Comprehensive Integrated Single-Cell Whole Transcriptome Analysis Revealed the p-EMT Tumor Cells—CAFs Communication in Oral Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Results
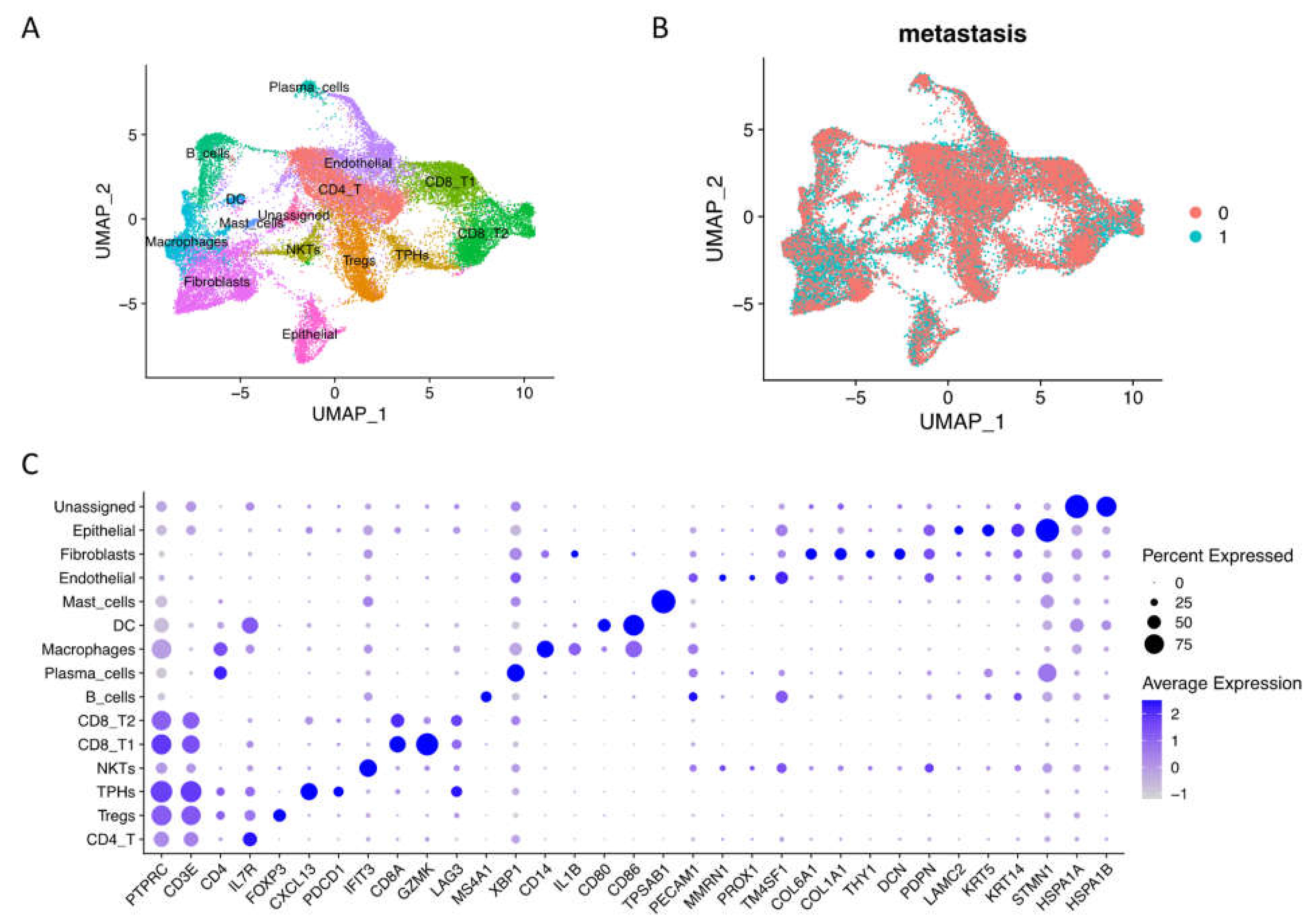
2.1. Integrated Single-Cell RNA Sequencing Mapped OSCC Microenvironment Cell Types across the Metastasis Condition
2.2. Cellular Community of OSCC Fibroblasts and Epithelial Cells at Single-Cell Level
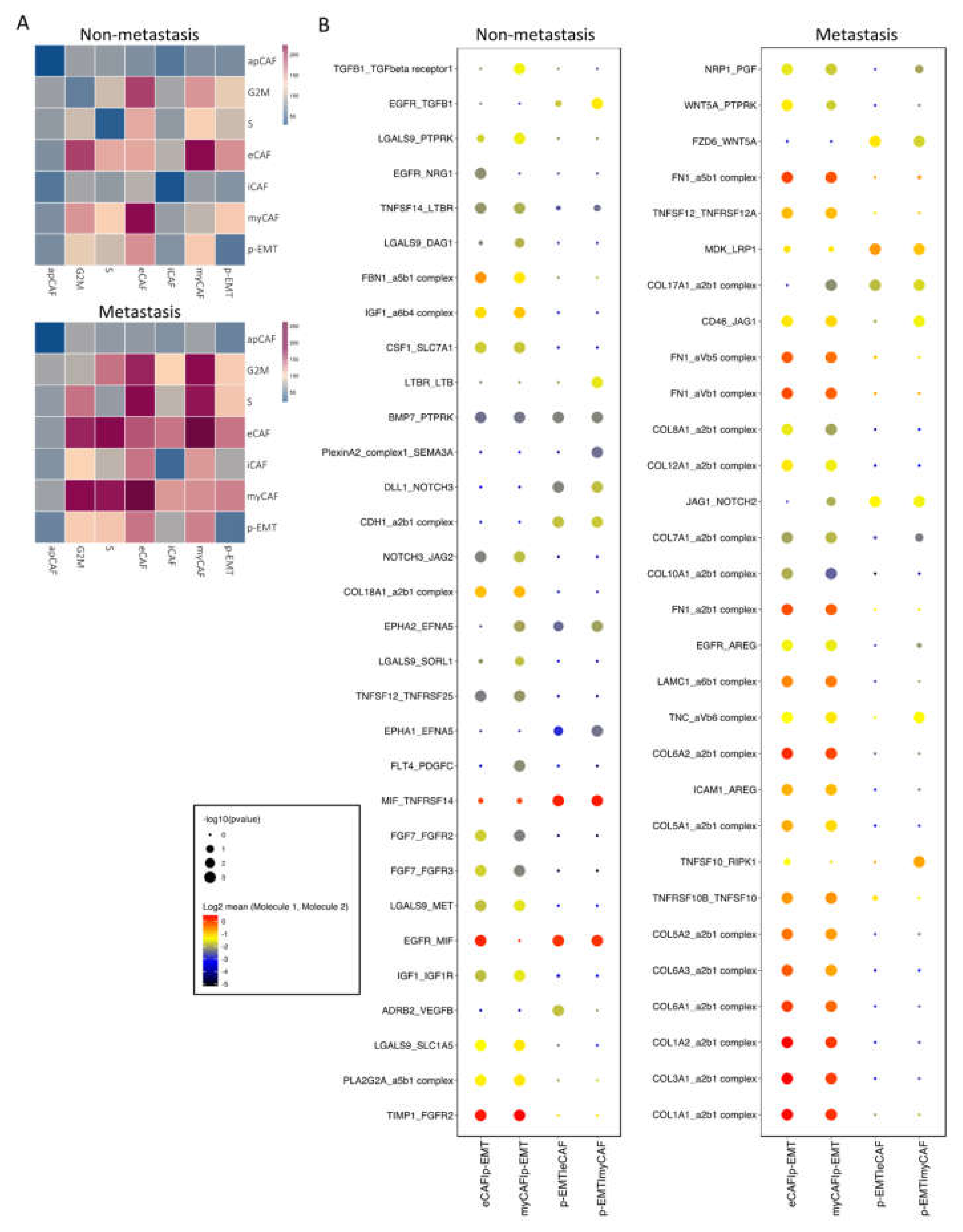
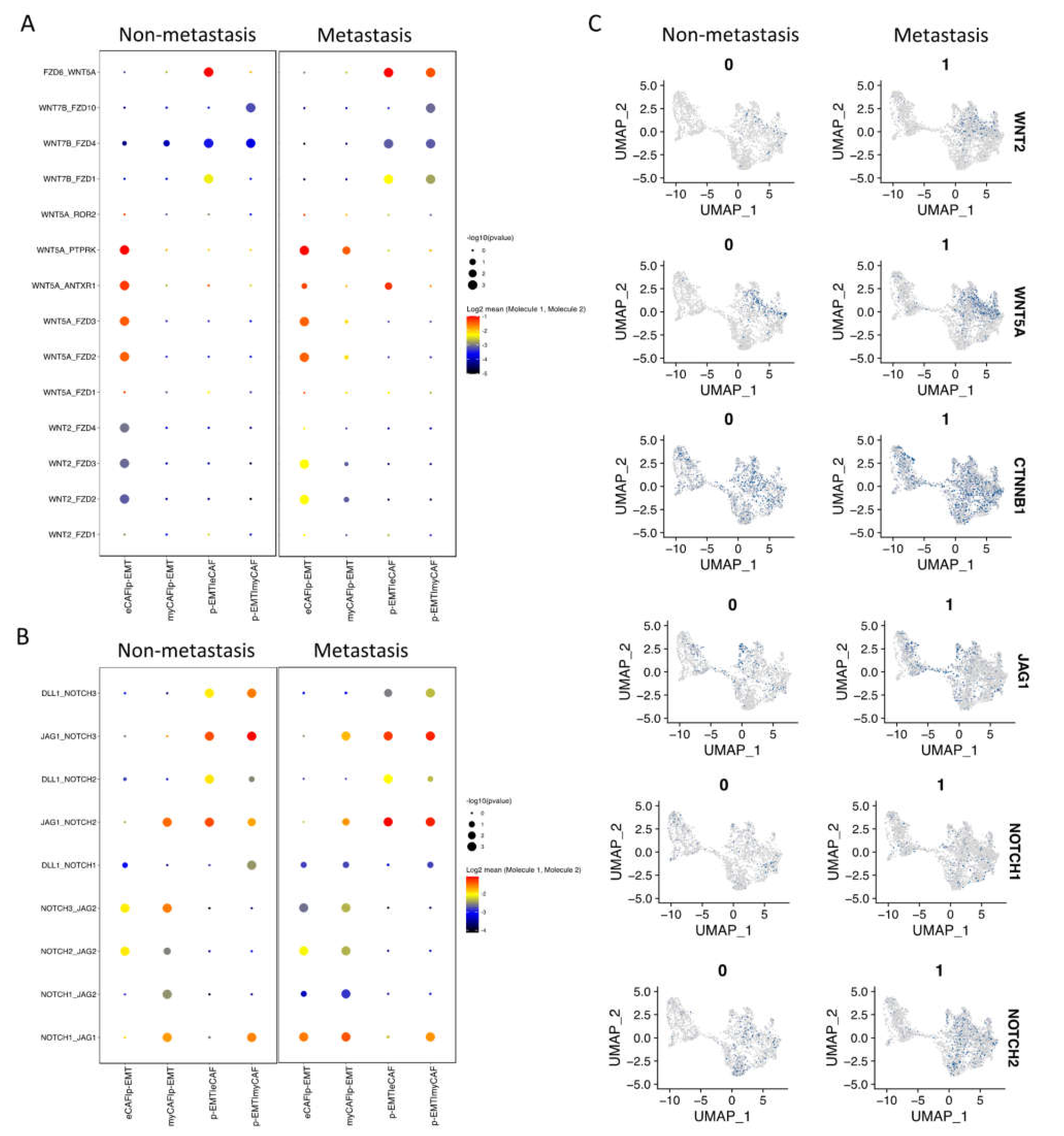
2.3. Cell–Cell Interaction of OSCC CAFs and p-EMT Tumor Cells in Non-Metastatsis and Metastasis Conditions
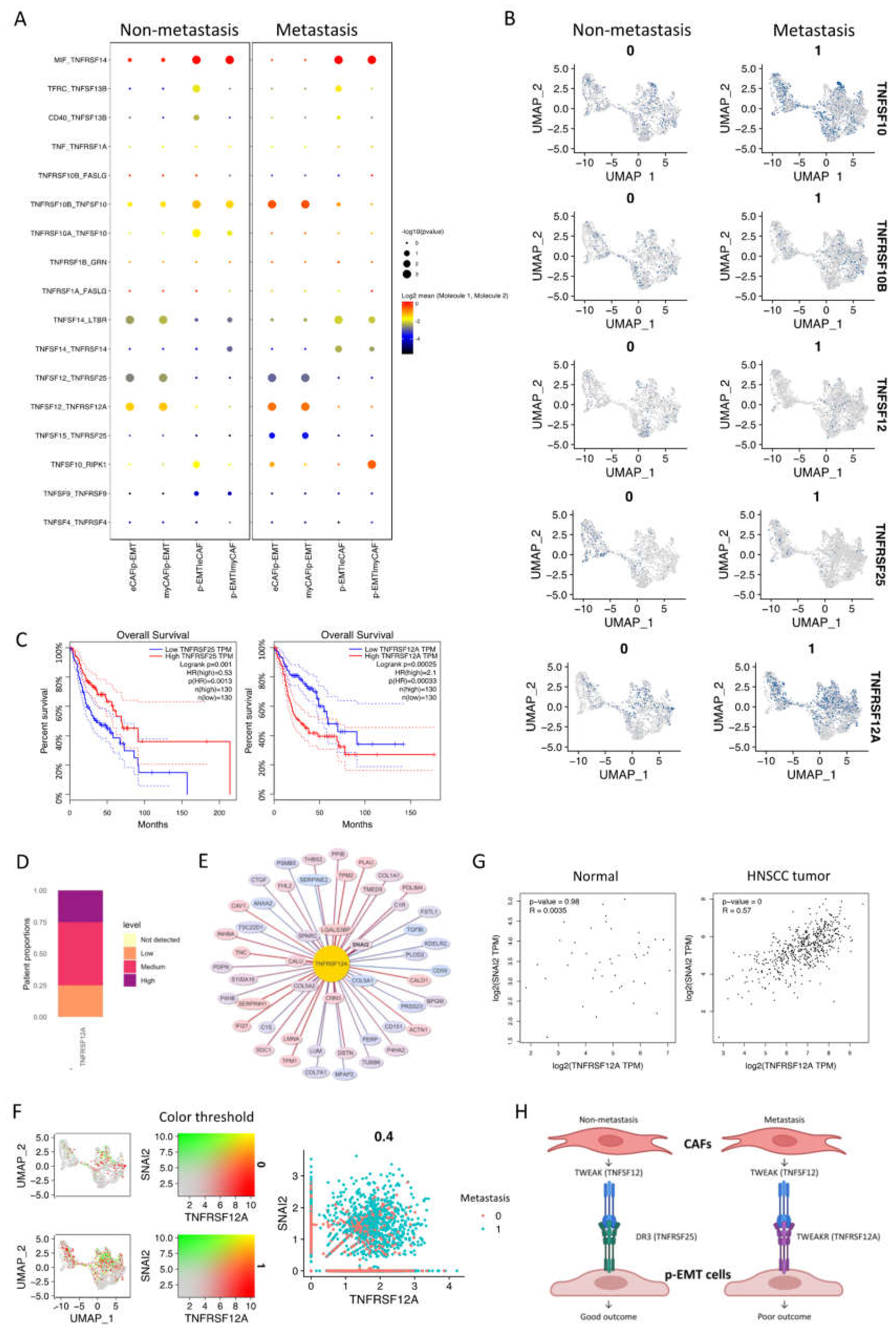
2.4. The p-EMT Tumor Cells—CAFs Communication Highlighted the Different Roles of TNFRSF25 in Non-Metastasis and TNFRSF12A in Non-Metastasis OSCC
3. Discussion
4. Materials and Methods
4.1. Data Obtaining and Preprocessing
4.2. Single-Cell RNA-seq Integration and Computational Methods
4.3. Enrichment and Cell–Cell Communication Analysis
4.4. Survival Analysis, Correlation Analysis and Images of Protein Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hsieh, Y.-P.; Wu, Y.-H.; Cheng, S.-M.; Lin, F.-K.; Hwang, D.-Y.; Jiang, S.-S.; Chen, K.-C.; Chen, M.-Y.; Chiang, W.-F.; Liu, K.-J.; et al. Single-Cell RNA Sequencing Analysis for Oncogenic Mechanisms Underlying Oral Squamous Cell Carcinoma Carcinogenesis with Candida albicans Infection. Int. J. Mol. Sci. 2022, 23, 4833. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.A.; Vacher, M.; Farah, C.S. Transcriptomic Biomarker Signatures for Discrimination of Oral Cancer Surgical Margins. Biomolecules 2022, 12, 464. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lesterhuis, W.J.; Bosco, A.; Millward, M.J.; Small, M.; Nowak, A.K.; Lake, R.A. Dynamic versus static biomarkers in cancer immune checkpoint blockade: Unravelling complexity. Nat. Rev. Drug Discov. 2017, 16, 264–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.Y.; Hsieh, Y.P.; Wang, Y.Y.; Hwang, D.Y.; Jiang, S.S.; Huang, W.T.; Chiang, W.F.; Liu, K.J.; Huang, T.T. Single-Cell Analysis of Different Stages of Oral Cancer Carcinogenesis in a Mouse Model. Int. J. Mol. Sci. 2020, 21, 8171. [Google Scholar] [CrossRef]
- Shalek, A.K.; Benson, M. Single-cell analyses to tailor treatments. Sci. Transl. Med. 2017, 9, 4730. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Han, C.; Wang, S.; Fang, P.; Ma, Z.; Xu, L.; Yin, R. Cancer-associated fibroblasts: An emerging target of anti-cancer immunotherapy. J. Hematol. Oncol. 2019, 12, 86. [Google Scholar] [CrossRef]
- Chen, X.; Song, E. Turning foes to friends: Targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 2019, 18, 99–115. [Google Scholar] [CrossRef]
- Lei, Y.; Yan, W.; Lin, Z.; Liu, J.; Tian, D.; Han, P. Comprehensive analysis of partial epithelial mesenchymal transition-related genes in hepatocellular carcinoma. J. Cell Mol. Med. 2021, 25, 448–462. [Google Scholar] [CrossRef]
- Kisoda, S.; Shao, W.; Fujiwara, N.; Mouri, Y.; Tsunematsu, T.; Jin, S.; Arakaki, R.; Ishimaru, N.; Kudo, Y. Prognostic value of partial EMT-related genes in head and neck squamous cell carcinoma by a bioinformatic analysis. Oral Dis. 2020, 26, 1149–1156. [Google Scholar] [CrossRef]
- Tyler, M.; Tirosh, I. Decoupling epithelial-mesenchymal transitions from stromal profiles by integrative expression analysis. Nat. Commun. 2021, 12, 2592. [Google Scholar] [CrossRef] [PubMed]
- Kurten, C.H.L.; Kulkarni, A.; Cillo, A.R.; Santos, P.M.; Roble, A.K.; Onkar, S.; Reeder, C.; Lang, S.; Chen, X.; Duvvuri, U.; et al. Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing. Nat. Commun. 2021, 12, 7338. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.A. T Cells That Help B Cells in Chronically Inflamed Tissues. Front. Immunol. 2018, 9, 1924. [Google Scholar] [CrossRef] [PubMed]
- Ohlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J.; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017, 214, 579–596. [Google Scholar] [CrossRef] [PubMed]
- Elyada, E.; Bolisetty, M.; Laise, P.; Flynn, W.F.; Courtois, E.T.; Burkhart, R.A.; Teinor, J.A.; Belleau, P.; Biffi, G.; Lucito, M.S.; et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019, 9, 1102–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, J.; Slowikowski, K.; Zhang, F. Single-cell transcriptomics in cancer: Computational challenges and opportunities. Exp. Mol. Med. 2020, 52, 1452–1465. [Google Scholar] [CrossRef]
- Liu, J.; Xu, T.; Jin, Y.; Huang, B.; Zhang, Y. Progress and Clinical Application of Single-Cell Transcriptional Sequencing Technology in Cancer Research. Front. Oncol. 2020, 10, 593085. [Google Scholar] [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e24. [Google Scholar] [CrossRef] [Green Version]
- Wacleche, V.S.; Wang, R.; Rao, D.A. Identification of T Peripheral Helper (Tph) Cells. Methods Mol. Biol. 2022, 2380, 59–76. [Google Scholar] [CrossRef]
- Yoshitomi, H.; Ueno, H. Shared and distinct roles of T peripheral helper and T follicular helper cells in human diseases. Cell Mol. Immunol. 2021, 18, 523–527. [Google Scholar] [CrossRef]
- Gu-Trantien, C.; Loi, S.; Garaud, S.; Equeter, C.; Libin, M.; de Wind, A.; Ravoet, M.; Le Buanec, H.; Sibille, C.; Manfouo-Foutsop, G.; et al. CD4(+) follicular helper T cell infiltration predicts breast cancer survival. J. Clin. Investig. 2013, 123, 2873–2892. [Google Scholar] [CrossRef] [PubMed]
- Gu-Trantien, C.; Migliori, E.; Buisseret, L.; de Wind, A.; Brohee, S.; Garaud, S.; Noel, G.; Dang Chi, V.L.; Lodewyckx, J.N.; Naveaux, C.; et al. CXCL13-producing TFH cells link immune suppression and adaptive memory in human breast cancer. JCI Insight 2017, 2, 91487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoersch, S.; Andrade-Navarro, M.A. Periostin shows increased evolutionary plasticity in its alternatively spliced region. BMC Evol. Biol. 2010, 10, 30. [Google Scholar] [CrossRef] [Green Version]
- Rotondo, J.C.; Bosi, S.; Bassi, C.; Ferracin, M.; Lanza, G.; Gafa, R.; Magri, E.; Selvatici, R.; Torresani, S.; Marci, R.; et al. Gene expression changes in progression of cervical neoplasia revealed by microarray analysis of cervical neoplastic keratinocytes. J. Cell. Physiol. 2015, 230, 806–812. [Google Scholar] [CrossRef]
- Costa, A.; Kieffer, Y.; Scholer-Dahirel, A.; Pelon, F.; Bourachot, B.; Cardon, M.; Sirven, P.; Magagna, I.; Fuhrmann, L.; Bernard, C.; et al. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell 2018, 33, 463–479.e410. [Google Scholar] [CrossRef] [Green Version]
- Sodergren, M.H.; Mangal, N.; Wasan, H.; Sadanandam, A.; Balachandran, V.P.; Jiao, L.R.; Habib, N. Immunological combination treatment holds the key to improving survival in pancreatic cancer. J. Cancer Res. Clin. Oncol. 2020, 146, 2897–2911. [Google Scholar] [CrossRef]
- Mace, T.A.; Ameen, Z.; Collins, A.; Wojcik, S.; Mair, M.; Young, G.S.; Fuchs, J.R.; Eubank, T.D.; Frankel, W.L.; Bekaii-Saab, T.; et al. Pancreatic cancer-associated stellate cells promote differentiation of myeloid-derived suppressor cells in a STAT3-dependent manner. Cancer Res. 2013, 73, 3007–3018. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Shi, R.; Xu, S.; Li, Y.; Zhang, H.; Liu, M.; Zhu, G.; Chen, C.; Pan, Z.; Liu, H.; et al. Identification of small proline-rich protein 1B (SPRR1B) as a prognostically predictive biomarker for lung adenocarcinoma by integrative bioinformatic analysis. Thorac. Cancer 2021, 12, 796–806. [Google Scholar] [CrossRef]
- Michifuri, Y.; Hirohashi, Y.; Torigoe, T.; Miyazaki, A.; Fujino, J.; Tamura, Y.; Tsukahara, T.; Kanaseki, T.; Kobayashi, J.; Sasaki, T.; et al. Small proline-rich protein-1B is overexpressed in human oral squamous cell cancer stem-like cells and is related to their growth through activation of MAP kinase signal. Biochem. Biophys. Res. Commun. 2013, 439, 96–102. [Google Scholar] [CrossRef]
- Pannone, G.; Santoro, A.; Feola, A.; Bufo, P.; Papagerakis, P.; Lo Muzio, L.; Staibano, S.; Ionna, F.; Longo, F.; Franco, R.; et al. The role of E-cadherin down-regulation in oral cancer: CDH1 gene expression and epigenetic blockage. Curr. Cancer Drug Targets 2014, 14, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Baumeister, P.; Zhou, J.; Canis, M.; Gires, O. Epithelial-to-Mesenchymal Transition-Derived Heterogeneity in Head and Neck Squamous Cell Carcinomas. Cancers 2021, 13, 5355. [Google Scholar] [CrossRef] [PubMed]
- Efremova, M.; Vento-Tormo, M.; Teichmann, S.A.; Vento-Tormo, R. CellPhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020, 15, 1484–1506. [Google Scholar] [CrossRef] [PubMed]
- Osathanon, T.; Nowwarote, N.; Pavasant, P. Expression and influence of Notch signaling in oral squamous cell carcinoma. J. Oral Sci. 2016, 58, 283–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, X.; Wu, H.; Xu, H.; Xiong, H.; Chu, Q.; Yu, S.; Wu, G.S.; Wu, K. Notch signaling: An emerging therapeutic target for cancer treatment. Cancer Lett. 2015, 369, 20–27. [Google Scholar] [CrossRef] [Green Version]
- Meurette, O.; Mehlen, P. Notch Signaling in the Tumor Microenvironment. Cancer Cell 2018, 34, 536–548. [Google Scholar] [CrossRef] [Green Version]
- Gan, R.H.; Wei, H.; Xie, J.; Zheng, D.P.; Luo, E.L.; Huang, X.Y.; Xie, J.; Zhao, Y.; Ding, L.C.; Su, B.H.; et al. Notch1 regulates tongue cancer cells proliferation, apoptosis and invasion. Cell Cycle 2018, 17, 216–224. [Google Scholar] [CrossRef] [Green Version]
- Reyes, M.; Flores, T.; Betancur, D.; Pena-Oyarzun, D.; Torres, V.A. Wnt/β-Catenin Signaling in Oral Carcinogenesis. Int. J. Mol. Sci. 2020, 21, 4682. [Google Scholar] [CrossRef]
- Xie, J.; Huang, L.; Lu, Y.G.; Zheng, D.L. Roles of the Wnt Signaling Pathway in Head and Neck Squamous Cell Carcinoma. Front Mol. Biosci. 2020, 7, 590912. [Google Scholar] [CrossRef]
- Baeten, J.T.; Lilly, B. Differential Regulation of NOTCH2 and NOTCH3 Contribute to Their Unique Functions in Vascular Smooth Muscle Cells. J. Biol. Chem. 2015, 290, 16226–16237. [Google Scholar] [CrossRef] [Green Version]
- Kayamori, K.; Katsube, K.; Sakamoto, K.; Ohyama, Y.; Hirai, H.; Yukimori, A.; Ohata, Y.; Akashi, T.; Saitoh, M.; Harada, K.; et al. NOTCH3 Is Induced in Cancer-Associated Fibroblasts and Promotes Angiogenesis in Oral Squamous Cell Carcinoma. PLoS ONE 2016, 11, e0154112. [Google Scholar] [CrossRef] [PubMed]
- Purow, B. Notch inhibition as a promising new approach to cancer therapy. Adv. Exp. Med. Biol. 2012, 727, 305–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, F.; Maillard, I. Therapeutic Targeting of Notch Signaling: From Cancer to Inflammatory Disorders. Front Cell Dev. Biol. 2021, 9, 649205. [Google Scholar] [CrossRef] [PubMed]
- Vellinga, T.T.; den Uil, S.; Rinkes, I.H.; Marvin, D.; Ponsioen, B.; Alvarez-Varela, A.; Fatrai, S.; Scheele, C.; Zwijnenburg, D.A.; Snippert, H.; et al. Collagen-rich stroma in aggressive colon tumors induces mesenchymal gene expression and tumor cell invasion. Oncogene 2016, 35, 5263–5271. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Knittel, J.G.; Yan, L.; Rueden, C.T.; White, J.G.; Keely, P.J. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008, 6, 11. [Google Scholar] [CrossRef] [Green Version]
- Winkles, J.A. The TWEAK-Fn14 cytokine-receptor axis: Discovery, biology and therapeutic targeting. Nat. Rev. Drug Discov. 2008, 7, 411–425. [Google Scholar] [CrossRef] [Green Version]
- Wang, E.C. On death receptor 3 and its ligands. Immunology 2012, 137, 114–116. [Google Scholar] [CrossRef]
- Perez, J.G.; Tran, N.L.; Rosenblum, M.G.; Schneider, C.S.; Connolly, N.P.; Kim, A.J.; Woodworth, G.F.; Winkles, J.A. The TWEAK receptor Fn14 is a potential cell surface portal for targeted delivery of glioblastoma therapeutics. Oncogene 2016, 35, 2145–2155. [Google Scholar] [CrossRef]
- Hu, G.; Zeng, W.; Xia, Y. TWEAK/Fn14 signaling in tumors. Tumour Biol. 2017, 39, 1010428317714624. [Google Scholar] [CrossRef] [Green Version]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e1821. [Google Scholar] [CrossRef]
- Hafemeister, C.; Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019, 20, 296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukasaki, M.; Asano, T.; Muro, R.; Huynh, N.C.; Komatsu, N.; Okamoto, K.; Nakano, K.; Okamura, T.; Nitta, T.; Takayanagi, H. OPG Production Matters Where It Happened. Cell Rep. 2020, 32, 108124. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blondel, V.D.; Guillaume, J.-L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, 2008, P10008. [Google Scholar] [CrossRef] [Green Version]
- Tsukasaki, M.; Huynh, N.C.; Okamoto, K.; Muro, R.; Terashima, A.; Kurikawa, Y.; Komatsu, N.; Pluemsakunthai, W.; Nitta, T.; Abe, T.; et al. Stepwise cell fate decision pathways during osteoclastogenesis at single-cell resolution. Nat. Metab. 2020, 2, 1382–1390. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Campbell, J.D.; Yau, C.; Bowlby, R.; Liu, Y.; Brennan, K.; Fan, H.; Taylor, A.M.; Wang, C.; Walter, V.; Akbani, R.; et al. Genomic, Pathway Network, and Immunologic Features Distinguishing Squamous Carcinomas. Cell Rep. 2018, 23, 194–212.e196. [Google Scholar] [CrossRef] [Green Version]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huynh, N.C.-N.; Huang, T.-T.; Nguyen, C.T.-K.; Lin, F.-K. Comprehensive Integrated Single-Cell Whole Transcriptome Analysis Revealed the p-EMT Tumor Cells—CAFs Communication in Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 6470. https://doi.org/10.3390/ijms23126470
Huynh NC-N, Huang T-T, Nguyen CT-K, Lin F-K. Comprehensive Integrated Single-Cell Whole Transcriptome Analysis Revealed the p-EMT Tumor Cells—CAFs Communication in Oral Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2022; 23(12):6470. https://doi.org/10.3390/ijms23126470
Chicago/Turabian StyleHuynh, Nam Cong-Nhat, Tze-Ta Huang, Chi Thi-Kim Nguyen, and Fang-Kuei Lin. 2022. "Comprehensive Integrated Single-Cell Whole Transcriptome Analysis Revealed the p-EMT Tumor Cells—CAFs Communication in Oral Squamous Cell Carcinoma" International Journal of Molecular Sciences 23, no. 12: 6470. https://doi.org/10.3390/ijms23126470





