Anti-Growth, Anti-Angiogenic, and Pro-Apoptotic Effects by CX-4945, an Inhibitor of Casein Kinase 2, on HuCCT-1 Human Cholangiocarcinoma Cells via Control of Caspase-9/3, DR-4, STAT-3/STAT-5, Mcl-1, eIF-2α, and HIF-1α
Abstract
:1. Introduction
2. Results
2.1. CX-4945 Inhibits Growth and Induces Apoptosis of HuCCT-1 Cells
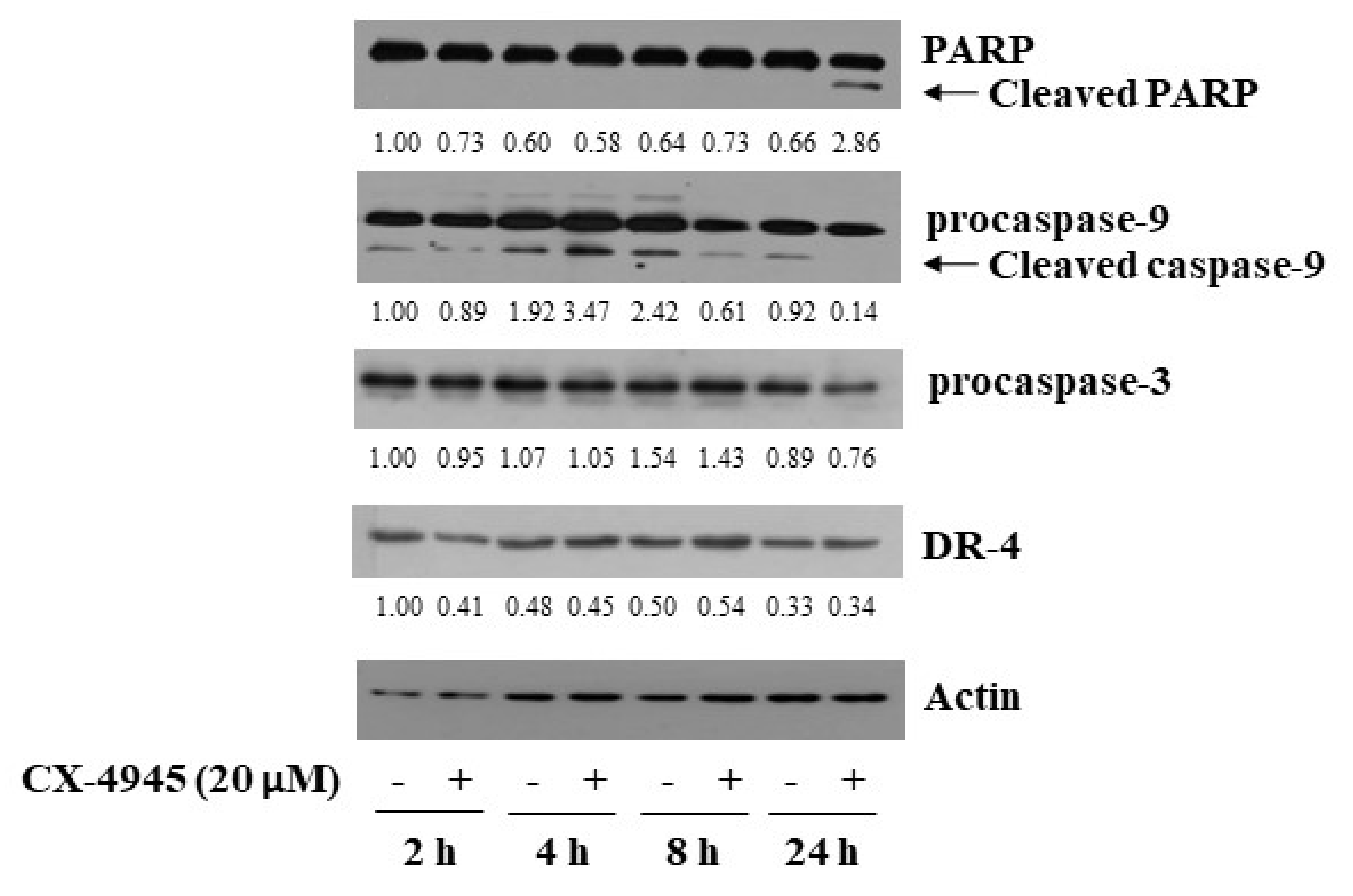
2.2. CX-4945 at 20 µM Induces PARP Cleavage, Activation of Caspase-9/3, and Up-Regulation of DR-4 in HuCCT-1 Cells
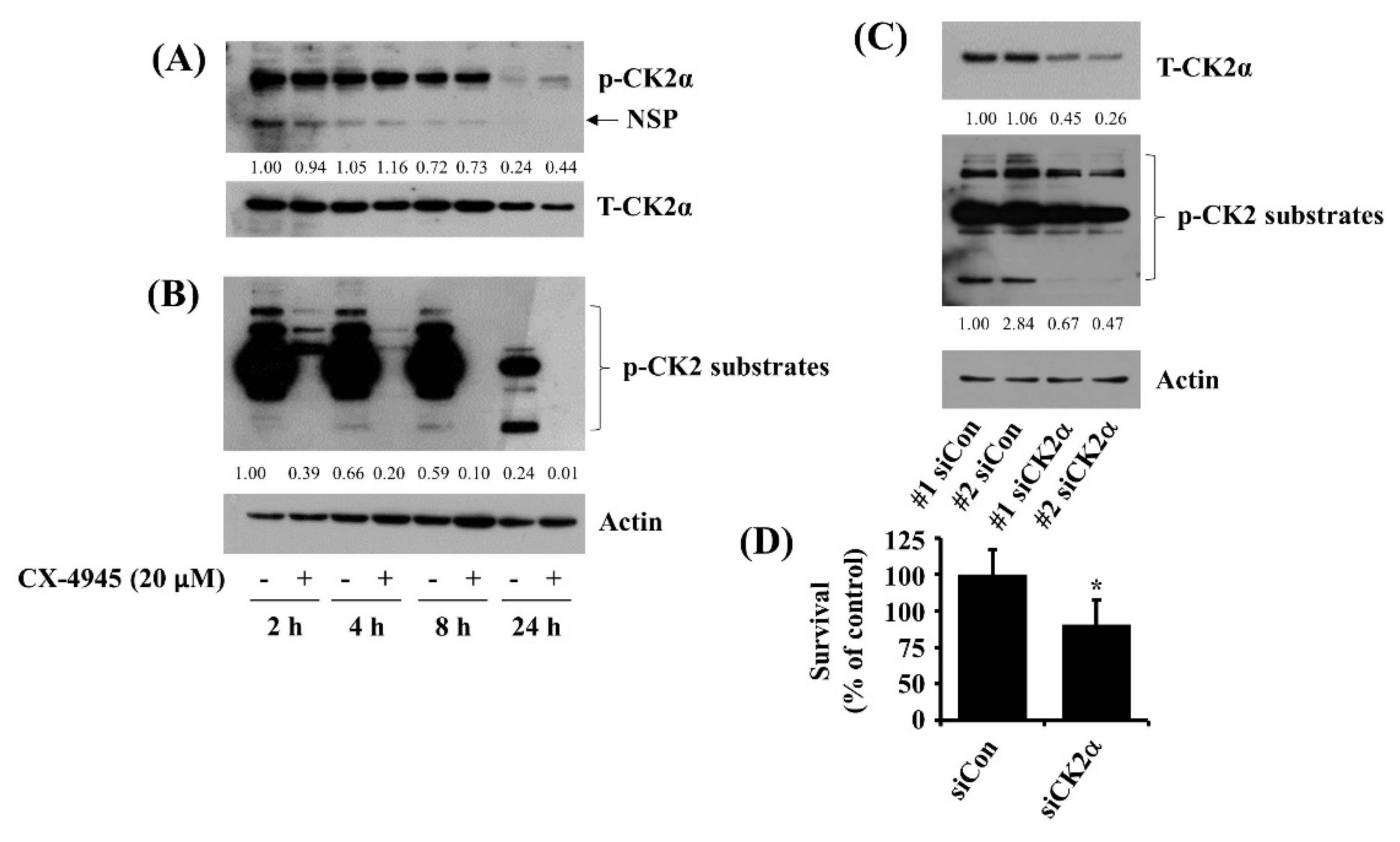
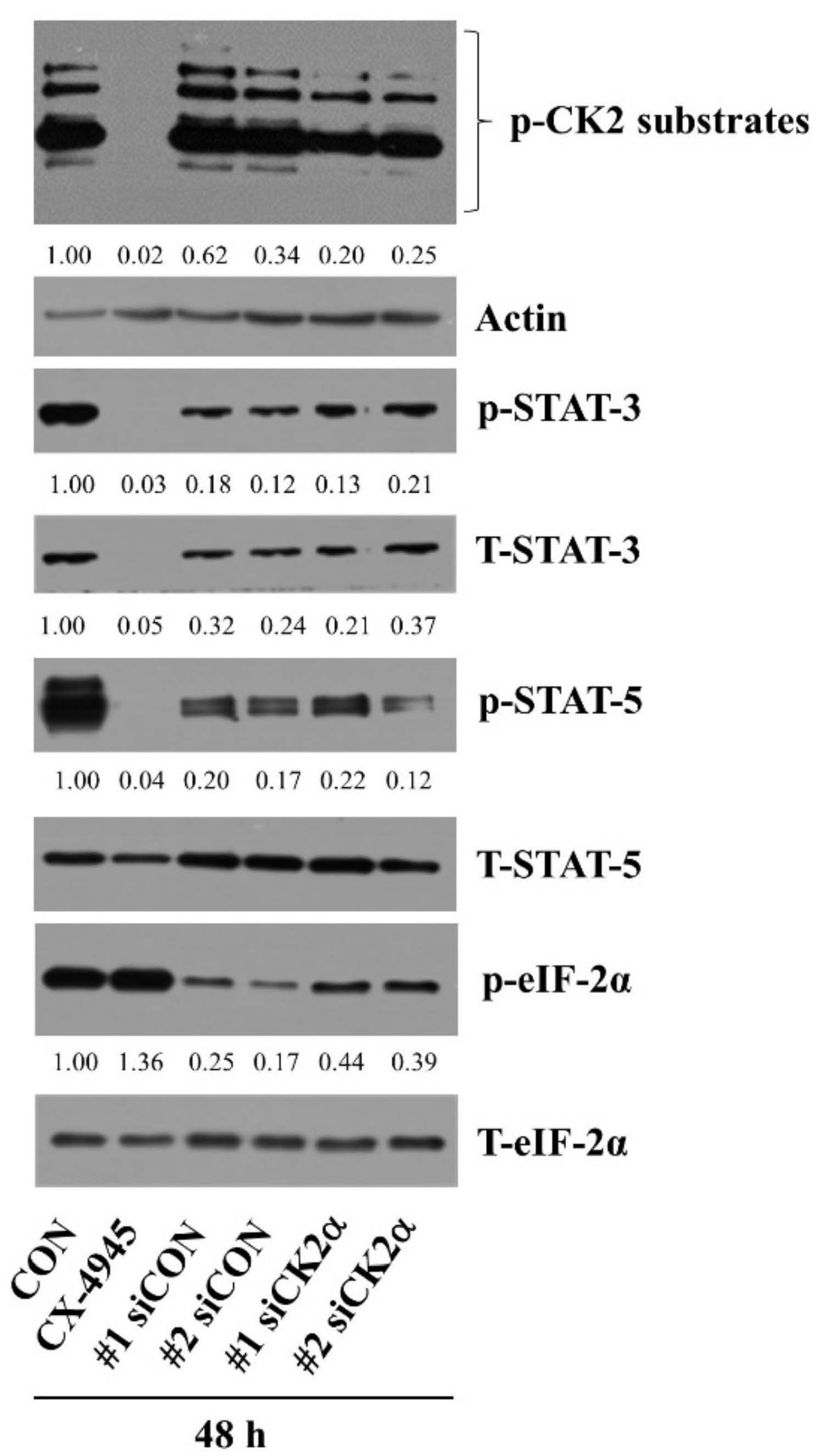
2.3. CX-4945 at 20 µM Vastly Reduces Phosphorylation Levels of CK2 Substrates and Knockdown of CK2 Leads to a Partial Reduction of HuCCT-1 Cell Survival
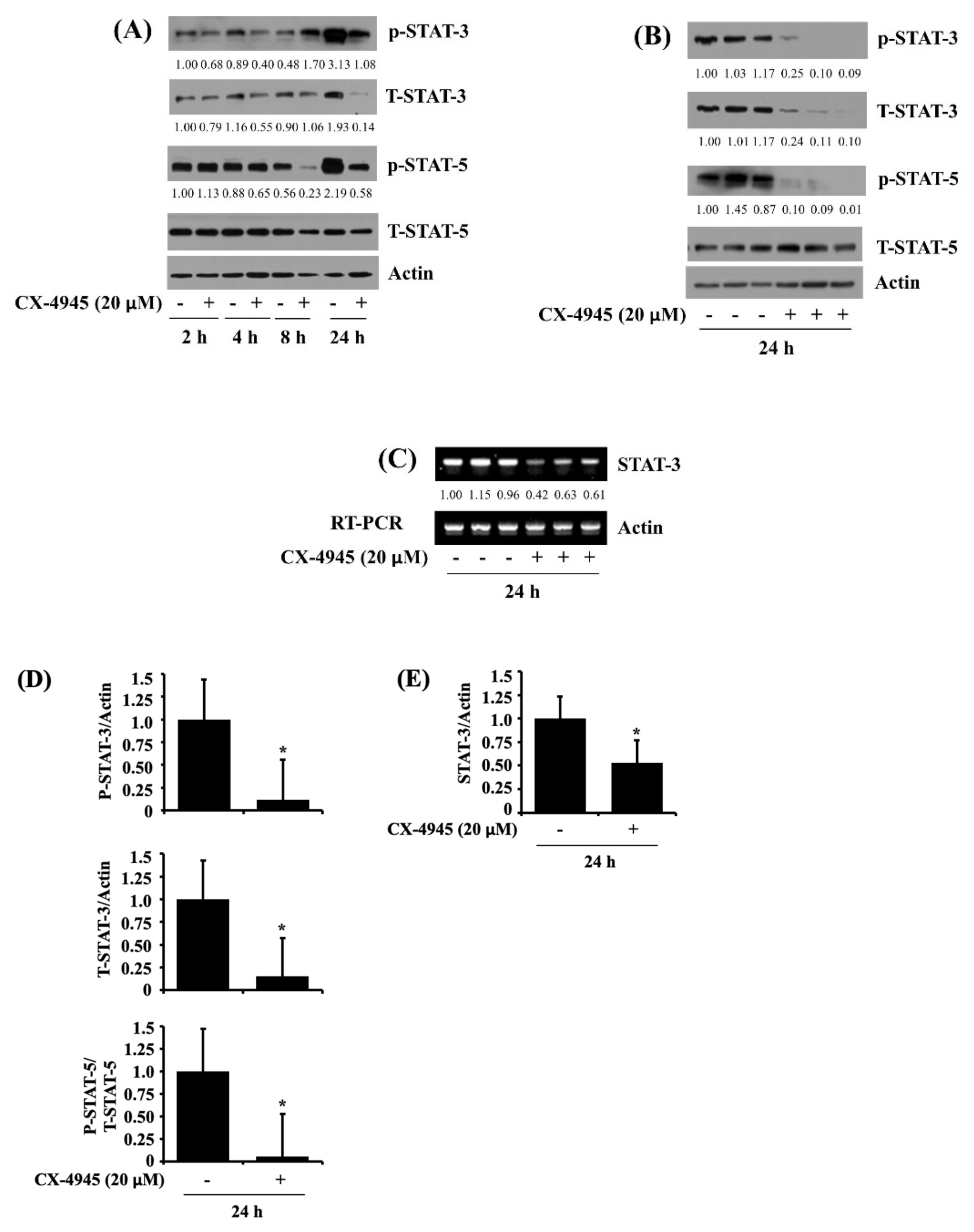
2.4. CX-4945 at 20 µM Reduces Phosphorylation and Expression Levels of STAT-3 and STAT-5 in HuCCT-1 Cells
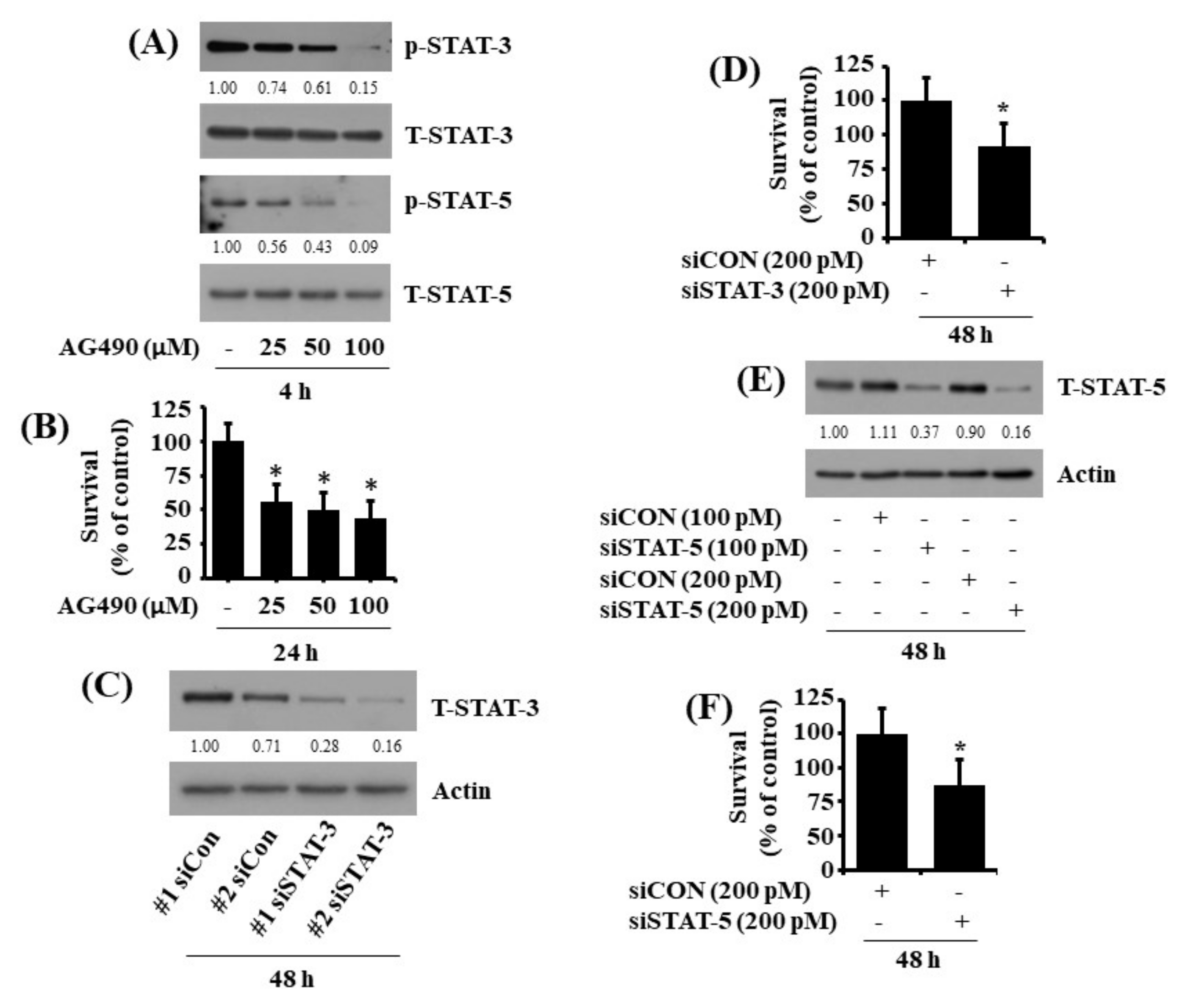
2.5. Pharmacological Inhibition or Respective Knockdown of STAT-3 and STAT-5 Leads to Reduction of HuCCT-1 Cell Survival
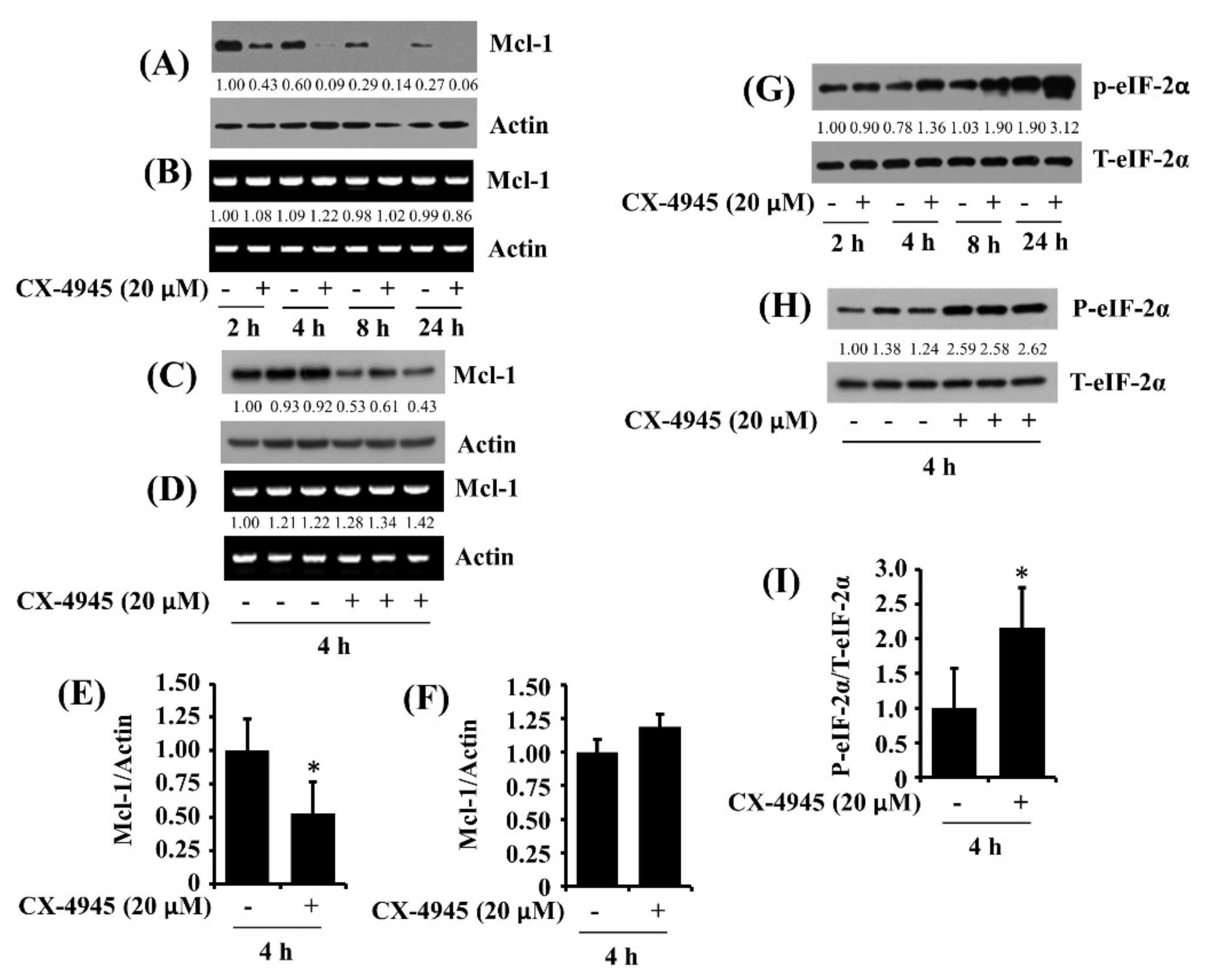
2.6. CX-4945 at 20 µM Reduces Protein Expression of Mcl-1 and Elevates Protein Phosphorylation of eIF-2α in HuCCT-1 Cells
2.7. CX-4945-Induced Decrease in the Expression and Phosphorylation of STAT-3/-5 Is the CK-2-Independent but CX-4945-Induced Increase in eIF-2α Phosphorylation Is the CK-2-Dependent in HuCCT-1 Cells
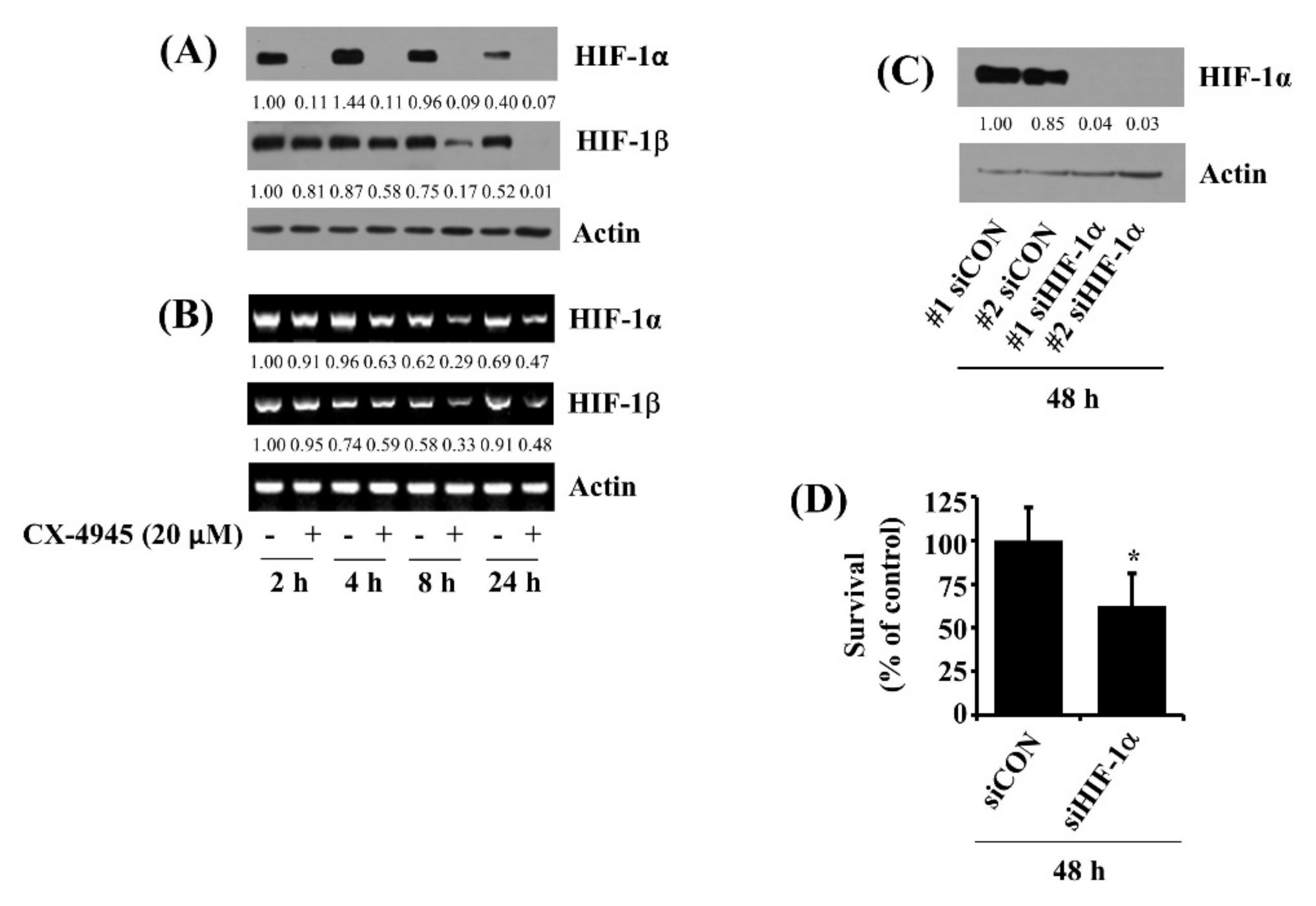
2.8. CX-4945 at 20 µM Down-Regulates HIF-1α in HuCCT-1 Cells Time-Differentially and Knockdown of HIF-1α Leads to a Significant Reduction of the Cell Survival
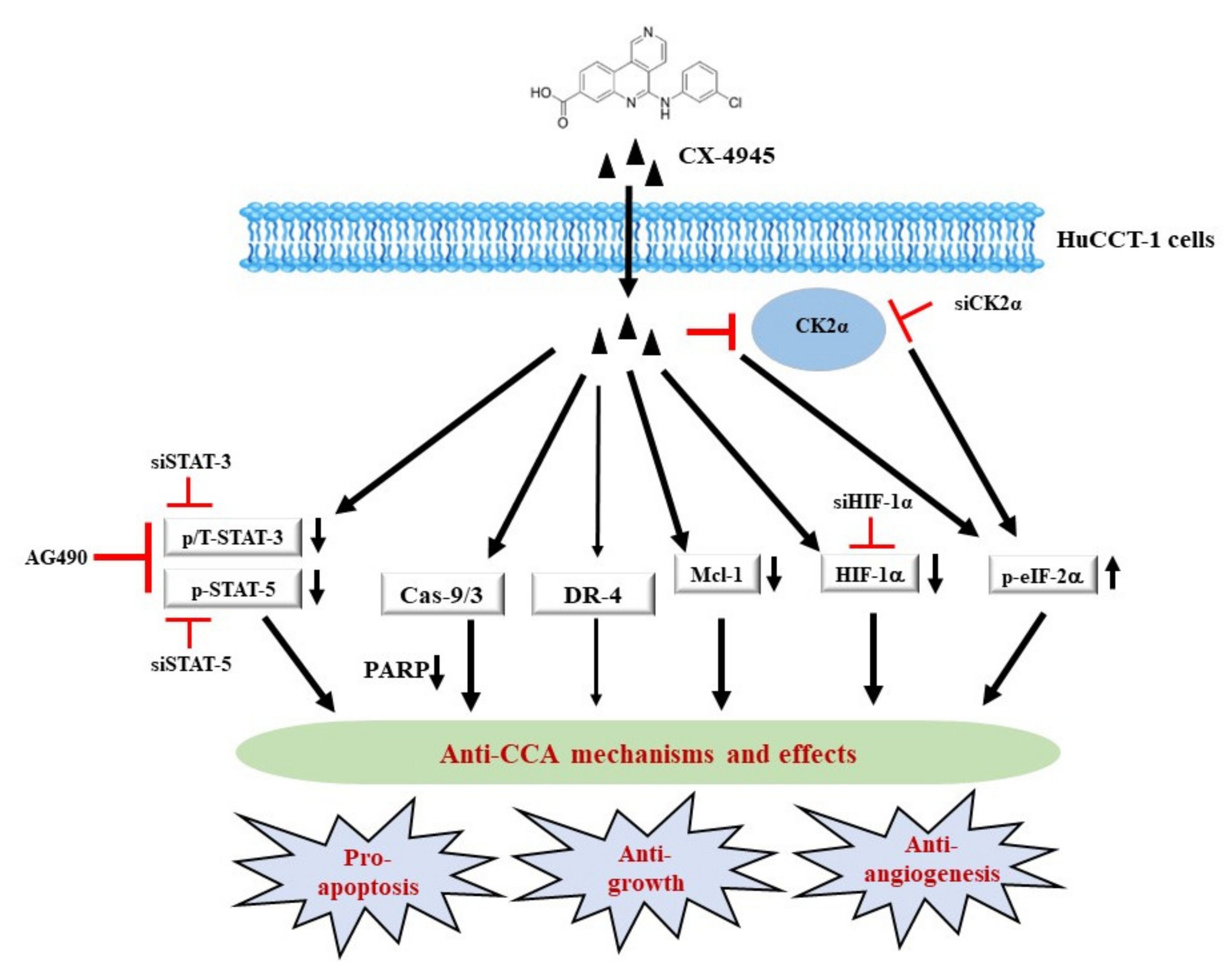
3. Discussion
4. Materials and Methods
4.1. Chemicals and Antibodies
4.2. Cell Culture
4.3. Cell Count Analysis
4.4. Colony Formation Assay
4.5. Measurement of Genomic DNA Fragmentation
4.6. Preparation of Protein Samples
4.7. Immunoblot Analysis
4.8. Small Interfering RNA (siRNA) Transfection
4.9. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
4.10. Small Interfering RNA (siRNA) Transfection
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sarcognato, S.; Sacchi, D.; Fassan, M.; Fabris, L.; Cadamuro, M.; Zanus, G.; Cataldo, I.; Capelli, P.; Baciorri, F.; Cacciatore, M.; et al. Cholangiocarcinoma. Pathologica 2021, 113, 158–169. [Google Scholar] [CrossRef]
- Banales, J.M.; Marin, J.J.G.; Lamarca, A. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588. [Google Scholar] [CrossRef]
- Mosadeghi, S.; Liu, B.; Bhuket, T.; Wong, R.J. Sex-specific and race/ethnicity-specific disparities in cholangiocarcinoma incidence and prevalence in the USA: An updated analysis of the 2000–2011 Surveillance, Epidemiology and End Results registry. Hepatol. Res. 2016, 46, 669–677. [Google Scholar] [CrossRef]
- Shimoda, M.; Kubota, K. Multi-disciplinary treatment for cholangiocellular carcinoma. World J. Gastroenterol. 2007, 13, n1500–n1504. [Google Scholar] [CrossRef] [Green Version]
- Hemming, A.W.; Reed, A.I.; Fujita, S.; Foley, D.P.; Howard, R.J. Surgical management of hilar cholangiocarcinoma. Ann. Surg. 2005, 241, 693–699; discussion 699–702. [Google Scholar] [CrossRef] [PubMed]
- Trembley, J.H.; Chen, Z.; Unger, G. Emergence of protein kinase CK2 as a key target in cancer therapy. Biofactors 2010, 36, 187–195. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Ahmad, K.A.; Ahmed, K. Role of protein kinase CK2 in the regulation of tumor necrosis factor-related apoptosis inducing ligand-induced apoptosis in prostate cancer cells. Cancer Res. 2006, 15, 2242–2249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, H.; Benveniste, E.N. The Immune Regulatory Role of Protein Kinase CK2 and Its Implications for Treatment of Cancer. Biomedicines 2021, 9, 1932. [Google Scholar] [CrossRef] [PubMed]
- Di Maira, G.; Gentilini, A.; Rovida, E.; Ottaviani, D.; Ruzzene, M.; Marra, F. Role of the protein kinase CK2 in the biology of cholangiocarcinoma cells. Dig. Liver Dis. 2016, 48, e20. [Google Scholar] [CrossRef]
- Di Maira, G.; Gentilini, A.; Pastore, M. The protein kinase CK2 contributes to the malignant phenotype of cholangiocarcinoma cells. Oncogenesis 2019, 8, 61. [Google Scholar] [CrossRef] [Green Version]
- Pierre, F.; Chua, P.C.; O’Brien, S.E.; Siddiqui-Jain, A.; Bourbon, P.; Haddach, M.; Michaux, J.; Nagasawa, J.; Schwaebe, M.K.; Stefan, E.; et al. Discovery and SAR of 5-(3-chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic acid (CX4945), the first clinical stage inhibitor of protein kinase CK2 for the treatment of cancer. J. Med. Chem. 2011, 54, 635–654. [Google Scholar] [CrossRef]
- Zakharia, K.; Miyabe, K.; Wang, Y.; Wu, D.; Moser, C.D.; Borad, M.J.; Roberts, L.R. Preclinical In Vitro and In Vivo Evidence of an Antitumor Effect of CX-4945, a Casein Kinase II Inhibitor, in Cholangiocarcinoma. Transl. Oncol. 2019, 12, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, P.S.; Gaston, K. Targeting protein kinase CK2 in the treatment of cholangiocarcinoma. Explor. Target. Anti-Tumor Ther. 2021, 2, 434–447. [Google Scholar] [CrossRef]
- Thomas, S.J.; Snowden, J.A.; Zeidler, M.P.; Danson, S.J. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer 2015, 113, 365–371. [Google Scholar] [CrossRef] [Green Version]
- Verhoeven, Y.; Tilborghs, S.; Jacobs, J.; de Waele, J.; Quatannens, D.; Deben, C.; Prenen, H.; Pauwels, P.; Trinh, X.B.; Wouters, A.; et al. The potential and controversy of targeting STAT family members in cancer. Semin. Cancer Biol. 2020, 60, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Baek, S.H.; Um, J.Y.; Shim, B.S.; Ahn, K.S. Resveratrol attenuates constitutive STAT3 and STAT5 activation through induction of PTPε and SHP-2 tyrosine phosphatases and potentiates sorafenib-induced apoptosis in renal cell carcinoma. BMC Nephrol. 2016, 17, 19. [Google Scholar] [CrossRef] [Green Version]
- Campbell, K.J.; Tait, S.W.G. Targeting BCL-2 regulated apoptosis in cancer. Open Biol. 2018, 8, 180002. [Google Scholar] [CrossRef]
- Zheng, Q.; Ye, J.; Cao, J. Translational regulator eIF2α in tumor. Tumour Biol. 2014, 35, 6255–6264. [Google Scholar] [CrossRef] [PubMed]
- Zimna, A.; Kurpisz, M. Hypoxia-Inducible Factor-1 in Physiological and Pathophysiological Angiogenesis: Applications and Therapies. Biomed. Res. Int. 2015, 2015, 549412. [Google Scholar] [CrossRef] [Green Version]
- Thongchot, S.; Yongvanit, P.; Loilome, W.; Seubwai, W.; Phunicom, K.; Tassaneeyakul, W.; Pairojkul, C.; Promkotra, W.; Techasen, A.; Namwat, N. High expression of HIF-1α, BNIP3 and PI3KC3: Hypoxia-induced autophagy predicts cholangiocarcinoma survival and metastasis. Asian Pac. J. Cancer Prev. 2014, 15, 5873–5878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borad, M.J.; Bai, L.; Chen, M.; Hubbard, J.M.; Mody, K.; Rha, S.Y.; Richards, D.A.; Davis, S.L.; Soong, J.; Huang, C.C.; et al. Silmitasertib (CX-4945) in combination with gemcitabine and cisplatin as first-line treatment for patients with locally advanced or metastatic cholangiocarcinoma: A phase Ib/II study. J. Clin. Oncol. 2021, 39, 312. [Google Scholar] [CrossRef]
- Kim, H.M.; Jeong, I.; Kim, H.J.; Kang, S.K.; Kwon, W.S.; Kim, T.S.; Park, K.H.; Jung, M.; Soong, J.; Lin, S.C.; et al. Casein Kinase 2 Inhibitor, CX-4945, as a Potential Targeted Anticancer Agent in Gastric Cancer. Anticancer Res. 2018, 38, 6171–6180. [Google Scholar] [CrossRef]
- Gray, G.K.; McFarland, B.C.; Rowse, A.L.; Gibson, S.A.; Benveniste, E.N. Therapeutic CK2 inhibition attenuates diverse prosurvival signaling cascades and decreases cell viability in human breast cancer cells. Oncotarget 2014, 5, 6484–6496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, D.W.; So, K.S.; Kim, S.C.; Park, K.M.; Lee, Y.J.; Kim, S.W.; Choi, C.M.; Rho, J.K.; Choi, Y.J.; Lee, J.C. Autophagy Induced by CX-4945, a Casein Kinase 2 Inhibitor, Enhances Apoptosis in Pancreatic Cancer Cell Lines. Pancreas 2017, 46, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.R.; Lucio, P.; Melao, A. Activity of the clinical-stage CK2-specific inhibitor CX-4945 against chronic lymphocytic leukemia. Leukemia 2014, 28, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Hengartner, M.O. The biochemistry of apoptosis. Nature 2000, 407, 770–776. [Google Scholar] [CrossRef]
- Pinna, L.A. Protein kinase CK2. Int. J. Biochem. Cell Biol. 1997, 29, 551–554. [Google Scholar] [CrossRef]
- Meggio, F.; Pinna, L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003, 17, 349–368. [Google Scholar] [CrossRef]
- Zhou, F.; Xu, J.; Ding, G.; Cao, L. Overexpressions of CK2[beta] and XIAP are associated with poor prognosis of patients with cholangiocarcinoma. Pathol. Oncol. Res. 2014, 20, 73–79. [Google Scholar] [CrossRef]
- Tolomeo, M.; Cascio, A. The Multifaced Role of STAT3 in Cancer and Its Implication for Anticancer Therapy. Int. J. Mol. Sci. 2021, 22, 603. [Google Scholar] [CrossRef] [PubMed]
- Dokduang, H.; Yongvanit, P.; Namwat, N.; Pairojkul, C.; Sangkhamanon, S.; Yageta, M.S.; Murakami, Y.; Loilome, W. Xanthohumol inhibits STAT3 activation pathway leading to growth suppression and apoptosis induction in human cholangiocarcinoma cells. Oncol. Rep. 2016, 35, 2065–2072. [Google Scholar] [CrossRef] [Green Version]
- Quotti, T.L.; Canovas, N.S.; Brancalion, A.; Doriguzzi, B.E.; Manni, S.; Mandato, E.; Zaffino, F.; Macaccaro, P.; Carrino, M.; Gianesin, K.; et al. Protein kinase CK2 regulates AKT, NF-κB and STAT3 activation, stem cell viability and proliferation in acute myeloid leukemia. Leukemia 2017, 31, 292–300. [Google Scholar] [CrossRef]
- Aparicio-Siegmund, S.; Sommer, J.; Monhasery, N. Inhibition of protein kinase II (CK2) prevents induced signal transducer and activator of transcription (STAT) 1/3 and constitutive STAT3 activation. Oncotarget 2014, 5, 2131–2148. [Google Scholar] [CrossRef] [Green Version]
- Kale, J.; Osterlund, E.; Andrews, D. BCL-2 family proteins: Changing partners in the dance towards death. Cell Death Differ. 2018, 25, 65–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Guo, M.; Wei, H.; Chen, Y. Targeting MCL-1 in cancer: Current status and perspectives. J. Hematol. Oncol. 2021, 14, 67. [Google Scholar] [CrossRef] [PubMed]
- Hao, P.; Yu, J.; Ward, R. Eukaryotic translation initiation factors as promising targets in cancer therapy. Cell Commun. Signal. 2020, 18, 175. [Google Scholar] [CrossRef]
- Wek, R.C. Role of eIF2α Kinases in Translational Control and Adaptation to Cellular Stress. Cold Spring Harb. Perspect. Biol. 2018, 10, a032870. [Google Scholar] [CrossRef]
- Intemann, J.; Saidu, N.E.; Schwind, L.; Montenarh, M. ER stress signaling in ARPE-19 cells after inhibition of protein kinase CK2 by CX-4945. Cell. Signal. 2014, 26, 1567–1575. [Google Scholar] [CrossRef]
- Féral, K.; Jaud, M.; Philippe, C.; di Bella, D.; Pyronnet, S.; Rouault-Pierre, K.; Mazzolini, L.; Touriol, C. ER Stress and unfolded protein response in leukemia: Friend, Foe, or Both? Biomolecules 2021, 11, 199. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.H.; Fang, W.G. Hypoxia-inducible factor-1 in tumour angiogenesis. World J. Gastroenterol. 2004, 10, 1082–1087. [Google Scholar] [CrossRef] [PubMed]
- Warfel, N.A.; El-Deiry, W.S. HIF-1 signaling in drug resistance to chemotherapy. Curr. Med. Chem. 2014, 21, 3021–3028. [Google Scholar] [CrossRef] [PubMed]
- Morine, Y.; Shimada, M.; Utsunomiya, T.; Imura, S.; Ikemoto, T.; Mori, H.; Hanaoka, J.; Kanamoto, M.; Iwahashi, S.; Miyake, H. Hypoxia inducible factor expression in intrahepatic cholangiocarcinoma. Hepatogastroenterology 2011, 58, 1439–1444. [Google Scholar] [CrossRef]
- Crowley, L.C.; Christensen, M.E.; Waterhouse, N.J. Measuring Survival of Adherent Cells with the Colony-Forming Assay. Cold Spring Harb. Protoc. 2016, 8, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, H.; Yadav, A.K.; Do, Y.; Heo, M.; Bishop-Bailey, D.; Lee, J.; Jang, B.C. Anti-survival and pro-apoptotic effects of meridianin C derivatives on MV4-11 human acute myeloid leukemia cells. Int. J. Oncol. 2020, 56, 368–378. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Yadav, A.K.; Han, J.-Y.; Ahn, K.S.; Jang, B.-C. Anti-Growth, Anti-Angiogenic, and Pro-Apoptotic Effects by CX-4945, an Inhibitor of Casein Kinase 2, on HuCCT-1 Human Cholangiocarcinoma Cells via Control of Caspase-9/3, DR-4, STAT-3/STAT-5, Mcl-1, eIF-2α, and HIF-1α. Int. J. Mol. Sci. 2022, 23, 6353. https://doi.org/10.3390/ijms23116353
Wang S, Yadav AK, Han J-Y, Ahn KS, Jang B-C. Anti-Growth, Anti-Angiogenic, and Pro-Apoptotic Effects by CX-4945, an Inhibitor of Casein Kinase 2, on HuCCT-1 Human Cholangiocarcinoma Cells via Control of Caspase-9/3, DR-4, STAT-3/STAT-5, Mcl-1, eIF-2α, and HIF-1α. International Journal of Molecular Sciences. 2022; 23(11):6353. https://doi.org/10.3390/ijms23116353
Chicago/Turabian StyleWang, Saini, Anil Kumar Yadav, Jin-Yi Han, Keun Soo Ahn, and Byeong-Churl Jang. 2022. "Anti-Growth, Anti-Angiogenic, and Pro-Apoptotic Effects by CX-4945, an Inhibitor of Casein Kinase 2, on HuCCT-1 Human Cholangiocarcinoma Cells via Control of Caspase-9/3, DR-4, STAT-3/STAT-5, Mcl-1, eIF-2α, and HIF-1α" International Journal of Molecular Sciences 23, no. 11: 6353. https://doi.org/10.3390/ijms23116353
APA StyleWang, S., Yadav, A. K., Han, J.-Y., Ahn, K. S., & Jang, B.-C. (2022). Anti-Growth, Anti-Angiogenic, and Pro-Apoptotic Effects by CX-4945, an Inhibitor of Casein Kinase 2, on HuCCT-1 Human Cholangiocarcinoma Cells via Control of Caspase-9/3, DR-4, STAT-3/STAT-5, Mcl-1, eIF-2α, and HIF-1α. International Journal of Molecular Sciences, 23(11), 6353. https://doi.org/10.3390/ijms23116353






