.N.; Teuscher, C. Histamine H₂ Receptor Signaling × Environment Interactions Determine Susceptibility to Experimental Allergic Encephalomyelitis. Fed. Am. Soc. Exp. Biol. 2014, 28, 1898–1909. [Google Scholar] [CrossRef] [Green Version]
- Lock, C.; Hermans, G.; Pedotti, R.; Brendolan, A.; Schadt, E.; Garren, H.; Langer-Gould, A.; Strober, S.; Cannella, B.; Allard, J.; et al. Gene-Microarray Analysis of Multiple Sclerosis Lesions Yields New Targets Validated in Autoimmune Encephalomyelitis. Nat. Med. 2002, 8, 500–508. [Google Scholar] [CrossRef]
- Costanza, M.; Di Dario, M.; Steinman, L.; Farina, C.; Pedotti, R. Gene Expression Analysis of Histamine Receptors in Peripheral Blood Mononuclear Cells from Individuals with Clinically-Isolated Syndrome and Different Stages of Multiple Sclerosis. J. Neuroimmunol. 2014, 277, 186–188. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.Z.; Gao, J.; Meeker, N.D.; Fillmore, P.D.; Tung, K.S.K.; Watanabe, T.; Zachary, J.F.; Offner, H.; Blankenhorn, E.P.; Teuscher, C. Identification of Bphs, an Autoimmune Disease Locus, as Histamine Receptor H1. Science 2002, 297, 620–623. [Google Scholar] [CrossRef] [Green Version]
- Noubade, R.; Milligan, G.; Zachary, J.F.; Blankenhorn, E.P.; del Rio, R.; Rincon, M.; Teuscher, C. Histamine Receptor H1 Is Required for TCR-Mediated P38 MAPK Activation and Optimal IFN-Gamma Production in Mice. J. Clin. Investig. 2007, 117, 3507–3518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimitriadou, V.; Pang, X.; Theoharides, T.C. Hydroxyzine Inhibits Experimental Allergic Encephalomyelitis (EAE) and Associated Brain Mast Cell Activation. Int. J. Immunopharmacol. 2000, 22, 673–684. [Google Scholar] [CrossRef]
- Pedotti, R.; De Voss, J.J.; Steinman, L.; Galli, S.J. Involvement of Both “allergic” and “Autoimmune” Mechanisms in EAE, MS and Other Autoimmune Diseases. Trends Immunol. 2003, 24, 479–484. [Google Scholar] [CrossRef]
- Logothetis, L.; Mylonas, I.A.; Baloyannis, S.; Pashalidou, M.; Orologas, A.; Zafeiropoulos, A.; Kosta, V.; Theoharides, T.C. A Pilot, Open Label, Clinical Trial Using Hydroxyzine in Multiple Sclerosis. Int. J. Immunopathol. Pharmacol. 2005, 18, 771–778. [Google Scholar] [CrossRef] [Green Version]
- Yong, H.Y.; McKay, K.A.; Daley, C.G.J.; Tremlett, H. Drug Exposure and the Risk of Multiple Sclerosis: A Systematic Review. Pharmacoepidemiol. Drug Saf. 2018, 27, 133–139. [Google Scholar] [CrossRef]
- Green, A.J.; Gelfand, J.M.; Cree, B.A.; Bevan, C.; Boscardin, W.J.; Mei, F.; Inman, J.; Arnow, S.; Devereux, M.; Abounasr, A.; et al. Clemastine Fumarate as a Remyelinating Therapy for Multiple Sclerosis (ReBUILD): A Randomised, Controlled, Double-Blind, Crossover Trial. Lancet 2017, 390, 2481–2489. [Google Scholar] [CrossRef] [Green Version]
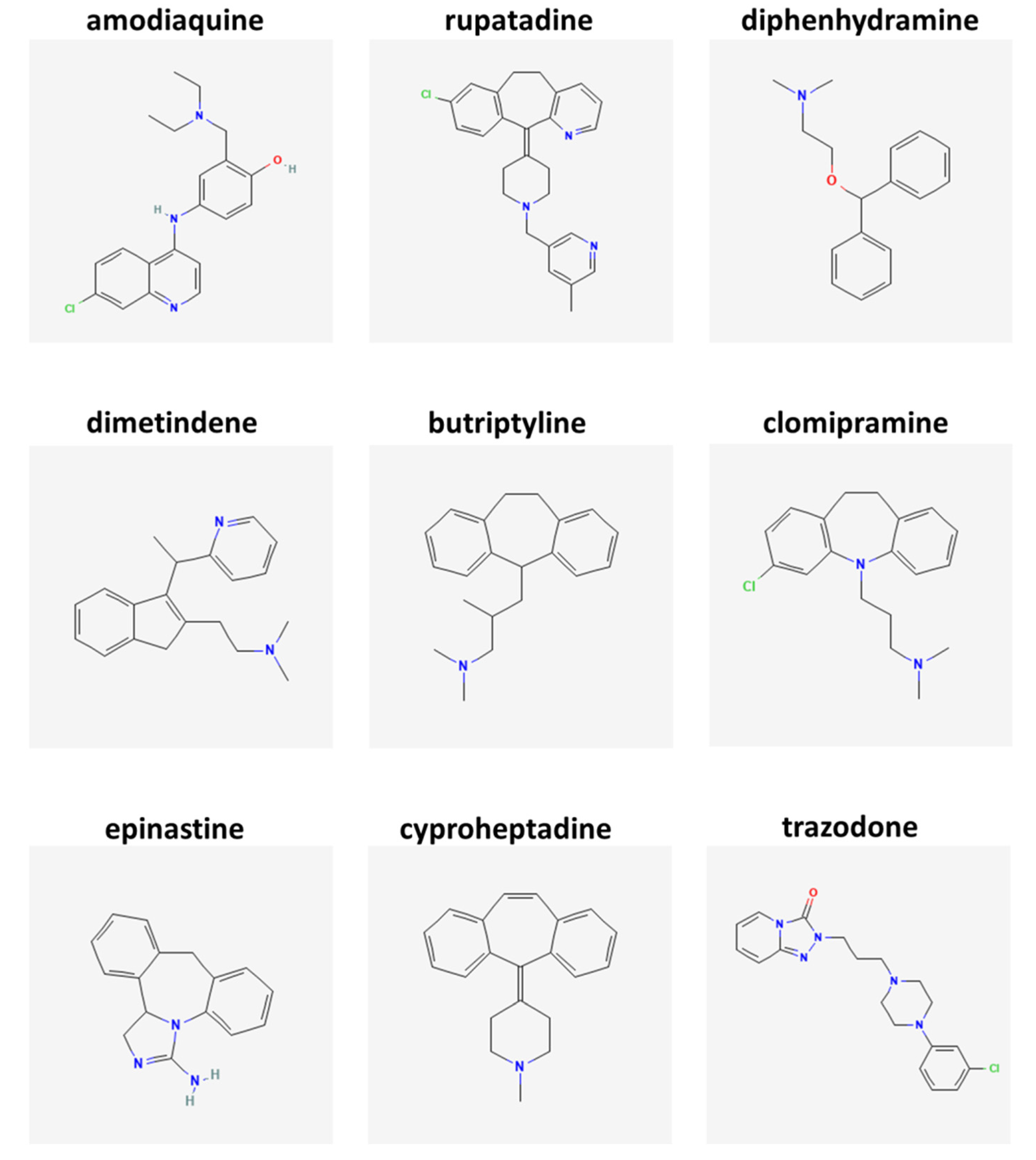
| Case | Age (Years) | Sex | Clinical Diagnosis | Disease Duration (Years) | Cause of Death | DTPI (h) |
|---|---|---|---|---|---|---|
| MS074 | 64 | F | SPMS | 36 | Gastrointestinal bleed/obstruction, aspiration pneumonia | 7 |
| MS076 | 49 | F | SPMS | 18 | Chronic renal failure, heart disease | 31 |
| MS114 | 52 | F | SPMS | 15 | Pneumonia, sepsis, pulmonary embolism | 12 |
| MS125 | 76 | F | SPMS | 31 | MS | 13 |
| MS163 | 45 | F | SPMS | 6 | Urinary tract infection, MS | 28 |
| DRUG | Similarity Value | p-Value | GSEA Score | Drug Bank Code | Target |
|---|---|---|---|---|---|
| Amodiaquine | 1 | 0.00806 | 3 | DB00613 | HNMT |
| Rupatadine | 0.62791 | 0.01412 | 3 | DB11614 | H1 receptor |
| Diphenhydramine | 0.62791 | 0.01571 | 2 | DB01075 | H1 receptor |
| Dimetindene | 0.62791 | 0.01654 | na | DB08801 | H1 receptor |
| Butriptyline | 0.50388 | 0.02857 | na | DB09016 | H1 receptor |
| Clomipramine | 0.37984 | 0.03232 | 3 | DB01242 | H1 receptor |
| Epinastine | 0.37984 | 0.0044 | 2 | DB00751 | H1 receptor |
| Cyproheptadine | 0.36213 | 0.01584 | 3 | DB00434 | H1 receptor |
| Trazodone | 0.36213 | 0.03221 | 3 | DB00656 | H1 receptor |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amadio, S.; Conte, F.; Esposito, G.; Fiscon, G.; Paci, P.; Volonté, C. Repurposing Histaminergic Drugs in Multiple Sclerosis. Int. J. Mol. Sci. 2022, 23, 6347. https://doi.org/10.3390/ijms23116347
Amadio S, Conte F, Esposito G, Fiscon G, Paci P, Volonté C. Repurposing Histaminergic Drugs in Multiple Sclerosis. International Journal of Molecular Sciences. 2022; 23(11):6347. https://doi.org/10.3390/ijms23116347
Chicago/Turabian StyleAmadio, Susanna, Federica Conte, Giorgia Esposito, Giulia Fiscon, Paola Paci, and Cinzia Volonté. 2022. "Repurposing Histaminergic Drugs in Multiple Sclerosis" International Journal of Molecular Sciences 23, no. 11: 6347. https://doi.org/10.3390/ijms23116347
APA StyleAmadio, S., Conte, F., Esposito, G., Fiscon, G., Paci, P., & Volonté, C. (2022). Repurposing Histaminergic Drugs in Multiple Sclerosis. International Journal of Molecular Sciences, 23(11), 6347. https://doi.org/10.3390/ijms23116347








