Effective Natural Killer Cell Degranulation Is an Essential Key in COVID-19 Evolution
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics, Lymphocyte Subpopulations, and Inflammatory Parameters
2.2. Innate Immune Profile in COVID-19
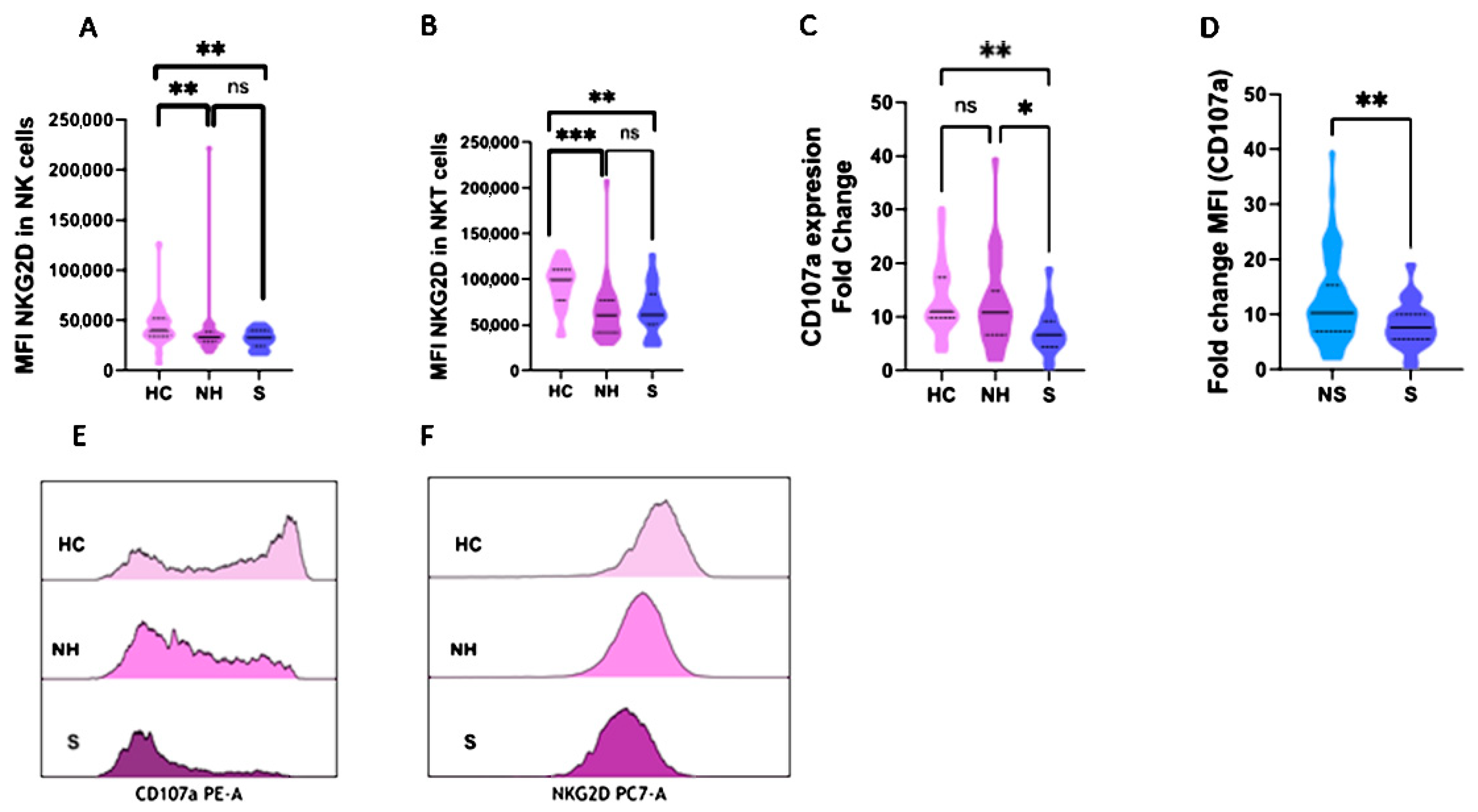
2.3. NKG2D and CD107a Expression in NK and NKT Cells
2.4. Granzyme A and B Studies in COVID-19 Patients
2.5. Multivariable Analysis
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Patients
4.3. Patients Classification
4.4. Study Definitions
4.5. Data Collection
4.6. Samples
4.7. Innate Cells Subsets
4.8. NK Degranulation Assay
4.9. Granzyme Evaluation
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bohn, M.K.; Hall, A.; Sepiashvili, L.; Jung, B.; Steele, S.; Adeli, K. Pathophysiology of COVID-19: Mechanisms Underlying Disease Severity and Progression. Physiology 2020, 35, 288–301. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Bangash, H.I.; Habiba, M.; Lei, Y.; Xie, T.; Sun, J.; Wei, Z.; Hong, Z.; Shao, L.; Zhang, Q. Immune dysregulation and system pathology in COVID-19. Virulence 2021, 12, 918–936. [Google Scholar] [CrossRef] [PubMed]
- Weiskopf, D.; Schmitz, K.S.; Raadsen, M.P.; Grifoni, A.; Okba, N.M.; Endeman, H.; van den Akker, J.P.C.; Molenkamp, R.; Koopmans, M.P.G.; van Gorp, E.C.M. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020, 5, eabd2071. [Google Scholar] [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Strålin, K.; Gorin, J.-B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020, 183, 158–168.e14. [Google Scholar] [CrossRef]
- Martín-Sánchez, E.; Garcés, J.J.; Maia, C.; Inogés, S.; de Cerio, A.L.-D.; Carmona-Torre, F.; Marin-Oto, M.; Alegre, F.; Molano, E.; Fernandez-Alonso, M.; et al. Immunological Biomarkers of Fatal COVID-19: A Study of 868 Patients. Front. Immunol. 2021, 12, 659018. [Google Scholar] [CrossRef]
- Jiang, Y.; Rubin, L.; Peng, T.; Liu, L.; Xing, X.; Lazarovici, P.; Zheng, W. Cytokine storm in COVID-19: From viral infection to immune responses, diagnosis and therapy. Int. J. Biol. Sci. 2022, 18, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2021, 93, 250–256. [Google Scholar] [CrossRef]
- Fajgenbaum, D.C.; June, C.H. Cytokine Storm. N. Engl. J. Med. 2020, 383, 2255–2273. [Google Scholar] [CrossRef]
- Petrey, A.C.; Qeadan, F.; Middleton, E.A.; Pinchuk, I.V.; Campbell, R.A.; Beswick, E.J. Cytokine release syndrome in COVID-19: Innate immune, vascular, and platelet pathogenic factors differ in severity of disease and sex. J. Leukoc. Biol. 2021, 109, 55–66. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, J.; Zhang, D.; Xu, Z.; Ji, J.; Wen, C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020, 11, 1708. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Callender, L.A.; Curran, M.; Bates, S.M.; Mairesse, M.; Weigandt, J.; Betts, C.J. The Impact of Pre-existing Comorbidities and Therapeutic Interventions on COVID-19. Front. Immunol. 2020, 11, 1991. [Google Scholar] [CrossRef] [PubMed]
- Gil-Etayo, F.J.; Suàrez-Fernández, P.; Cabrera-Marante, O.; Arroyo, D.; Garcinuño, S.; Naranjo, L.; Pleguezuelo, D.E.; Allende, L.M.; Mancebo, E.; Lalueza, A.; et al. T-Helper Cell Subset Response Is a Determining Factor in COVID-19 Progression. Front. Cell. Infect. Microbiol. 2021, 11, 624483. [Google Scholar] [CrossRef]
- Gil-Etayo, F.J.; Garcinuño, S.; Utrero-Rico, A.; Cabrera-Marante, O.; Arroyo-Sanchez, D.; Mancebo, E.; Pleguezuelo, D.E.; Rodríguez-Frías, E.; Allende, L.M.; Morales-Pérez, P.; et al. An Early Th1 Response Is a Key Factor for a Favorable COVID-19 Evolution. Biomedicines 2022, 10, 296. [Google Scholar] [CrossRef] [PubMed]
- Kubelkova, K.; Macela, A. Innate Immune Recognition: An Issue More Complex Than Expected. Front. Cell. Infect. Microbiol. 2019, 9, 241. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, G.; D’Osualdo, A.; Schubert, D.A.; Weber, A.; Bruscia, E.M.; Hartl, D. Cellular Innate Immunity: An Old Game with New Players. J. Innate Immun. 2017, 9, 111–125. [Google Scholar] [CrossRef]
- Hinks, T.S.C.; Zhang, X.-W. MAIT Cell Activation and Functions. Front. Immunol. 2020, 11, 1014. [Google Scholar] [CrossRef]
- Melandri, D.; Zlatareva, I.; Chaleil, R.A.; Dart, R.J.; Chancellor, A.; Nussbaumer, O.; Polyakova, O.; Roberts, N.A.; Wesch, D.; Kabelitz, D.; et al. The γδTCR combines innate immunity with adaptive immunity by utilizing spatially distinct regions for agonist selection and antigen responsiveness. Nat. Immunol. 2018, 19, 1352–1365. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Espinosa, G.; Lalueza, A.; Bravo-Gallego, L.Y.; Diaz-Simón, R.; Garcinuño, S.; Gil-Etayo, J.; Moises, J.; Naranjo, L.; Prieto-González, S.; et al. Beta-2-Glycoprotein-I Deficiency Could Precipitate an Antiphospholipid Syndrome-like Prothrombotic Situation in Patients with Coronavirus Disease. ACR Open Rheumatol. 2021, 3, 267–276. [Google Scholar] [CrossRef]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.-H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370, eabd4585. [Google Scholar] [CrossRef]
- Ma, L.; Li, Q.; Cai, S.; Peng, H.; Huyan, T.; Yang, H. The role of NK cells in fighting the virus infection and sepsis. Int. J. Med. Sci. 2021, 18, 3236–3248. [Google Scholar] [CrossRef]
- Waggoner, S.N.; Reighard, S.D.; Gyurova, I.E.; Cranert, S.A.; Mahl, S.E.; Karmele, E.P.; McNally, J.P.; Moran, M.T.; Brooks, T.R.; Yaqoob, F.; et al. Roles of natural killer cells in antiviral immunity. Curr. Opin. Virol. 2016, 16, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Moretta, L.; Moretta, A. Unravelling natural killer cell function: Triggering and inhibitory human NK receptors. EMBO J. 2004, 23, 255–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudreau, J.E.; Hsu, K.C. Natural Killer Cell Education and the Response to Infection and Cancer Therapy: Stay Tuned. Trends Immunol. 2018, 39, 222–239. [Google Scholar] [CrossRef] [PubMed]
- Tremblay-McLean, A.; Bruneau, J.; Lebouché, B.; Lisovsky, I.; Song, R.; Bernard, N.F. Expression Profiles of Ligands for Activating Natural Killer Cell Receptors on HIV Infected and Uninfected CD4+ T Cells. Viruses 2017, 9, 295. [Google Scholar] [CrossRef]
- Murugin, V.V.; Zuikova, I.N.; Murugina, N.E.; Shulzhenko, A.E.; Pinegin, B.V.; Pashenkov, M.V. Reduced degranulation of NK cells in patients with frequently recurring herpes. Clin. Vaccine Immunol. 2011, 18, 1410–1415. [Google Scholar] [CrossRef] [Green Version]
- Stinchcombe, J.C.; Griffiths, G.M. Secretory Mechanisms in Cell-Mediated Cytotoxicity. Annu. Rev. Cell Dev. Biol. 2007, 23, 495–517. [Google Scholar] [CrossRef] [PubMed]
- Matsuda-Lennikov, M.; Biancalana, M.; Zou, J.; Ravell, J.C.; Zheng, L.; Kanellopoulou, C.; Jiang, P.; Notarangelo, G.; Jing, H.; Masutani, E.; et al. Magnesium transporter 1 (MAGT1) deficiency causes selective defects in N-linked glycosylation and expression of immune-response genes. J. Biol. Chem. 2019, 294, 13638–13656. [Google Scholar] [CrossRef]
- Blommaert, E.; Cherepanova, N.A.; Staels, F.; Wilson, M.P.; Gilmore, R.; Schrijvers, R.; Jaeken, J.; Foulquier, F.; Matthijs, G. Lack of NKG2D in MAGT1-deficient patients is caused by hypoglycosylation. Hum. Genet. 2022, in press. [Google Scholar] [CrossRef]
- Lee, H.; Kim, H.S.; Lee, J.M.; Park, K.H.; Choi, A.R.; Yoon, J.H.; Ryu, H.; Oh, E.J. Natural Killer Cell Function Tests by Flowcytometry-Based Cyto-toxicity and IFN-γ Production for the Diagnosis of Adult Hemophagocytic Lymphohistiocytosis. Int. J. Mol. Sci. 2019, 20, 5413. [Google Scholar] [CrossRef] [Green Version]
- La Rosée, P.; Horne, A.; Hines, M.; von Bahr Greenwood, T.; Machowicz, R.; Berliner, N.; Birndt, S.; Gil-Herrera, J.; Girschikofsky, M.; Jordan, M.B.; et al. Recommendations for the management of hemophagocytic lymphohistiocytosis in adults. Blood 2019, 133, 2465–2477. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, T. Secondary haemophagocytic lymphohistiocytosis: Experience from the Uppsala University Hospital. Upsala J. Med. Sci. 2015, 120, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Passarelli, C.; Civino, A.; Rossi, M.N.; Cifaldi, L.; Lanari, V.; Moneta, G.M.; Caiello, I.; Bracaglia, C.; Montinaro, R.; Novelli, A.; et al. IFNAR2 Deficiency Causing Dysregulation of NK Cell Functions and Presenting with Hemophagocytic Lymphohistiocytosis. Front. Genet. 2020, 11, 937. [Google Scholar] [CrossRef]
- Opoka-Winiarska, V.; Grywalska, E.; Rolinski, J. Could hemophagocytic lymphohistiocytosis be the core issue of severe COVID-19 cases? BMC Med. 2020, 18, 214. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Rivas, M.; Corbella, X.; Formiga, F.; Fernández, E.M.; Escalante, M.D.M.; Fernández, I.B.; Fernández, F.A.; Del Corral-Beamonte, E.; Lalueza, A.; Virto, A.P.; et al. Risk Categories in COVID-19 Based on Degrees of Inflammation: Data on More Than 17,000 Patients from the Spanish SEMI-COVID-19 Registry. J. Clin. Med. 2021, 10, 2214. [Google Scholar] [CrossRef]
- Rosado, F.G.; Kim, A.S. Hemophagocytic lymphohistiocytosis: An update on diagnosis and pathogenesis. Am. J. Clin. Pathol. 2013, 139, 713–727. [Google Scholar] [CrossRef]
- Kaya, H.; Kaji, M.; Usuda, D. Soluble interleukin-2 receptor levels on admission associated with mortality in coronavirus disease. Int. J. Infect. Dis. 2021, 105, 522–524. [Google Scholar] [CrossRef]
- Jianguo, L.; Zhixuan, Z.; Rong, L.; Xiaodong, S. Ruxolitinib in Alleviating the Cytokine Storm of Hemophagocytic Lymphohistiocytosis. Pediatrics 2020, 146, e20191301. [Google Scholar] [CrossRef]
- Hersperger, A.R.; Martin, J.N.; Shin, L.Y.; Sheth, P.M.; Kovacs, C.M.; Cosma, G.L.; Makedonas, G.; Pereyra, F.; Walker, B.D.; Kaul, R.; et al. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood 2011, 117, 3799–3808. [Google Scholar] [CrossRef] [Green Version]
- Sultana, M.A.; Du, A.; Carow, B.; Angbjär, C.M.; Weidner, J.M.; Kanatani, S.; Fuks, J.M.; Muliaditan, T.; James, J.; Mansfield, I.O.; et al. Downmodulation of Effector Functions in NK Cells upon Toxoplasma gondii Infection. Infect. Immun. 2017, 85, e00069-17. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Yang, L.M.; Pei, S.F.; Chong, Y.Z.; Guo, Y.; Gao, X.L.; Tang, Q.Y.; Li, Y.; Feng, F.M. CRP, SAA, LDH, and DD predict poor prognosis of coronavirus disease (COVID-19): A meta-analysis from 7739 patients. Scand. J. Clin. Lab. Investig. 2021, 81, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Wang, Q.; Zhang, D.; Ding, J.; Huang, Q.; Tang, Y.Q.; Wang, Q.; Miao, H. Lymphopenia predicts disease severity of COVID-19: A descriptive and predictive study. Signal Transduct. Target. Ther. 2020, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, S.S.; Kim, T.Y.; Lee, D.G.; Kim, D.W. Lymphopenia as a Biological Predictor of Outcomes in COVID-19 Patients: A Nationwide Cohort Study. Cancers 2021, 13, 471. [Google Scholar] [CrossRef]
- Kjer-Nielsen, L.; Patel, O.; Corbett, A.J.; Le Nours, J.; Meehan, B.; Liu, L.; Bhati, M.; Chen, Z.; Kostenko, L.; Reantragoon, R.; et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 2012, 491, 717–723. [Google Scholar] [CrossRef]
- Van Wilgenburg, B.; Scherwitzl, I.; Hutchinson, E.C.; Leng, T.; Kurioka, A.; Kulicke, C.; de Lara, C.; Cole, S.; Vasanawathana, S.; Limpitikul, W.; et al. MAIT cells are activated during human viral infections. Nat. Commun. 2016, 7, 11653. [Google Scholar] [CrossRef] [Green Version]
- Cano, V.; March, C.; Insua, J.L.; Aguiló, N.; Llobet, E.; Moranta, D.; Regueiro, V.; Brennan, G.P.; Millan-Lou, M.I.; Martin, C.; et al. Klebsiella pneumoniae survives within macrophages by avoiding delivery to lysosomes. Cell. Microbiol. 2015, 17, 1537–1560. [Google Scholar] [CrossRef] [Green Version]
- Parrot, T.; Gorin, J.-B.; Ponzetta, A.; Maleki, K.T.; Kammann, T.; Emgård, J.; Perez-Potti, A.; Sekine, T.; Rivera-Ballesteros, O.; The Karolinska COVID-19 Study Group; et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci. Immunol. 2020, 5, eabe1670. [Google Scholar] [CrossRef]
- Björkström, N.K.; Strunz, B.; Ljunggren, H.-G. Natural killer cells in antiviral immunity. Nat. Rev. Immunol. 2021, 22, 112–123. [Google Scholar] [CrossRef]
- Osman, M.; Faridi, R.M.; Sligl, W.; Shabani-Rad, M.-T.; Dharmani-Khan, P.; Parker, A.; Kalra, A.; Tripathi, M.B.; Storek, J.; Tervaert, J.W.C.; et al. Impaired natural killer cell counts and cytolytic activity in patients with severe COVID-19. Blood Adv. 2020, 4, 5035–5039. [Google Scholar] [CrossRef]
- van Eeden, C.; Khan, L.; Osman, M.S.; Tervaert, J.W.C. Natural Killer Cell Dysfunction and Its Role in COVID-19. Int. J. Mol. Sci. 2020, 21, 6351. [Google Scholar] [CrossRef]
- Kärre, K. NK Cells, MHC Class I Molecules and the Missing Self. Scand. J. Immunol. 2002, 55, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L. Up on the tightrope: Natural killer cell activation and inhibition. Nat. Immunol. 2008, 9, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L. NKG2D Receptor and Its Ligands in Host Defense. Cancer Immunol. Res. 2015, 3, 575–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Osterburg, A.R.; Flury, J.; Huang, S.; McCormack, F.X.; Cormier, S.; Borchers, M.T. NKG2D Regulation of Lung Pathology and Dendritic Cell Function Following Respiratory Syncytial Virus Infection. J. Infect. Dis. 2018, 218, 1822–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Kaer, L.; Parekh, V.V.; Wu, L. The Response of CD1d-Restricted Invariant NKT Cells to Microbial Pathogens and Their Products. Front. Immunol. 2015, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Krzewski, K.; Coligan, J.E. Human NK cell lytic granules and regulation of their exocytosis. Front. Immunol. 2012, 3, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigón, L.; Fuertes, D.; García-Pérez, J.; Torres, M.; Rodríguez-Mora, S.; Mateos, E.; Corona, M.; Saez-Marín, A.J.; Malo, R.; Navarro, C.; et al. Impaired Cytotoxic Response in PBMCs from Patients with COVID-19 Admitted to the ICU: Biomarkers to Predict Disease Severity. Front. Immunol. 2021, 12, 665329. [Google Scholar] [CrossRef]
- Krämer, B.; Knoll, R.; Bonaguro, L.; ToVinh, M.; Raabe, J.; Astaburuaga-García, R.; Schulte-Schrepping, J.; Kaiser, K.M.; Rieke, G.J.; Bischoff, J.; et al. Early IFN-α signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity 2021, 54, 2650–2669.e14. [Google Scholar] [CrossRef]
- Channappanavar, R.; Perlman, S. Pathogenic human coronavirus infections: Causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017, 39, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, A.; Salvati, L.; Maggi, L.; Capone, M.; Vanni, A.; Spinicci, M.; Mencarini, J.; Caporale, R.; Peruzzi, B.; Antonelli, A.; et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Investig. 2020, 130, 4694–4703. [Google Scholar] [CrossRef]
- Strowig, T.; Brilot, F.; Munz, C. Noncytotoxic functions of NK cells: Direct pathogen restriction and assistance to adaptive im-munity. J. Immunol. 2008, 180, 7785–7791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Dong, X.; Zhao, L.; Wang, X.; Wang, Y.; Yang, X.; Wang, H.; Zhao, W. Natural killer cells regulate Th1/Treg and Th17/Treg balance in chlamydial lung infection. J. Cell. Mol. Med. 2016, 20, 1339–1351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, L.; Gao, X.; Joyee, A.G.; Zhao, L.; Qiu, H.; Yang, M.; Fan, Y.; Wang, S.; Yang, X. NK Cells Promote Type 1 T Cell Immunity through Modulating the Function of Dendritic Cells during Intracellular Bacterial Infection. J. Immunol. 2011, 187, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Zenarruzabeitia, O.; Astarloa-Pando, G.; Terrén, I.; Orrantia, A.; Pérez-Garay, R.; Seijas-Betolaza, I.; Nieto-Arana, J.; Imaz-Ayo, N.; Pérez-Fernández, S.; Arana-Arri, E.; et al. T Cell Activation, Highly Armed Cytotoxic Cells and a Shift in Monocytes CD300 Receptors Expression Is Characteristic of Patients with Severe COVID-19. Front. Immunol. 2021, 12, 655934. [Google Scholar] [CrossRef] [PubMed]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef]
- Grossman, W.J.; Verbsky, J.W.; Tollefsen, B.L.; Kemper, C.; Atkinson, J.P.; Ley, T.J. Differential expression of granzymes A and B in human cytotoxic lymphocyte subsets and T regulatory cells. Blood 2004, 104, 2840–2848. [Google Scholar] [CrossRef]
- Bratke, K.; Kuepper, M.; Bade, B.; Virchow, J.C.; Luttmann, W. Differential expression of human granzymes A, B, and K in natural killer cells and during CD8+ T cell differentiation in peripheral blood. Eur. J. Immunol. 2005, 35, 2608–2616. [Google Scholar] [CrossRef]
- Van Daalen, K.R.; Reijneveld, J.F.; Bovenschen, N. Modulation of Inflammation by Extracellular Granzyme A. Front. Immunol. 2020, 11, 931. [Google Scholar] [CrossRef]
- Wilson, J.A.C.; Prow, N.; Schroder, W.A.; Ellis, J.; Cumming, H.E.; Gearing, L.J.; Poo, Y.S.; Taylor, A.; Hertzog, P.; Di Giallonardo, F.; et al. RNA-Seq analysis of chikungunya virus infection and identification of granzyme A as a major promoter of arthritic inflammation. PLoS Pathog. 2017, 13, e1006155. [Google Scholar] [CrossRef] [Green Version]
- Schanoski, A.S.; Le, T.T.; Kaiserman, D.; Rowe, C.; Prow, N.A.; Barboza, D.D.; Santos, C.A.; Zanotto, P.M.A.; Magalhães, K.G.; Aurelio, L.; et al. Granzyme A in Chikungunya and Other Arboviral Infections. Front. Immunol. 2019, 10, 3083. [Google Scholar] [CrossRef] [Green Version]
- Froelich, C.J.; Pardo, J.; Simon, M.M. Granule-associated serine proteases: Granzymes might not just be killer proteases. Trends Immunol. 2009, 30, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Wensink, A.C.; Hack, C.E.; Bovenschen, N. Granzymes Regulate Proinflammatory Cytokine Responses. J. Immunol. 2015, 194, 491–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeglinski, M.R.; Granville, D.J. Granzymes in cardiovascular injury and disease. Cell. Signal. 2020, 76, 109804. [Google Scholar] [CrossRef] [PubMed]
- El-Mesallamy, H.O.; Hamdy, N.M.; El-Etriby, A.K.; Wasfey, E.F. Plasma Granzyme B in ST Elevation Myocardial Infarction versus Non-ST Elevation Acute Coronary Syndrome: Comparisons with IL-18 and Fractalkine. Mediat. Inflamm. 2013, 2013, 343268. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, C.M.; Ang, L.S.; Boivin, W.A.; Cooper, D.M.; Williams, S.J.; Zhao, H.; Hendel, A.; Folkesson, M.; Swedenborg, J.; Allard, M.F.; et al. Perforin-Independent Extracellular Granzyme B Activity Contributes to Abdominal Aortic Aneurysm. Am. J. Pathol. 2010, 176, 1038–1049. [Google Scholar] [CrossRef] [PubMed]
- Choy, J.; Cruz, R.P.; Kerjner, A.; Geisbrecht, J.; Sawchuk, T.; Fraser, S.A.; Hudig, D.; Bleackley, R.C.; Jirik, F.R.; McManus, B.M.; et al. Granzyme B Induces Endothelial Cell Apoptosis and Contributes to the Development of Transplant Vascular Disease. Am. J. Transpl. 2005, 5, 494–499. [Google Scholar] [CrossRef]
- Zmievskaya, E.; Valiullina, A.; Ganeeva, I.; Petukhov, A.; Rizvanov, A.; Bulatov, E. Application of CAR-T Cell Therapy beyond Oncology: Autoimmune Diseases and Viral Infections. Biomedicines 2021, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-García, R.; Lermo-Rojo, S.; Martinez-Lostao, L.; Mancebo, E.; Mora-Díaz, S.; Paz-Artal, E.; Ruiz-Contreras, J.; Anel, A.; Gonzalez-Granado, L.I.; Allende, L.M. A case of partial dedicator of cytokinesis 8 deficiency with altered effector phenotype and impaired CD8+ and natural killer cell cytotoxicity. J. Allergy Clin. Immunol. 2014, 134, 218–221.e7. [Google Scholar] [CrossRef]



| Healthy Controls; n = 24 | COVID-19 Population; n = 99 | ||||
|---|---|---|---|---|---|
| Variables | Median | IQR | Median | IQR | p-Value |
| % CD3+ | 69.9 | 62.2–73.2 | 62.7 | 51.8–68.8 | 0.004 |
| % CD4+ in CD3+ | 65 | 59.2–68 | 60.1 | 54–69.2 | 0.262 |
| % CD8+ in CD3+ | 31.8 | 23.3–38.2 | 36 | 28.7–42.9 | 0.222 |
| % NK | 9.2 | 7.2–14.9 | 14.3 | 8.5–19.6 | 0.051 |
| % NK CD56 brigth | 0.55 | 0.35–0.8 | 0.4 | 0.2–0.6 | 0.016 |
| % NK CD56 dim | 8.7 | 6.6–14.4 | 13.6 | 8.2–19 | 0.039 |
| % NKT | 4.6 | 1.7–9.8 | 4.9 | 2.9–7.5 | 0.904 |
| % MAIT | 2.85 | 1.6–4.2 | 0.9 | 0.4–2.3 | 0.001 |
| % MAIT in CD4+ T cells | 0.4 | 0.3–0.9 | 0.4 | 0.2–0.8 | 0.419 |
| % MAIT in CD8+ T cells | 4.6 | 2.2–11 | 1.8 | 1.7–4.3 | 0.001 |
| MFI NKG2D in NK cells | 39,192 | 34,876–50,420 | 32,256 | 27,210–39,459 | <0.001 |
| MFI NKG2D in NKT cells | 99,577 | 81,873–107,068 | 62,247 | 45,737–82,792 | <0.001 |
| % TCR gd | 4 | 2.7–10.1 | 3.8 | 2.2–6 | 0.275 |
| CD107a Fold Change in NK cells | 11 | 9.8–17.4 | 10 | 6.4–13.7 | 0.13 |
| Non-Hospitalized; n = 38 | Hospitalized COVID-19; n = 61 | ||
|---|---|---|---|
| Variables | Median | Median | p-Value |
| Male (%) | 18 (47%) | 36 (59%) | 0.26 |
| Female (%) | 20 (53%) | 25 (41%) | |
| Age (Years) | 43 (32–50) | 53 (38–62) | 0.001 |
| Lymphocytes (cells/uL) | 1300 (1000–1600) | 900 (600–1425) | 0.002 |
| Neutrophils (×103 cells/uL) | 3.8 (2.5–5.3) | 5 (3.7–7.2) | 0.019 |
| CD3+ T lymphocytes (%) | 64.2 (59.7–73.2) | 58.1 (48–67.4) | 0.004 |
| CRP (mg/dL) | 1.18 (0.4–2.8) | 7.44 (2.1–11.3) | <0.001 |
| LDH (U/L) | 261 (213–31) | 359 (314–428) | <0.001 |
| DD (ng/dL; n = 64) | 516 (387–645) (n = 17) | 674(241–1429) (n = 47) | 0.024 |
| Variables | Univariant | Multivariant | ||||
|---|---|---|---|---|---|---|
| OR | OR 95% IC | p-Value | OR | OR 95% IC | p-Value | |
| (A) NH vs. H | ||||||
| Lymphocytes | 0.28 | 0.11–0.69 | 0.005 | 0.34 | 0.12–0.95 | 0.041 |
| %CD3+ | 0.42 | 0.21–0.86 | 0.017 | 0.53 | 0.23–1.2 | 0.133 |
| MFI NKG2D in NKT | 2 | 1.1–3.6 | 0.022 | 2.02 | 1.1–3.9 | 0.033 |
| Area Under the ROC Curve | 0.779 | 0.683–0.856 | ||||
| (B) NS vs. S | ||||||
| %CD3+ | 0.48 | 0.24–0.96 | 0.036 | 0.53 | 0.25–1.1 | 0.083 |
| MFI NKG2D in NKT | 2 | 1.01–3.8 | 0.036 | 2.22 | 1.12–4.4 | 0.022 |
| CD107a expression in NK (MFI Fold change) | 0.88 | 0.8–0.97 | 0.015 | 0.87 | 0.78–0.98 | 0.021 |
| Area Under the ROC Curve | 0.752 | 0.655–0.834 | ||||
| (C) NH vs. M | ||||||
| Lymphocytes | 0.26 | 0.1–0.71 | 0.008 | 0.26 | 0.08–0.81 | 0.017 |
| MFI NKG2D in NKT | 1.87 | 0.95–3.65 | 0.065 | 1.93 | 0.9–4.13 | 0.089 |
| Area Under the ROC Curve | 0.803 | 0.695–0.886 | ||||
| (D) NH vs. S | ||||||
| MFI NKG2D in NKT | 2.77 | 1.28–5.99 | 0.009 | 3.51 | 1.44–8.53 | 0.005 |
| CD107a expression in NK (MFI Fold change) | 0.89 | 0.8–0.99 | 0.031 | 0.86 | 0.75–0.99 | 0.047 |
| Area Under the ROC Curve | 0.84 | 0.729–0.918 | ||||
| (E) A vs. S | ||||||
| Lymphocytes | 0.23 | 0.05–1.02 | 0.053 | 0.14 | 0.02–0.87 | 0.032 |
| CD107a expression in NK (MFI Fold change) | 1.42 | 1.1–1.81 | 0.005 | 0.84 | 0.72–0.98 | 0.033 |
| Area Under the ROC Curve | 0.808 | 0.627–0.927 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcinuño, S.; Gil-Etayo, F.J.; Mancebo, E.; López-Nevado, M.; Lalueza, A.; Díaz-Simón, R.; Pleguezuelo, D.E.; Serrano, M.; Cabrera-Marante, O.; Allende, L.M.; et al. Effective Natural Killer Cell Degranulation Is an Essential Key in COVID-19 Evolution. Int. J. Mol. Sci. 2022, 23, 6577. https://doi.org/10.3390/ijms23126577
Garcinuño S, Gil-Etayo FJ, Mancebo E, López-Nevado M, Lalueza A, Díaz-Simón R, Pleguezuelo DE, Serrano M, Cabrera-Marante O, Allende LM, et al. Effective Natural Killer Cell Degranulation Is an Essential Key in COVID-19 Evolution. International Journal of Molecular Sciences. 2022; 23(12):6577. https://doi.org/10.3390/ijms23126577
Chicago/Turabian StyleGarcinuño, Sara, Francisco Javier Gil-Etayo, Esther Mancebo, Marta López-Nevado, Antonio Lalueza, Raquel Díaz-Simón, Daniel Enrique Pleguezuelo, Manuel Serrano, Oscar Cabrera-Marante, Luis M. Allende, and et al. 2022. "Effective Natural Killer Cell Degranulation Is an Essential Key in COVID-19 Evolution" International Journal of Molecular Sciences 23, no. 12: 6577. https://doi.org/10.3390/ijms23126577






